The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Growth Conditions
2.2. dsRNA Extraction and Purification
2.3. dsRNA-Seq Library Preparation and High-Throughput Sequencing
2.4. Bioinformatics Analysis
2.5. Phylogenetic Analysis
2.6. PCR Validation of Assembled ssDNA Viral Contigs
3. Results
3.1. Extraction of dsRNA and Analysis of Electrophoretic Profiles
3.2. High-Throughput Sequencing and Reconstruction of Viral Genomes
3.2.1. Mitoviridae and Splipalmiviridae
3.2.2. Botourmiaviridae
3.2.3. Hypoviridae
3.2.4. Fusariviridae
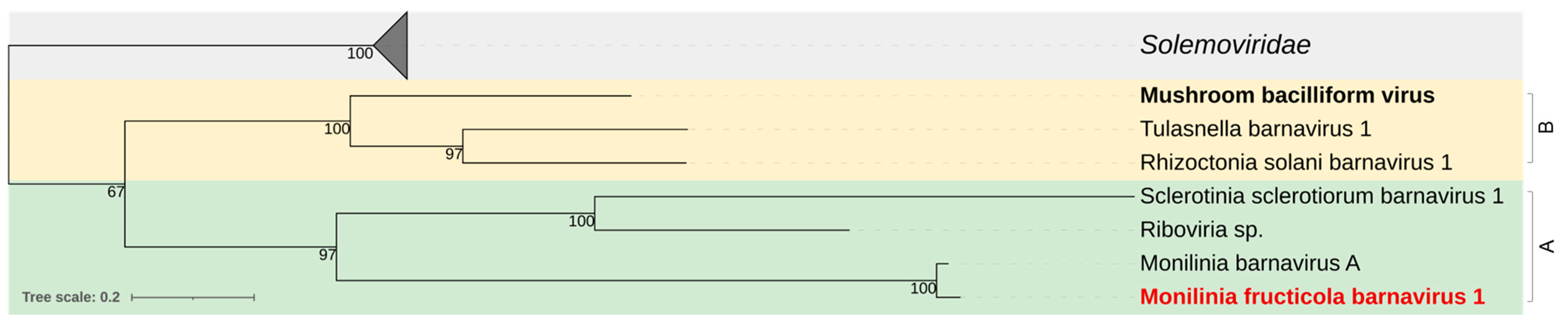
3.2.5. Barnaviridae
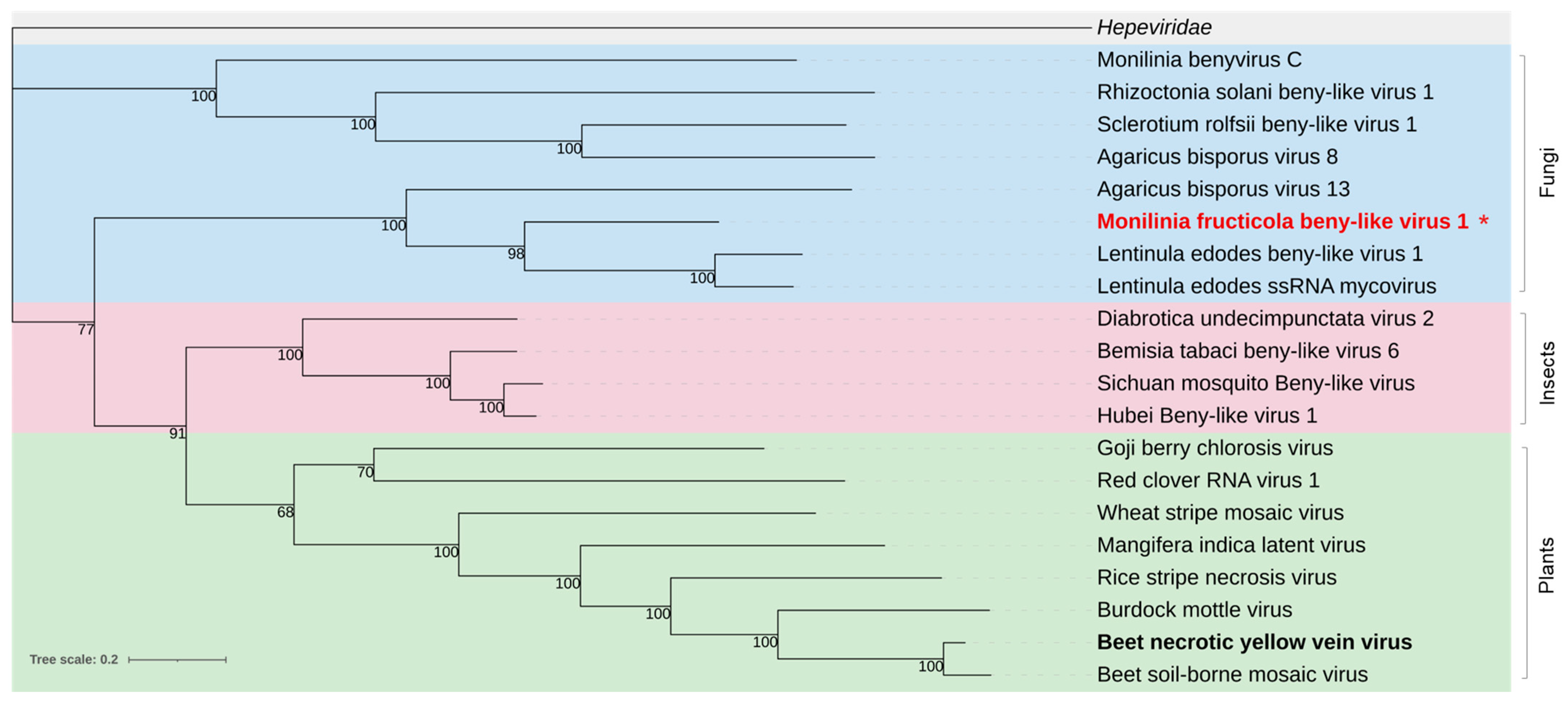
3.2.6. Benyviridae
3.2.7. Parvoviridae
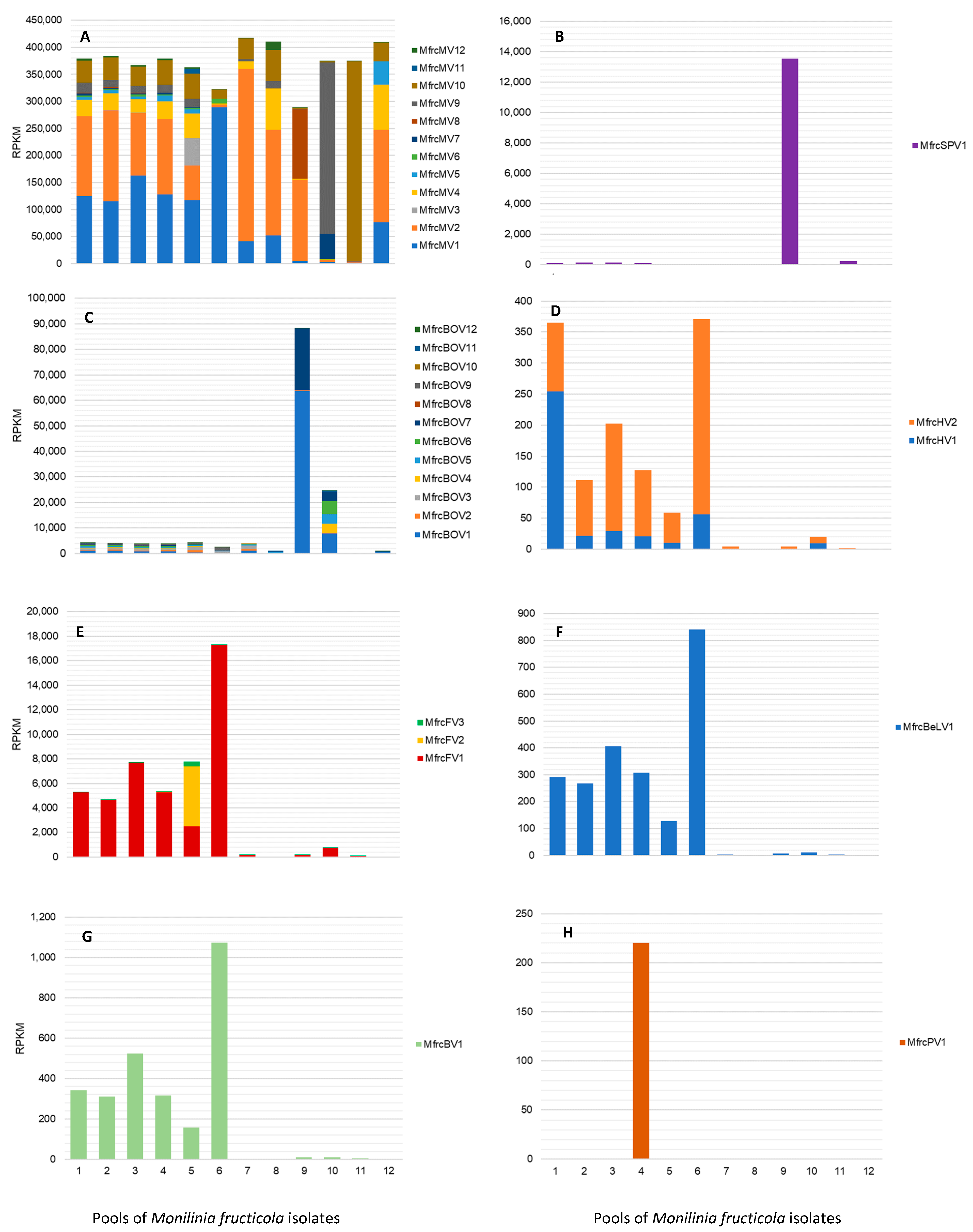
3.2.8. Frequencies of Mycoviral Sequences in M. fructicola dsRNAs
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Hillman, B.I.; Annisa, A.; Suzuki, N. Viruses of plant-interacting fungi. Adv. Virus Res. 2018, 100, 99–116. [Google Scholar] [CrossRef]
- Uchida, K.; Sakuta, K.; Ito, A.; Takahashi, Y.; Katayama, Y.; Omatsu, T.; Mizutani, T.; Arie, T.; Komatsu, K.; Fukuhara, T.; et al. Two novel endornaviruses co-infecting a Phytophthora pathogen of Asparagus officinalis modulate the developmental stages and fungicide sensitivities of the host oomycete. Front. Microbiol. 2021, 12, 122. [Google Scholar] [CrossRef]
- Sutela, S.; Forgia, M.; Vainio, E.J.; Chiapello, M.; Daghino, S.; Vallino, M.; Martino, E.; Girlanda, M.; Perotto, S.; Turina, M. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020, 6, veaa076. [Google Scholar] [CrossRef]
- Crucitti, D.; Chiapello, M.; Oliva, D.; Forgia, M.; Turina, M.; Carimi, F.; La Bella, F.; Pacifico, D. Identification and molecular characterization of novel mycoviruses in Saccharomyces and non-Saccharomyces yeasts of oenological interest. Viruses 2022, 14, 52. [Google Scholar] [CrossRef]
- Deakin, G.; Dobbs, E.; Bennett, J.M.; Jones, I.M.; Grogan, H.M.; Burton, K.S. Multiple viral infections in Agaricus bisporus-characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci. Rep. 2017, 7, 2469. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.H.; Fujita, M.; Chiba, S.; Hyodo, K.; Andika, I.B.; Suzuki, N.; Kondo, H. Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 2019, 533, 125–136. [Google Scholar] [CrossRef]
- Myers, J.M.; Bonds, A.E.; Clemons, R.A.; Thapa, N.A.; Simmons, D.R.; Carter-House, D.; Ortanez, J.; Liu, P.; Miralles-Durán, A.; Desirò, A.; et al. Survey of early-diverging lineages of fungi reveals abundant and diverse mycoviruses. mBio 2020, 11, e02027-20. [Google Scholar] [CrossRef]
- Sato, Y.; Castón, J.R.; Suzuki, N. The biological attributes, genome architecture and packaging of diverse multi-component fungal viruses. Curr. Opin. Virol. 2018, 33, 55–65. [Google Scholar] [CrossRef]
- Son, M.; Yu, J.; Kim, K.H. Five questions about mycoviruses. PLoS Pathog. 2015, 11, e1005172. [Google Scholar] [CrossRef]
- Kondo, H.; Chiba, S.; Toyoda, K.; Suzuki, N. Evidence for negative-strand RNA virus infection in fungi. Virology 2013, 435, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Padilla, A.; Rodríguez-Romero, J.; Gómez-Cid, I.; Pacifico, D.; Ayllón, M.A. Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus. mBio 2021, 12, e03705-20. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Ayllón, M.A.; Turina, M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020, 6, veaa058. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Choi, G.H.; Dawe, A.L.; Churbanov, A.; Smith, M.L.; Milgroom, M.G.; Nuss, D.L. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 2012, 190, 113–127. [Google Scholar] [CrossRef] [Green Version]
