Molecular and Pathogenic Characterization of Fusarium Species Associated with Corm Rot Disease in Saffron from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolation and Morphological Characterization
2.2. DNA Sequencing and Molecular Phylogenetic Analysis
2.3. Pathogenicity Studies
3. Results
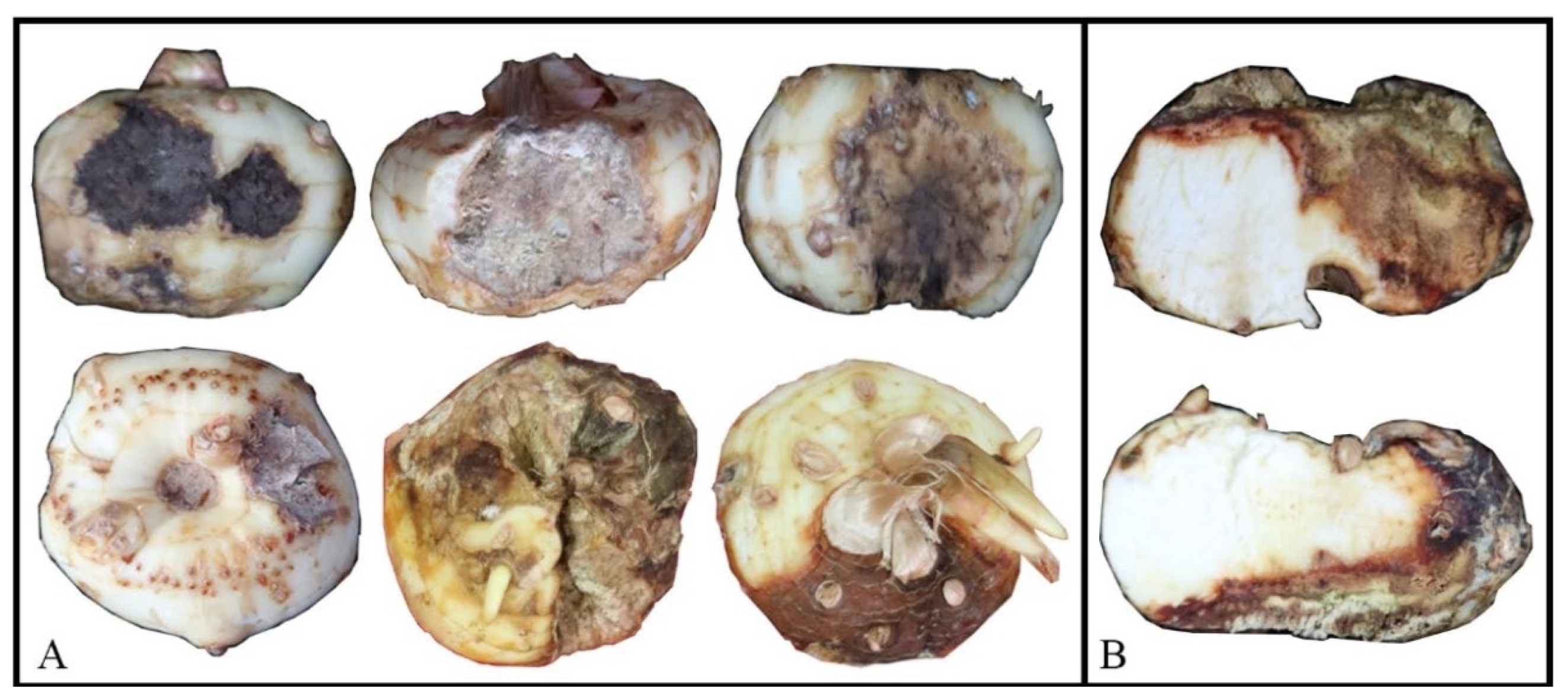
3.1. Field Survey, Disease Symptoms, and Pathogen Isolations
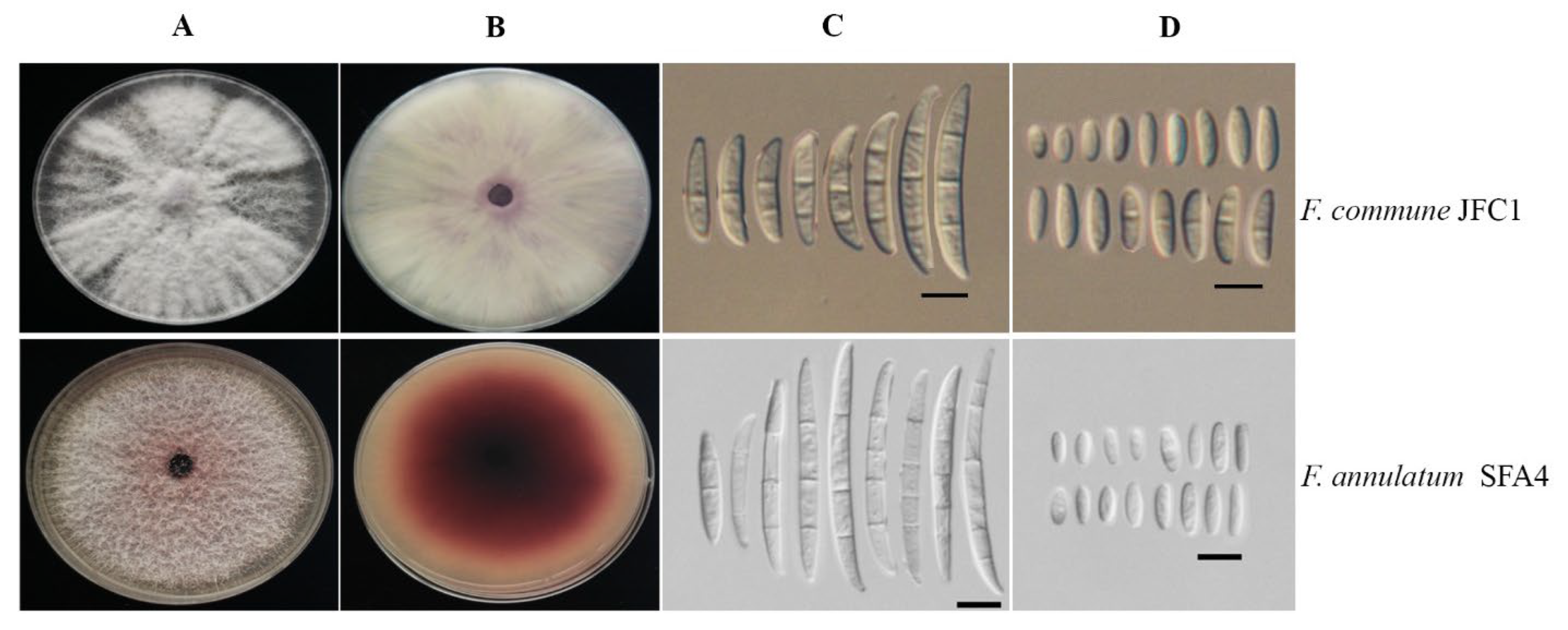
3.2. Morphological Identification
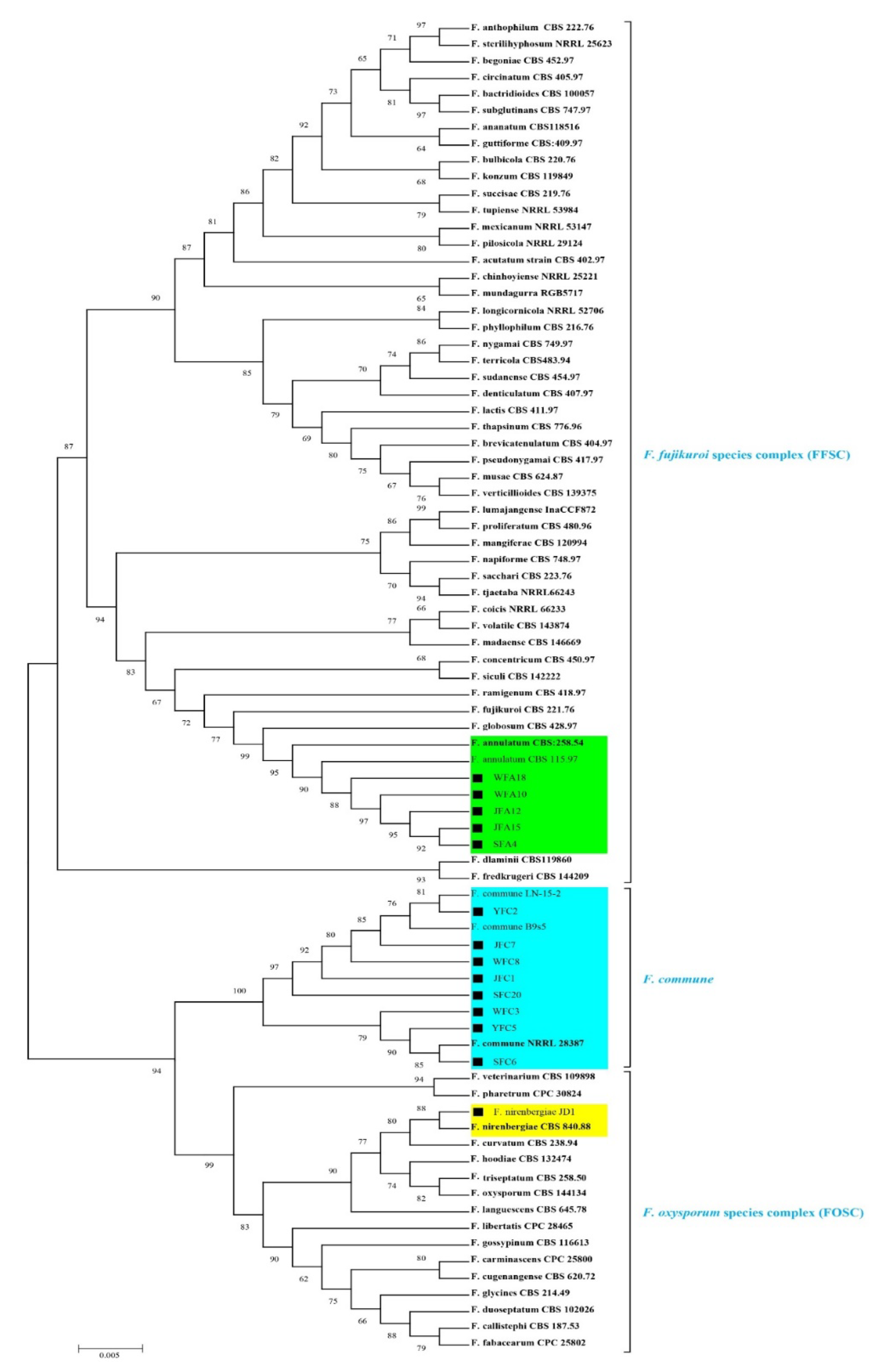
3.3. Molecular Characterization and Phylogeny
3.4. Pathogenicity Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Pandita, D. Saffron (Crocus sativus L.): Phytochemistry, therapeutic significance and omics-based biology. In Medicinal and Aromatic Plants; Academic Press: Cambridge, MA, USA, 2021; Volume 14, pp. 325–396. [Google Scholar]
- Shokrpour, M. Saffron (Crocus sativus L.) breeding: Opportunities and challenges. In Advances in Plant Breeding Strategies: Industrial and Food Crops; Springer: Berlin/Heidelberg, Germany, 2019; Volume 17, pp. 675–706. [Google Scholar]
- Singh, A.K.; Chand, D. Indigenous knowledge and ethnobotany associated with saffron (Crocus sativus L.) in Kashmir. Indian J. Plant Gene. Res. 2005, 18, 191–195. [Google Scholar]
- Golmohammadi, F. Saffron and its farming, economic importance, export, medicinal characteristics and various uses in South Khorasan Province-East of Iran. Int. J. Farming Allied Sci. 2014, 3, 566–596. [Google Scholar]
- Yamamoto, W.; Omatsu, T.; Takami, K. Studies on the corm rots of Crocus sativus L. on saprophytic propagation of Sclerotinia gladioli and Fusarium oxysporum f. sp. gladioli on various plants and soils. Sci. Rep. Hyogo Univ. Agric. 1954, 1, 64–70. [Google Scholar]
- Palmero, D.; Rubiomoraga, A.; Galvezpaton, L.; Nogueras, J.; Abato, C.; Gomezgomez, L.; Ahrazem, O. Pathogenicity and genetic diversity of Fusarium oxysporum isolates from corms of Crocus sativus. Ind. Crop. Prod. 2014, 61, 186–192. [Google Scholar] [CrossRef]
- Najari, G.H.; Nourollahi, K.H.; Piri, M. The first report of (Fusarium oxysporum) causal agent of wild saffron corm rot disease in Iran. Saffron Agron. Technol. 2018, 6, 119–123. [Google Scholar]