- García-Pedrajas, M.D.; Cañizares, M.C.; Sarmiento-Villamil, J.L.; Jacquat, A.G.; Dambolena, J.S. Mycoviruses in biological control: From basic research to field implementation. Phytopathology 2019, 109, 1828–1839. [Google Scholar] [CrossRef]
- Yu, X.; Li, B.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Yi, X.; Jiang, D. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. USA 2013, 110, 1452–1457. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, M.E.; MacDiarmid, R.M. A mechanically transmitted DNA mycovirus is targeted by the defence machinery of its host, Botrytis cinerea. Viruses 2021, 13, 1315. [Google Scholar] [CrossRef]
- Liu, S.; Xie, J.; Cheng, J.; Li, B.; Chen, T.; Fu, Y.; Li, G.; Wang, M.; Jin, H.; Wan, H.; et al. Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. USA 2016, 113, 12803–12808. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Cheng, J.; Fu, Y.; Chen, T.; Jiang, D.; Ghabrial, S.A.; Xie, J. Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog. 2017, 13, e1006234. [Google Scholar] [CrossRef] [Green Version]
- Hyder, R.; Pennanen, T.; Hamberg, L.; Vainio, E.J.; Piri, T.; Hantula, J. Two viruses of Heterobasidion confer beneficial, cryptic or detrimental effects to their hosts in different situations. Fungal Ecol. 2013, 6, 387–396. [Google Scholar] [CrossRef]
- Zhu, J.Z.; Zhu, H.J.; Da Gao, B.; Zhou, Q.; Zhong, J. Diverse, novel mycoviruses from the virome of a hypovirulent Sclerotium rolfsii strain. Front. Plant Sci. 2018, 9, 1738. [Google Scholar] [CrossRef] [Green Version]
- Osaki, H.; Sasaki, A.; Nomiyama, K.; Tomioka, K. Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 2016, 52, 835–847. [Google Scholar] [CrossRef]
- Dobbs, E.; Deakin, G.; Bennett, J.; Fleming-Archibald, C.; Jones, I.; Grogan, H.; Burton, K. Viral interactions and pathogenesis during multiple viral infections in Agaricus bisporus. mBio 2021, 12, e03470-20. [Google Scholar] [CrossRef]
- Buck, K. Molecular variability of viruses of fungi. In Molecular Variability of Fungal Pathogens; Bridge, P.D., Couteaudier, Y., Clackson, J.M., Eds.; CAB International: Wallingford, UK, 1998; pp. 53–72. [Google Scholar]
- Schmitt, M.; Breinig, F. Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef]
- Ahn, I.P.; Lee, Y.H. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant-Microbe Interact. 2001, 14, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.P.; Lo, G.C.S.; Chow, F.W.N.; Fan, R.Y.Y.; Cai, J.J.; Yuen, K.Y.; Woo, P.C.Y. Novel Partitivirus enhances virulence of and causes aberrant gene expression in Talaromyces marneffei. mBio 2018, 9, e00947-18. [Google Scholar] [CrossRef] [Green Version]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef]
- Fuke, K.; Takeshita, K.; Aoki, N.; Fukuhara, T.; Egusa, M.; Kodama, M.; Moriyama, H. The presence of double-stranded RNAs in Alternaria alternata Japanese pear pathotype is associated with morphological changes. J. Gen. Plant Pathol. 2011, 77, 248–252. [Google Scholar] [CrossRef]
- Kotta-Loizou, I. Mycoviruses and their role in fungal pathogenesis. Curr. Opin. Microbiol. 2021, 63, 10–18. [Google Scholar] [CrossRef]