- Gupta, V.; Sharma, A.; Rai, P.K.; Gupta, S.K.; Singh, B.; Sharma, S.K. Corm rot of saffron: Epidemiology and management. Agronomy 2021, 11, 339. [Google Scholar] [CrossRef]
- Mirghasempour, S.A.; Studholme, D.J.; Chen, W.; Cui, D.; Mao, B. Identification and characterization of Fusarium nirenbergiae associated with saffron corm rot disease. Plant Dis. 2022, 106, 486–495. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef]
- Kumar, S. Molecular taxonomy, diversity, and potential applications of genus Fusarium. In Industrially Important Fungi for Sustainable Development; Springer: Berlin/Heidelberg, Germany, 2021; pp. 277–293. [Google Scholar]
- Lombard, L.; Sandoval-Denis, M.; Lamprecht, S.C.; Crous, P.W. Epitypification of Fusarium oxysporum: Clearing the taxonomic chaos. Persoonia 2019, 43, 1–47. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Dongzhen, F.; Xilin, L.; Xiaorong, C.; Wenwu, Y.; Yunlu, H.; Yi, C.; Jia, C.; Zhimin, L.; Litao, G.; Tuhong, W.; et al. Fusarium species and Fusarium oxysporum species complex genotypes associated with yam wilt in south-central China. Front. Microbiol. 2020, 11, 1964. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Al-Hatmi, A.M.; Aoki, T.; Brankovics, B.; Cano-Lira, J.F.; Coleman, J.J.; de Hoog, G.S.; Di Pietro, A.; Frandsen, R.J.; Geiser, D.M.; et al. No to Neocosmospora: Phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus Fusarium. Msphere 2020, 5, e00810-20. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Ramos, J.F. Fusarium and Fusariosis, reference module in biomedical sciences. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- O’Donnell, K.; Whitaker, B.; Laraba, I.; Proctor, R.; Brown, D.; Broders, K.; Kim, H.S.; McCormick, S.P.; Busman, M.; Aoki, T.; et al. DNA sequence-based identification of Fusarium: A work in progress. Plant Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Sandoval-Denis, M.; Lombard, L.; Visagie1, C.M.; Wingfield1, B.D.; Crous, P.W. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 2021, 46, 129–162. [Google Scholar] [CrossRef]