- van der Lende, T.R.; Harmsen, M.C.; Go, S.J.; Wessels, J.G.; Wessel, J.G. Double-stranded RNA mycoviruses in mycelium of Pleurotus ostreatus. FEMS Microbiol. Lett. 1995, 125, 51–56. [Google Scholar] [CrossRef]
- Magae, Y. Molecular characterization of a novel mycovirus in the cultivated mushroom, Lentinula edodes. Virol. J. 2012, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Li, X.; Kotta-Loizou, I.; Dong, K.; Li, S.; Ni, D.; Hong, N.; Wang, G.; Xu, W. A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 2021, 15, 1893–1906. [Google Scholar] [CrossRef]
- Márquez, L.M.; Redman, R.S.; Rodriguez, R.J.; Roossinck, M.J. A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 2007, 315, 513–515. [Google Scholar] [CrossRef] [Green Version]
- Feau, N.; Dutech, C.; Brusini, J.; Rigling, D.; Robin, C. Multiple introductions and recombination in Cryphonectria hypovirus 1: Perspective for a sustainable biological control of chestnut blight. Evol. Appl. 2014, 7, 580–596. [Google Scholar] [CrossRef]
- Brasier, C.M. Viruses as biological control agents of the dutch elm disease fungus Ophiostoma novo-ulmi. In The Elms; Dunn, C.P., Ed.; Springer: Boston, MA, USA, 2000; pp. 201–212. [Google Scholar] [CrossRef]
- Aihara, M.; Urayama, S.; Le, M.T.; Katoh, Y.; Higashiura, T.; Fukuhara, T.; Arie, T.; Teraoka, T.; Komatsu, K.; Moriyama, H. Infection by Magnaporthe oryzae chrysovirus 1 strain A triggers reduced virulence and pathogenic race conversion of its host fungus, Magnaporthe oryzae. J. Gen. Plant Pathol. 2018, 84, 92–103. [Google Scholar] [CrossRef]
- Grasse, W.; Zipper, R.; Totska, M.; Spring, O. Plasmopara halstedii virus causes hypovirulence in Plasmopara halstedii, the downy mildew pathogen of the sunflower. Fungal Genet. Biol. 2013, 57, 42–47. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, M.; Chen, Q.; Zhu, M.; Zhou, E. A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 2014, 456–457, 220–226. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Salaipeth, L.; Lin, Y.H.; Sasaki, A.; Kanematsu, S.; Suzuki, N. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 2009, 83, 12801–12812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, K.B.; Holcomb, E.E.; Allscheid, R.L.; Carrington, J.C. Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS ONE 2019, 14, e0219207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzano, S.Y.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016, 90, 6846–6863. [Google Scholar] [CrossRef] [Green Version]
- Donaire, L.; Ayllon, M.A. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol. Plant Pathol. 2017, 18, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Jurvansuu, J.; Streng, J.; Rajamäki, M.L.; Hantula, J.; Valkonen, J.P. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J. Gen. Virol. 2015, 96, 714–725. [Google Scholar] [CrossRef] [Green Version]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Lichou, J.; Myrin, J.F.; Breniaux, D. First report of brown rot caused by Monilinia fructicola in peach orchards in France. Phytoma 2002, 47, 22. [Google Scholar]
- Villarino, M.; Egüen, B.; Lamarca, N.; Segarra, J.; Usall, J.; Melgarejo, P.; De Cal, A. Occurrence of Monilinia laxa and M. fructigena after introduction of M. fructicola in peach orchards in Spain. Eur. J. Plant Pathol. 2013, 137, 835–845. [Google Scholar] [CrossRef]