- Rampersad, S.N. Pathogenomics and management of Fusarium diseases in plants. Pathogens 2020, 1, 340. [Google Scholar] [CrossRef] [PubMed]
- McTaggart, A.R.; James, T.Y.; Shivas, R.G.; Drenth, A.; Wingfield, B.D.; Summerell, B.A.; Duong, T.A. Population genomics reveals historical and ongoing recombination in the Fusarium oxysporum species complex. Stud. Mycol. 2021, 99, 100132. [Google Scholar] [CrossRef]
- Bedoya, E.T.; Bebber, D.; Studholme, D.J. Taxonomic revision of the banana Fusarium wilt TR4 pathogen is premature. Phytopathology 2021, 12, 2141–2145. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Knyaz, M.; Li, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Skovgaard, K.; Rosendahl, S.; O’Donnell, K.; Nirenberg, H.I. Fusarium commune is a new species identified by morphological and molecular phylogenetic data. Mycologia 2003, 95, 630–636. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, H.; van der Lee, T.A.J.; Waalwijk, C.; van Diepeningen, A.D.; Feng, J.; Brankovics, B.; Chen, W. Population genomic analysis reveals a highly conserved mitochondrial genome in Fusarium asiaticum. Front. Microbiol. 2020, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Husna, A.; Zakaria, L.; Mohamed-Nor, N.M.I. Fusarium commune associated with wilt and root rot disease in rice. Plant Pathol. 2020, 70, 123–132. [Google Scholar] [CrossRef]
- Stewart, J.E.; Kim, M.S.; James, R.L.; Dumroese, R.K.; Klopfenstein, N.B. Molecular characterization of Fusarium oxysporum and Fusarium commune isolates from a conifer nursery. Phytopathology 2006, 96, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Babadoost, M. Occurrence of Fusarium commune and F. oxysporum in horseradish roots. Plant Dis. 2013, 97, 453–460. [Google Scholar] [CrossRef]
- Dobbs, J.; Kim, M.S.; Dudley, N.; Jones, T.; Yeh, A.; Dumroese, R.K.; Cannon, P.; Hauff, R.; Klopfenstein, N.; Wright, S.; et al. Fusarium spp. diversity associated with symptomatic Acacia koa in Hawai‘i. For. Pathol. 2021, 51, 12713. [Google Scholar] [CrossRef]
- Zhong, Q.; Xiao, Y.S.; He, B.; Cao, Z.H.; Shou, Z.G.; Zhong, J.; Zhu, J.Z. First report of Fusarium commune causing stem rot of tobacco (Nicotiana tabacum) in Hunan province, China. Plant Dis. 2020, 10, 1568. [Google Scholar] [CrossRef]
- Simpson, A.C.; Urbaniak, C.; Bateh, J.R.; Singh, N.K.; Wood, J.M.; Debieu, M.; O’Hara, N.B.; Houbraken, J.; Mason, C.E.; Venkateswaran, K. Draft genome sequences of fungi isolated from the international Space Station during the microbial tracking-2 experiment. Microbiol. Resour. Announc. 2021, 10, e0075121. [Google Scholar] [CrossRef]
- Bustamante, M.I.; Elfar, K.; Smith, R.; Bettiga, L.; Tian, T.; Torres-Londoño, G.A.; Eskalen, A. First report of Fusarium annulatum associated with young vine decline in California. Plant Dis. 2022, 3. [Google Scholar] [CrossRef]
- Lin, S.; Taylor, N.J.; Peduto, H.F. Identification and characterization of fungal pathogens causing fruit rot of deciduous holly. Plant Dis. 2018, 102, 2430–2445. [Google Scholar] [CrossRef]
- Okello, P.N.; PetrovićK, K.B.; Mathew, F.M. Eight species of Fusarium cause root rot of corn (Zea mays) in South Dakota. Plant Health Prog. 2019, 20, 38–43. [Google Scholar] [CrossRef]
- Kamali-Sarvestani, S.; Mostowfizadeh-Ghalamfarsa, R.; Salmaninezhad, F.; Cacciola, S.O. Fusarium and Neocosmospora species associated with rot of Cactaceae and other succulent plants. J. Fungi 2022, 8, 364. [Google Scholar] [CrossRef]
- Swett, C.L.; Science, P.; Architecture, L.; Park, C.; Sharon, C. Evidence for a hemibiotrophic association of the pitch canker pathogen Fusarium circinatum with Pinus radiata. Plant Dis. 2016, 100, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Asselin, J.; Bonasera, J.M.; Beer, S.V. PCR primers for detection of Pantoea ananatis, Burkholderia spp., and Enterobacter sp. from onion. Plant Dis. 2016, 100, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Degani, O.; Movshowitz, D.; Dor, S.; Meerson, A.; Goldblat, Y.; Rabinovitz, O. Evaluating azoxystrobin seed coating against maize late wilt disease using a sensitive qPCR-based method. Plant Dis. 2019, 103, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.H.; Barbedo, J.G.A.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.K. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2020, 2, 9. [Google Scholar] [CrossRef]