- Abate, D.; Pastore, C.; Gerin, D.; De Miccolis Angelini, R.M.; Rotolo, C.; Pollastro, S.; Faretra, F. Characterization of Monilinia spp. populations on stone fruit in South Italy. Plant Dis. 2018, 102, 1708–1717. [Google Scholar] [CrossRef] [Green Version]
- Tsai, P.; Pearson, M.; Beever, R. Mycoviruses in Monilinia fructicola. Mycol. Res. 2004, 108, 907–912. [Google Scholar] [CrossRef]
- Tran, T.T.; Li, H.; Nguyen, D.Q.; Jones, M.G.; Wylie, S.J. Co-infection with three mycoviruses stimulates growth of a Monilinia fructicola isolate on nutrient medium but does not induce hypervirulence in a natural host. Viruses 2019, 11, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.; Back, C.G.; Choi, H.; Cho, W.K. Comparative microbiome study of mummified peach fruits by metagenomics and metatranscriptomics. Plants 2020, 9, 1052. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Choi, H.; Chu, H.; Cho, W.K. Identification of viruses from fungal transcriptomes. BioRxiv, 2020; preprint. [Google Scholar] [CrossRef]
- De Miccolis Angelini, R.M.; Abate, D.; Rotolo, C.; Gerin, D.; Pollastro, S.; Faretra, F. De novo assembly and comparative transcriptome analysis of Monilinia fructicola, Monilinia laxa and Monilinia fructigena, the causal agents of brown rot on stone fruits. BMC Genom. 2018, 19, 436. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, K.E.; Srb, A.M. Heterokaryosis and parasexuality in the fungus Ascochyta imperfecta. Am. J. Bot. 1965, 52, 72–81. [Google Scholar] [CrossRef]
- Beadle, G.W.; Tatum, E.L. Neurospora. II. Methods of producing and detecting mutations concerned with nutritional requirements. Am. J. Bot. 1945, 32, 678–686. [Google Scholar] [CrossRef]
- Morris, T.J.; Dodds, J.A. Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 1979, 69, 854–858. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Sato, Y.; Shahi, S.; Telengech, P.; Hisano, S.; Cornejo, C.; Rigling, D.; Kondo, H.; Suzuki, N. A new tetra-segmented splipalmivirus with divided RdRP domains from Cryphonectria naterciae, a fungus found on chestnut and cork oak trees in Europe. Virus Res. 2022, 307, 198606. [Google Scholar] [CrossRef] [PubMed]
- Ayllón, M.A.; Turina, M.; Xie, J.; Nerva, L.; Marzano, S.L.; Donaire, L.; Jiang, D. ICTV Virus taxonomy profile: Botourmiaviridae. J. Gen. Virol. 2020, 101, 454–455. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Ghabrial, S.A.; Kim, K.H.; Pearson, M.; Marzano, S.Y.L.; Yaegashi, H.; Xie, J.; Guo, L.; Kondo, H.; Koloniuk, I.; et al. ICTV virus taxonomy profile: Hypoviridae. J. Gen. Virol. 2018, 99, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, M.E.; Pearson, M.N. Characterisation of a novel hypovirus from Sclerotinia sclerotiorum potentially representing a new genus within the Hypoviridae. Virology 2014, 464–465, 441–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Wu, S.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. Molecular characterization of two positive-strand RNA viruses co-infecting a hypovirulent strain of Sclerotinia sclerotiorum. Virology 2014, 464–465, 450–459. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Shen, G.; Wang, J.; Liu, M.; Bian, Y.; Xu, Z. Mycoviral diversity and characteristics of a negative-stranded RNA virus LeNSRV1 in the edible mushroom Lentinula edodes. Virology 2021, 555, 89–101. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. Ictv Report Consortium. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant virus metagenomics: Advances in virus discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [Green Version]