- Kim, J.H.; Kang, M.R.; Kim, H.K.; Lee, S.H.; Lee, T.; Yun, S.H. Population structure of the Gibberella fujikuroi species complex associated with rice and corn in Korea. J. Plant Pathol. 2012, 28, 357–363. [Google Scholar] [CrossRef]
- Zainudin, N.; Razak, A.A.; Salleh, B. Bakanae diseases of rice in Malaysia and Indonesia: Etiology of the causal agent based on morphological, physiological and pathogenicity characteristics. J. Plant Prot. Res. 2008, 48, 476–485. [Google Scholar]
- Eğerci, Y.; Kınay-Teksür, P.; Uysal-Morca, A. First report of Bakanae disease caused by Fusarium proliferatum on rice in Turkey. J. Plant Dis. Prot. 2021, 128, 577–582. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]

| Province | City | Geographic Coordinates | Sample No. | F. nirenbergiae a | F. commune a | F. annulatum a |
|---|---|---|---|---|---|---|
| Zhejiang | Jiande | 119.5998° E 29.2401° N | 45 | 58 | 26 | 19 |
| Wuyi | 119.8165° E 28.8926° N | 25 | 37 | 15 | 12 | |
| Shanghai | Chong Ming Dao | 121.3975° E 31.6227° N | 25 | 31 | 11 | 8 |
| Yunnan | Shangri-la | 99.7060° E 27.8230° N | 25 | 26 | 18 | - |
| Total | - | - | 120 | 152 | 70 | 39 |
| Frequency (%) | - | - | - | 58.2 | 26.8 | 14.9 |
| Origin | Species | Representative Isolate | GenBank Accession Number | ||
|---|---|---|---|---|---|
| ITS | tef1 | rpb2 | |||
| Jiande | F. commune | JFC1 | MZ313313 | MZ338563 | MZ338571 |
| JFC7 | MZ313314 | MZ338564 | MZ338572 | ||
| F. annulatum | JFA12 | MZ313130 | MZ338578 | MZ338573 | |
| JFA15 | MZ313131 | MZ338579 | MZ338574 | ||
| Chong Ming Dao | F. commune | SFC6 | MZ318050 | MZ338561 | MZ338569 |
| SFC20 | MZ318051 | MZ338562 | MZ338570 | ||
| F. annulatum | SFA4 | MZ313134 | MZ338582 | MZ338577 | |
| Wuyi | F. commune | WFC3 | MZ313309 | MZ338557 | MZ338565 |
| WFC8 | MZ313310 | MZ338558 | MZ338566 | ||
| F. annulatum | WFA10 | MZ313132 | MZ338580 | MZ338575 | |
| WFA18 | MZ313133 | MZ338581 | MZ338576 | ||
| Shangri-la | F. commune | YFC2 | MZ313311 | MZ338559 | MZ338567 |
| YFC5 | MZ313312 | MZ338560 | MZ338568 | ||
| Species | Culture Accession | GenBank Accession | |
|---|---|---|---|
| rpb2 | tef1 | ||
| Fusarium carminascens | CPC 144738 T | MH484937 | MH485028 |
| F. contaminatum | CBS 111552 T | MH484901 | MH484992 |
| F. pharetrum | CBS 144751T | MH484952 | MH485043 |
| F. veterinarium | CBS 109898 T | MH484899 | MH484990 |
| F. cugenangense | CBS 620.72 | MH484879 | MH484970 |
| F. curvatum | CBS 238.94 T | MH484893 | MH484984 |
| F. fabacearum | CPC 25802 T | MH484939 | MH485030 |
| F. glycines | CBS 144746 T | MH484942 | MH485033 |
| F. gossypinum | CBS 116613 T | MH484909 | MH485000 |
| F. languescens | CBS 645.78 T | MH484880 | MH484971 |
| F. libertatis | CPC 28465 T | MH484944 | MH485035 |
| F. nirenbergiae | CBS 840.88 T | MH484887 | MH484978 |
| JD1 | MT864705 | MT814630 | |
| JD2 | MT864708 | MT814633 | |
| JD3 | MT864711 | MT814629 | |
| JD4 | MT864712 | MT814625 | |
| SH1 | MT864704 | MT814627 | |
| SH2 | MT864706 | MT814631 | |
| BZ1 | MT864700 | MT814622 | |
| BZ3 | MT864701 | MT814623 | |
| BZ4 | MT864702 | MT814624 | |
| WY5 | MT864713 | MT814635 | |
| WY9 | MT864709 | MT814634 | |
| WY11 | MT864710 | MT814628 | |
| GY2 | MT864703 | MT814626 | |
| GY6 | MT864707 | MT814632 | |
| F. oxysporum | CBS 144134 ET | MH484953 | MH485044 |
| F. hoodiae | CBS 132474 T | MH484929 | MH485020 |
| F. duoseptatum | CBS 102026 T | MH484896 | MH484987 |
| F. callistephi | CBS 187.53 T | MH484875 | MH484966 |
| F. triseptatum | CBS 258.50 T | MH484910 | MH485001 |
| F. languescens | CBS 645.78 T | MH484880 | MH484971 |
| F. elaeidis | CBS 217.49 T | MH484870 | MH484961 |
| F. commune | NRRL 28387 | HM068356 | HM057338 |
| LN-15-2 | MH716813 | MH716809 | |
| B9s5 | MN892350 | MK560330 | |
| NRRL 38348 | - | FJ985389 | |
| NRRL28058 | - | AF324333 | |
| CBS 110090 T | MW934368 | - | |
| F. acutatum | CBS 402.97 T | MW402768 | MW402125 |
| F. agapanthi | NRRL 54463 T | KU900625 | KU900630 |
| F. ananatum | CBS 118516 T | LT996137 | LT996091 |
| F. andiyazi | CBS 119857 T | LT996138 | MN193854 |
| F. annulatum | CBS 115.97 | MW402785 | MW401973 |