- Marzano, S.Y.L.; Domier, L.L. Novel mycoviruses discovered from metatranscriptomics survey of soybean phyllosphere phytobiomes. Virus Res. 2016, 213, 332–342. [Google Scholar] [CrossRef] [Green Version]
- Bartholomäus, A.; Wibberg, D.; Winkler, A.; Pühler, A.; Schlüter, A.; Varrelmann, M. Deep sequencing analysis reveals the mycoviral diversity of the virome of an avirulent isolate of Rhizoctonia solani AG-2-2 IV. PLoS ONE 2016, 11, e0165965. [Google Scholar] [CrossRef] [Green Version]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, K.N.; Liang, Z.; Lipton, H.L. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA ciruses. J. Virol. 2015, 89, 9383–9392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, F.; Xie, J.; Cheng, S.; You, M.P.; Barbetti, M.J.; Jia, J.; Wang, Q.; Cheng, J.; Fu, Y.; Chen, T.; et al. Virome characterization of a collection of S. sclerotiorum from Australia. Front. Microbiol. 2018, 8, 2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolja, V.V.; Koonin, E.V. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 2011, 1, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Mu, F.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. A single ssRNA segment encoding RdRp is sufficient for replication, infection, and transmission of ourmia-like virus in fungi. Front. Microbiol. 2020, 11, 379. [Google Scholar] [CrossRef] [Green Version]
- Hillman, B.I.; Cai, G. The family Narnaviridae: Simplest of RNA viruses. Adv. Virus Res. 2013, 86, 149–176. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, L.; Li, G.; Jiang, D.; Ghabrial, S.A. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 2010, 406, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, M.E.; Pearson, M.N. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 2013, 441, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Picarelli, M.A.S.; Forgia, M.; Rivas, E.B.; Nerva, L.; Chiapello, M.; Turina, M.; Colariccio, A. Extreme diversity of mycoviruses present in isolates of Rhizoctonia solani AG2-2 LP from Zoysia japonica from Brazil. Front. Cell. Infect. Microbiol. 2019, 9, 244. [Google Scholar] [CrossRef] [Green Version]
- Shahi, S.; Eusebio-Cope, A.; Kondo, H.; Hillman, B.I.; Suzuki, N. Investigation of host range of and host defense against a mitochondrially replicating mitovirus. J. Virol. 2019, 93, e01503-18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zou, Q.; Dai, Z.; Hong, N.; Wang, G.; Wang, L. Four novel mycoviruses from the hypovirulent Botrytis cinerea SZ-2-3y isolate from Paris polyphylla: Molecular characterisation and mitoviral sequence transboundary entry into plants. Viruses 2022, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A.; Warner, B.E.; Yerramsetty, P. Widespread mitovirus sequences in plant genomes. PeerJ 2015, 3, e876. [Google Scholar] [CrossRef] [Green Version]