| CBS 258.54 T | MT010983 | MT010994 | |
| F. anthophilum | CBS 222.76 ET | MW402811 | MW402114 |
| F. bactridioides | CBS 100057 T | MN534235 | MN533993 |
| F. begoniae | CBS 452.97 T | MN534243 | MN533994 |
| F. brevicatenulatum | CBS 404.97 T | MN534295 | MN533995 |
| F. bulbicola | CBS 220.76 T | MW402767 | KF466415 |
| F. xyrophilum | NRRL 62721 T | MN193905 | - |
| F. chinhoyiense | NRRL 25221 T | MN534262 | MN534050 |
| F. subglutinans | CBS 747.97 NT | MW402773 | MW402150 |
| F. circinatum | CBS 405.97 T | MN534252 | MN533997 |
| F. coicis | NRRL 66233 T | KP083274 | KP083251 |
| F. concentricum | CBS 450.97 T | JF741086 | AF160282 |
| F. denticulatum | CBS 407.97 T | MN534274 | MN534000 |
| F. dlaminii | CBS 119860 T | KU171701 | MW401995 |
| F. echinatum | CBS 146497 T | - | MW834273 |
| F. fredkrugeri | CBS 144209 T | LT996147 | LT996097 |
| F. fujikuroi | CBS 221.76 T | KU604255 | MN534010 |
| F. globosum | CBS 428.97 T | KF466406 | KF466417 |
| F. guttiforme | CBS 409.97 T | MT010967 | MT010999 |
| F. konzum | CBS 119849 T | MW402733 | LT996098 |
| F. lactis | CBS 411.97 ET | MN534275 | MN193862 |
| F. longicornicola | NRRL 52706 T | JF741114 | JF740788 |
| F. lumajangense | InaCCF872 T | LS479850 | LS479441 |
| F. madaense | CBS 146669 T | MW402764 | MW402098 |
| F. mangiferae | CBS 120994 T | MN534271 | MN534017 |
| F. mexicanum | NRRL 53147 T | MN724973 | GU737282 |
| F. mundagurra | RGB5717 T | KP083276 | KP083256 |
| F. musae | CBS 624.87 T | MW402772 | FN552086 |
| F. napiforme | CBS 748.97 T | MN534291 | MN193863 |
| F. nygamai | CBS 749.97 T | EF470114 | MW402151 |
| F. ophioides | CBS 118512 T | MN534303 | MN534022 |
| F. phyllophilum | CBS 216.76 T | KF466410 | MN193864 |
| F. pilosicola | NRRL 29124 T | MN534248 | MN534055 |
| F. proliferatum | CBS 480.96 ET | MN534272 | MN534059 |
| F. pseudonygamai | CBS 417.97 T | MN534285 | AF160263 |
| F. ramigenum | CBS 418.97 T | KF466412 | KF466423 |
| F. sacchari | CBS 223.76 ET | JX171580 | MW402115 |
| F. siculi | CBS 142222 T | LT746327 | LT746214 |
| F. succisae | CBS 219.76 ET | MW402766 | AF160291 |
| F. sudanense | CBS 454.97 T | MN534278 | MN534037 |
| F. terricola | CBS 483.94 T | LT996156 | MN534042 |
| F.thapsinum | CBS 776.96 T | MN534289 | MN534044 |
| F. tjaetaba | NRRL 66243 T | KP083275 | KP083263 |
| F. verticillioides | CBS 139375 T | MW402802 | MW402068 |
| F. volatile | CBS 143874 T | LR596006 | LR596007 |
| F. werrikimbe | CBS 125535 T | MN534304 | MW928846 |
| F. prieskaense | CBS 146498 T | - | MW834275 |
| F. pseudoanthophilum | CBS 414.97 T | MT010980 | MT011006 |
| F. ficicrescens | CBS 125178 T | KT154002 | MT011004 |
| F. pseudocircinatum | CBS 449.97 T | MT010968 | MT011003 |
| F. foetens | CBS 110286 T | MW928825 | MT011001 |
| F. pininemorale | CMW 25243 T | MN534250 | MN534026 |
| F. inflexum | NRRL 20433 T | JX171583 | AF008479 |
| F. sterilihyphosum | NRRL 25623 T | MN193897 | MN193869 |
| F. xylarioides | NRRL 25486 T | - | AY707136 |
| F. hostae | NRRL 29889 T | JX171640 | - |
| F. ramigenum | CBS 418.97 T | MT010975 | MT011012 |
| F. redolens | NRRL 25600 T | MT409443 | - |
| F. udum | BBA 65058 T | KY498875 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirghasempour, S.A.; Studholme, D.J.; Chen, W.; Zhu, W.; Mao, B. Molecular and Pathogenic Characterization of Fusarium Species Associated with Corm Rot Disease in Saffron from China. J. Fungi 2022, 8, 515. https://doi.org/10.3390/jof8050515
Mirghasempour SA, Studholme DJ, Chen W, Zhu W, Mao B. Molecular and Pathogenic Characterization of Fusarium Species Associated with Corm Rot Disease in Saffron from China. Journal of Fungi. 2022; 8(5):515. https://doi.org/10.3390/jof8050515
Chicago/Turabian StyleMirghasempour, Seyed Ali, David J. Studholme, Weiliang Chen, Weidong Zhu, and Bizeng Mao. 2022. "Molecular and Pathogenic Characterization of Fusarium Species Associated with Corm Rot Disease in Saffron from China" Journal of Fungi 8, no. 5: 515. https://doi.org/10.3390/jof8050515
APA StyleMirghasempour, S. A., Studholme, D. J., Chen, W., Zhu, W., & Mao, B. (2022). Molecular and Pathogenic Characterization of Fusarium Species Associated with Corm Rot Disease in Saffron from China. Journal of Fungi, 8(5), 515. https://doi.org/10.3390/jof8050515