- Nibert, M.L.; Vong, M.; Fugate, K.K.; Debat, H.J. Evidence for contemporary plant mitoviruses. Virology 2018, 518, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Zhu, J.Z.; Gao, B.D.; Zhu, H.J.; Zhou, Q.; Zhong, J. Characterization of a novel ourmia-like mycovirus infecting Magnaporthe oryzae and implications for viral diversity and evolution. Viruses 2019, 11, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhang, Y.; Wan, X.; She, Y.; Li, M.; Xi, H.; Xie, J.; Wen, C. A novel ourmia-like mycovirus confers hypovirulence-associated traits on Fusarium oxysporum. Front. Microbiol. 2020, 11, 569869. [Google Scholar] [CrossRef] [PubMed]
- Ohkita, S.; Lee, Y.; Nguyen, Q.; Ikeda, K.; Suzuki, N.; Nakayashiki, H. Three ourmia-like viruses and their associated RNAs in Pyricularia oryzae. Virology 2019, 534, 25–35. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Esmael, A.; Duan, J.; Bian, X.; Jia, J.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D.; et al. Four novel botourmiaviruses co-infecting an isolate of the rice blast fungus Magnaporthe oryzae. Viruses 2020, 12, 1383. [Google Scholar] [CrossRef]
- Marzano, S.Y.L.; Hobbs, H.A.; Nelson, B.D.; Hartman, G.L.; Eastburn, D.M.; McCoppin, N.K.; Domier, L.L. Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J. Virol. 2015, 89, 5060–5071. [Google Scholar] [CrossRef] [Green Version]
- Hao, F.; Ding, T.; Wu, M.; Zhang, J.; Yang, L.; Chen, W.; Li, G. Two novel hypovirulence-associated mycoviruses in the phytopathogenic fungus Botrytis cinerea: Molecular characterization and suppression of infection cushion formation. Viruses 2018, 10, 254. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Fu, Y.; Jiang, D.; Mu, F.; Cheng, J.; Lin, Y.; Li, B.; Marzano, S.-Y.L.; Xie, J. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol. 2021, 7, veab032. [Google Scholar] [CrossRef]
- Gilmer, D.; Ratti, C.; Consortium, I.R. ICTV virus taxonomy profile: Benyviridae. J. Gen. Virol. 2017, 98, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Kolliopoulou, A.; Taning, C.N.T.; Smagghe, G.; Swevers, L. Viral delivery of dsRNA for control of insect agricultural pests and vectors of human disease: Prospects and challenges. Front. Physiol. 2017, 8, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
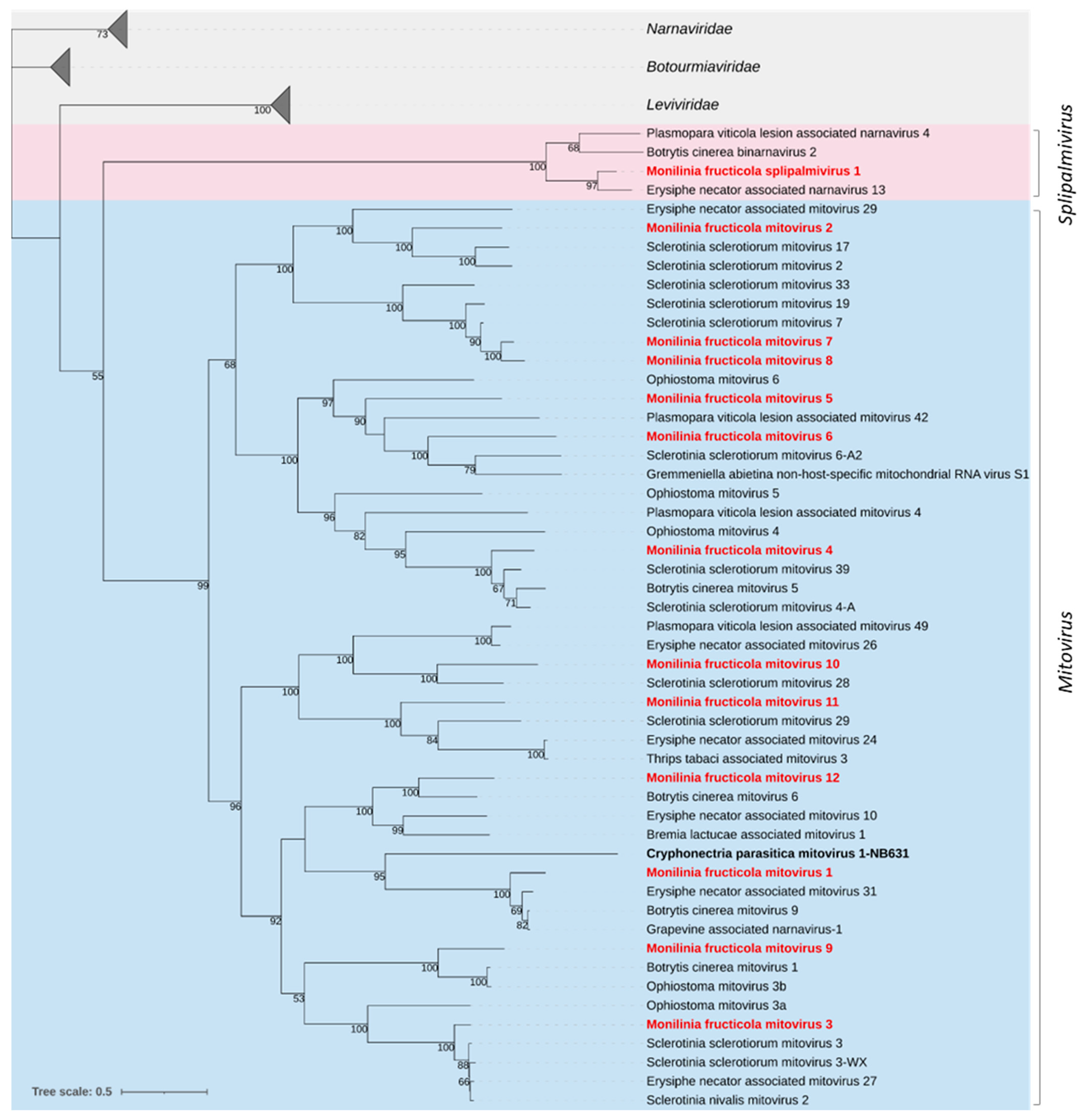
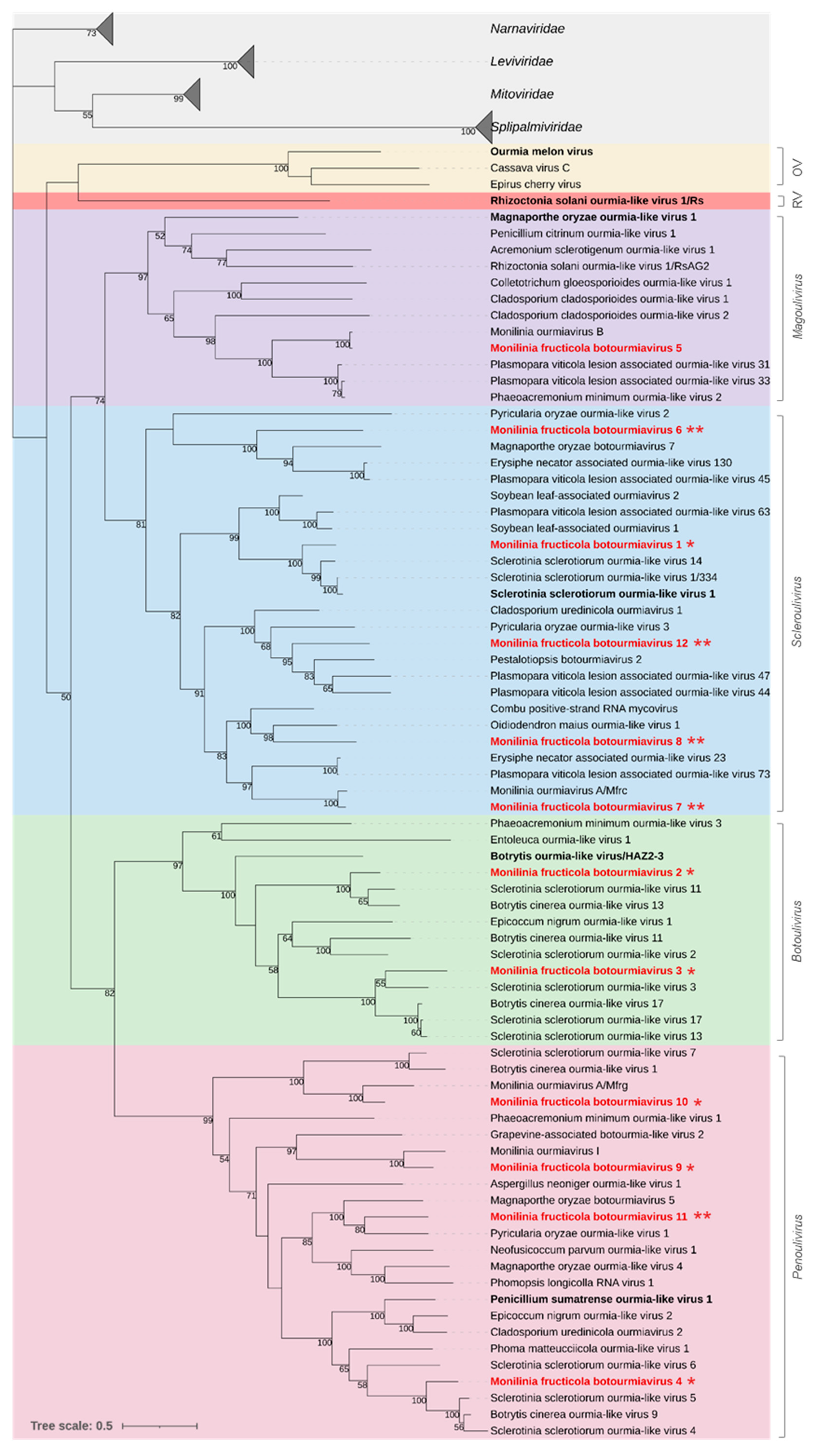
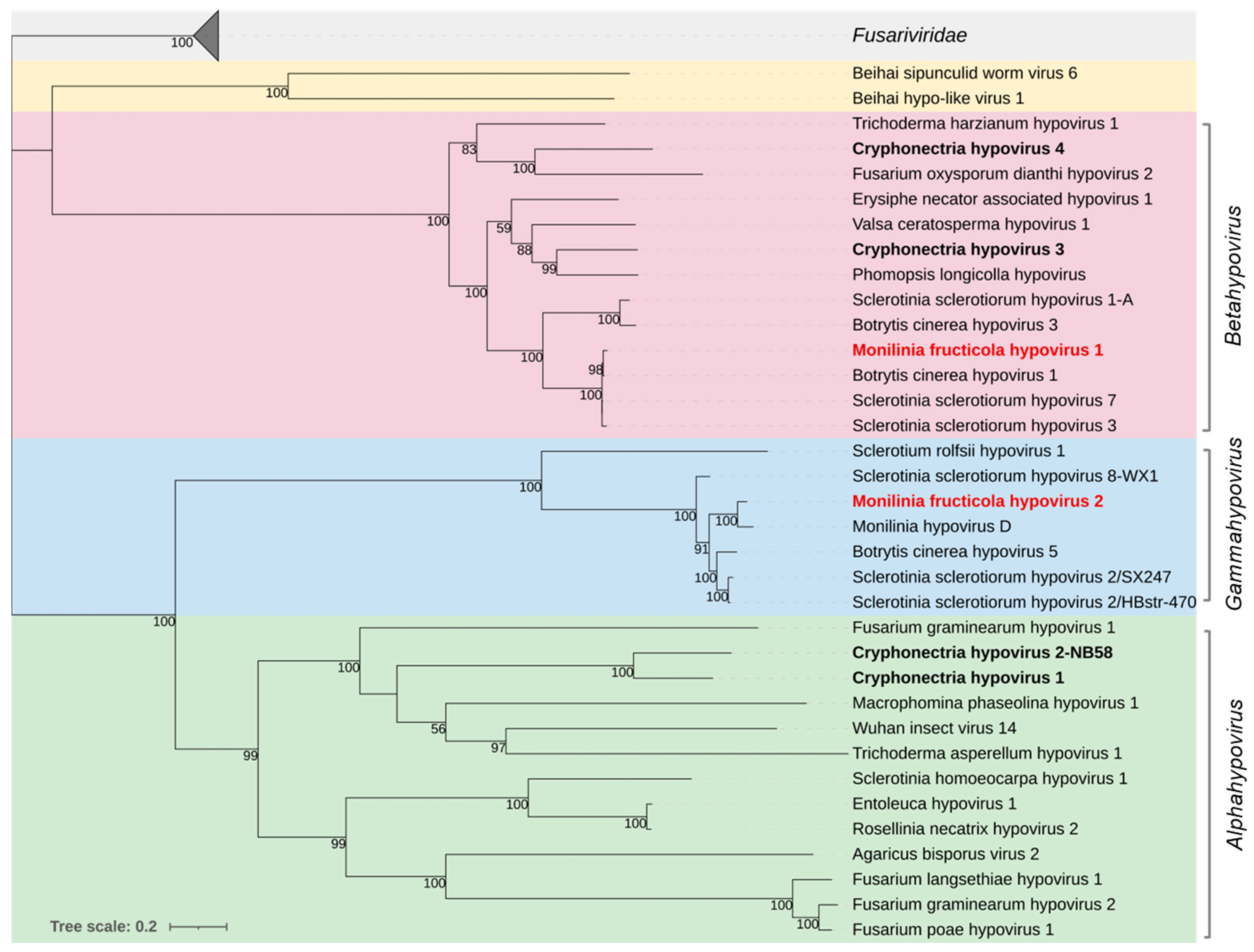
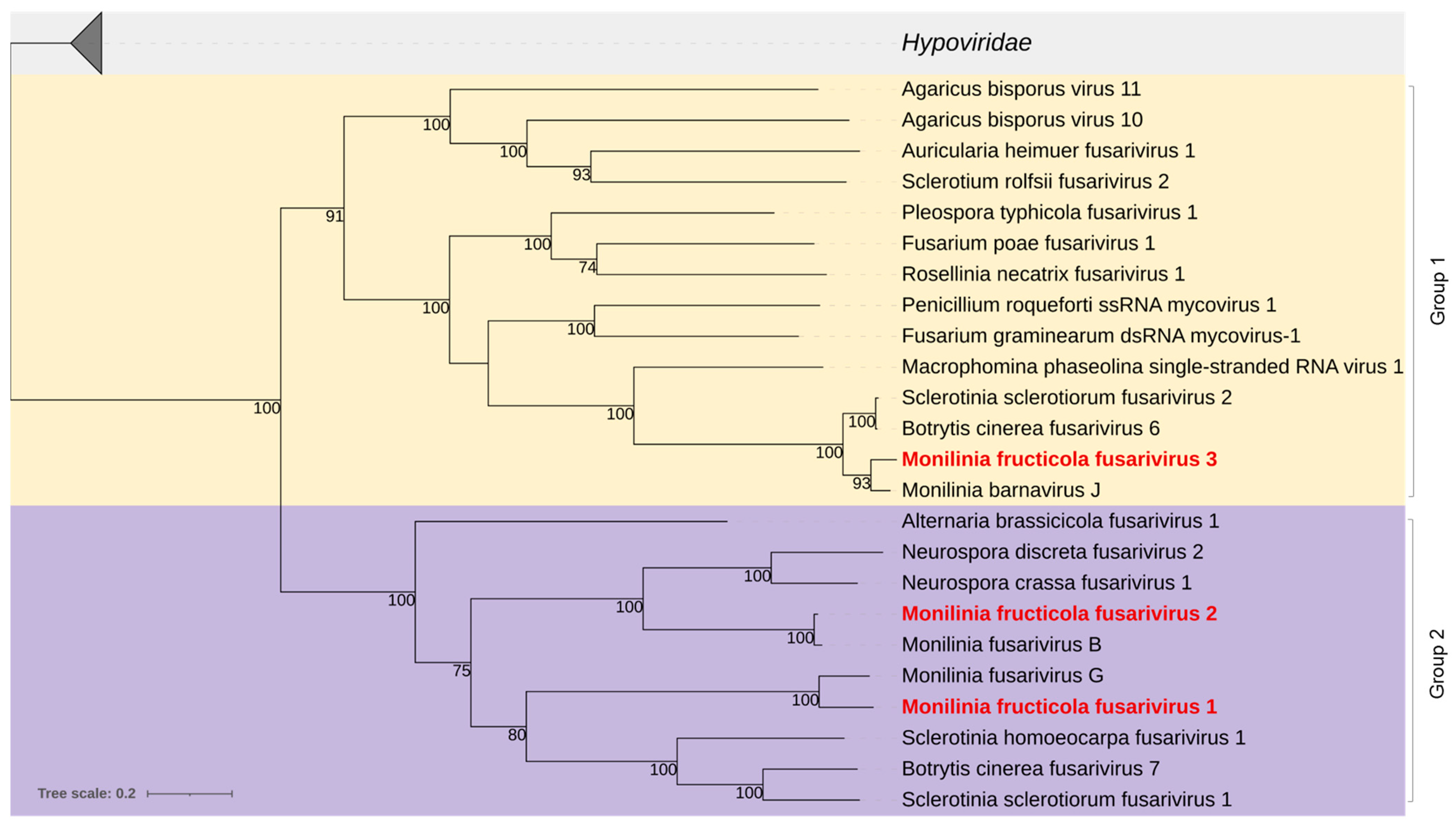
| Pool No. | Isolate Code | Original Code | Location | Host Plant | Year | Provided by |
|---|---|---|---|---|---|---|
| 1 | Mfrc426 | LSV M 177 | France (Provence-Alpes-Cotes d’Azur) | Apricot | 2010 | C. Guinet, France |
| Mfrc427 | LSV M 178 | France (Provence-Alpes-Cotes d’Azur) | Cherry | 2008 | ||
| Mfrc428 | LSV M 1061 | France (Provence-Alpes-Cotes d’Azur) | Peach | 2010 | ||
| Mfrc429 | LSV M 1062 | France (Provence-Alpes-Cotes d’Azur) | Peach | 2010 | ||
| Mfrc430 | LSV M 1063 | France (Provence-Alpes-Cotes d’Azur) | Peach | 2010 | ||
| 2 | Mfrc362 | F5 | Greece (Imathia, Central Macedonia) | Peach | Unknown | G. Karaoglanidis, Greece |
| Mfrc363 | F15 | Greece (Imathia, Central Macedonia) | Peach | Unknown | ||
| Mfrc364 | A6 | Greece (Pella, Central Macedonia) | Peach | Unknown | ||
| Mfrc365 | A7 | Greece (Pella, Central Macedonia) | Peach | Unknown | ||
| 3 | Mfrc416 | 2709 | New Zealand (Central Otago) | Apricot | 1969 | B. Weir, New Zealand |
| Mfrc418 | 7640 | New Zealand (Royal Oak—Auckland) | Peach | Unknown | ||
| Mfrc419 | 7641 | New Zealand (Whenuapai—Auckland) | Peach | 1979 | ||
| Mfrc420 | 7642 | New Zealand (Hastings—Hawke’s Bay) | Peach | 1979 | ||
| Mfrc424 | 20117 | New Zealand (Christchurch, Mid Canterbury) | Plum | 2014 | ||
| 4 | Mfrc407 | I/1 NPUD | Serbia (Udovice, Smederevo) | Nectarine | 2012 | J. Hrustic, Serbia |
| Mfrc408 | I/1 SPOS | Serbia (Osečina, Kolubara) | Plum | 2013 | ||
| Mfrc410 | I/2 TPST | Serbia (Smederevo) | Cherry | 2014 | ||
| Mfrc412 | 8 TP/28 | Serbia (Požarevac, Braničevo) | Cherry | 2015 | ||
| Mfrc414 | 27 BP/12 | Serbia (Golobok, Smederevska Palanka) | Peach | 2016 | ||
| 5 | Mfrc354 | M4.C7.14 | USA (Clemson, South Carolina) | Peach | 2016 | G. Schnabel, USA |
| Mfrc355 | M2.C1.1 | USA (Clemson, South Carolina) | Peach | 2016 | ||
| Mfrc356 | Z3.C10.5 | USA (Sandy Springs, South Carolina) | Peach | 2016 | ||
| Mfrc357 | Z4.C3.3 | USA (Sandy Springs, South Carolina) | Peach | 2016 | ||
| Mfrc358 | Z1.C6.4 | USA (Sandy Springs, South Carolina) | Peach | 2016 | ||
| 6 | Mfrc391 | 21C | Spain (Lleida, Alcarràs) | Unknown | 2013 | A. De Cal, Spain |
| Mfrc393 | 23C | Spain (Lleida, Alcarràs) | Unknown | 2013 | ||
| Mfrc394 | 34C | Spain (Lleida, Albesa) | Unknown | 2009 | ||
| Mfrc396 | 37C | Spain (Lleida, Alfarràs) | Unknown | 2009 | ||
| Mfrc397 | 43C | Spain | Unknown | 2011 | ||
| 7 | Mfrc435 | cfcc 80267 | China (Linyi City, Shandong Province) | Peach | 2005 | L.-Y. Guo, China |
| Mfrc436 | cfcc 80268 | China (Yantai City, Shandong Province) | Peach | 2005 | ||
| Mfrc437 | cfcc 80248 | China (Chaoyang, Pechino, Beijing) | Peach | 2005 | ||
| Mfrc438 | cfcc 80519 | China (Fangshan, Pechino, Beijing) | Plum | 2005 | ||
| 8 | Mfrc106 | C1 | Italy (Bisceglie, Apulia) | Cherry | 2014 | Our fungal collection, Italy |
| Mfrc123 | C18 | Italy (Bisceglie, Apulia) | Cherry | 2014 | ||
| Mfrc301 | M16 | Italy (Gioia del Colle, Apulia) | Cherry | 2014 | ||
| Mfrc322 | O17 | Italy (Gioia del Colle, Apulia) | Cherry | 2014 | ||
| Mfrc376 | 5 | Italy (Cerignola, Apulia) | Peach | 2015 | ||
| 9 | Mfrc148 | E3 | Italy (Tursi, Basilicata) | Plum | 2014 | Our fungal collection, Italy |
| Mfrc150 | E5 | Italy (Tursi, Basilicata) | Plum | 2014 | ||
| Mfrc247 | L2 | Italy (Policoro, Basilicata) | Flat peach | 2014 | ||
| Mfrc261 | L16 | Italy (Policoro, Basilicata) | Flat peach | 2014 | ||
| Mfrc373 | 2 | Italy (Loconia, Basilicata) | Peach | 2014 | ||
| 10 | Mfrc350 | VA bf P12m | USA (Oak Grove, Virginia) | Plum | 2012 | G. Schnabel, USA |
| Mfrc352 | BMPC 10 | USA (Byron, Georgia) | Peach | 2006 | ||
| Mfrc401 | cc866 | USA | Plum | 1995 | G.C.M. Van Leeuwen, The Netherlands | |
| Mfrc402 | cc867 | USA | Plum | 1995 | ||
| Mfrc534 | F534 | USA | Apricot | Unknown | F. Nigro, Italy | |
| 11 | Mfrc331 | 03-K47 | USA (Parlier, California) | Peach | 2002 | T. Michailides, USA |
| Mfrc347 | NY 9C | USA (Geneva, New York) | Cherry | 2007 | G. Schnabel, USA | |
| Mfrc351 | VA bf P16s | USA (Oak Grove, Virginia) | Plum | 2012 | ||
| Mfrc404 | dar27031 | Australia | Peach | 1995 | G.C.M. Van Leeuwen, The Netherlands | |
| Mfrc405 | NZ 2.89 | New Zealand | Plum | 1995 | ||
| 12 | Mfrc77 | A11 | Italy (Caserta, Campania) | Cherry | 2014 | Our fungal collection, Italy |
| Mfrc78 | A12 | Italy (Caserta, Campania) | Cherry | 2014 | ||
| Mfrc395 | 35C | Spain (Lleida, Albesa) | Unknown | 2009 | A. De Cal, Spain | |
| Mfrc399 | 45C | Spain | Unknown | 2011 | ||
| Mfrc415 | 3 VP/1L | Serbia (Šabac, Mačva) | Sour cherry | 2016 | J. Hrustic, Serbia |
| Pool | Total No. of Paired End (PE) Reads | PE Reads Filtered for Quality (QS ≥ 30) | GC% | ||
|---|---|---|---|---|---|
| Total No. | Nonredundant | ||||
| No. | % | ||||
| 1 | 21,633,810 | 21,483,924 | 15,579,294 | 72.5 | 29 |
| 2 | 20,710,678 | 20,562,344 | 17,423,394 | 84.7 | 34 |
| 3 | 22,653,808 | 22,481,652 | 12,863,610 | 83.9 | 34 |
| 4 | 31,855,794 | 31,616,498 | 26,697,938 | 84.4 | 29 |
| 5 | 21,628,744 | 21,482,266 | 16,170,072 | 75.3 | 30 |
| 6 | 106,258,396 | 105,125,504 | 26,667,956 | 25.4 | 40 |
| 7 | 85,651,472 | 84,768,112 | 21,327,038 | 25.2 | 41 |
| 8 | 87,698,960 | 86,789,590 | 21,053,874 | 24.2 | 44 |
| 9 | 80,371,552 | 79,533,392 | 20,619,356 | 25.9 | 47 |
| 10 | 66,807,410 | 66,139,758 | 24,827,978 | 37.5 | 43 |
| 11 | 62,570,222 | 61,947,854 | 24,285,582 | 39.2 | 43 |
| 12 | 72,161,200 | 71,415,466 | 15,575,196 | 21.8 | 40 |
| Pool | De Novo Assembly | No. of Putative Viral Contigs Selected by BLASTP Analysis | |||
|---|---|---|---|---|---|
| Contigs (No.) | Mapped Reads | Total | Nonredundant | ||
| (No.) ¥ | (%) | ||||
| 1 | 48,825 | 19,461,803 | 90.59 | 1527 | 128 |
| 2 | 34,131 | 17,854,177 | 86.83 | 1661 | 127 |
| 3 | 53,832 | 19,461,803 | 86.57 | 1612 | 111 |
| 4 | 49,458 | 28,213,117 | 89.24 | 1773 | 138 |
| 5 | 61,986 | 17,595,205 | 81.91 | 1510 | 193 |
| 6 | 43,774 | 103,533,111 | 98.49 | 2058 | 588 |
| 7 | 46,702 | 82,089,562 | 96.84 | 551 | 508 |
| 8 | 28,049 | 84,503,137 | 97.37 | 163 | 106 |
| 9 | 42,092 | 78,390,075 | 98.56 | 553 | 67 |
| 10 | 43,095 | 63,088,469 | 95.39 | 1725 | 145 |
| 11 | 29,999 | 59,945,146 | 96.77 | 1622 | 106 |
| 12 | 31,740 | 68,867,450 | 96.43 | 1308 | 93 |
| Virus | Genome (nt) | Protein (aa) | Similarity with Viral Sequences (BLASTP) | |||
|---|---|---|---|---|---|---|
| Mycovirus | Coverage | E-Value | Identity | |||
| Mitoviridae | ||||||
| MfrcMV1 | +ssRNA (2658) | RdRp (721) | Botrytis cinerea mitovirus 9 | 100% | 0 | 81.97% |
| MfrcMV2 | +ssRNA (2298) | RdRp (683) | Sclerotinia sclerotiorum mitovirus 2 | 97% | 0 | 63.34% |
| MfrcMV3 | +ssRNA (2657) | RdRp (712) | Sclerotinia sclerotiorum mitovirus 3-WX | 100% | 0 | 92.13% |
| MfrcMV4 | +ssRNA (2441) | RdRp (731) | Sclerotinia sclerotiorum mitovirus 39 | 97% | 0 | 84.11% |
| MfrcMV5 | +ssRNA (2430) | RdRp (698) | Ophiostoma mitovirus 6 | 94% | 3 × 10−162 | 42.28% |
| MfrcMV6 | +ssRNA (2341) | RdRp (710) | Sclerotinia sclerotiorum mitovirus 46 | 98% | 0 | 69.86% |
| MfrcMV7 | +ssRNA (2718) | RdRp (689) | Sclerotinia sclerotiorum mitovirus 7 | 100% | 0 | 99.13% |
| MfrcMV8 | +ssRNA (2758) | RdRp (689) | Sclerotinia sclerotiorum mitovirus 19 | 99% | 0 | 90.96% |
| MfrcMV9 | +ssRNA (2438) | RdRp (738) | Botrytis cinerea mitovirus 1 | 100% | 0 | 79.76% |
| MfrcMV10 | +ssRNA (2670) | RdRp (794) | Sclerotinia sclerotiorum mitovirus 28 | 97% | 0 | 70.82% |
| MfrcMV11 | +ssRNA (2816) | RdRp (866) | Sclerotinia sclerotiorum mitovirus 29 | 99% | 0 | 64.60% |
| MfrcMV12 | +ssRNA (2401) | RdRp (708) | Botrytis cinerea mitovirus 6 | 99% | 0 | 78.25% |
| Splipalmiviridae | ||||||
| MfrcSPV1 | +ssRNA (2466) | RdRp (785) | Erysiphe necator associated narnavirus 13 | 100% | 0 | 77.23% |
| +ssRNA (2123) 1 | Hypothetical protein (697) | Botrytis cinerea binarnavirus 2 | 93% | 0 | 47.87% | |
| Botourmiaviridae | ||||||
| MfrcBOV1 | +ssRNA (2830) | RdRp (684) | Sclerotinia sclerotiorum ourmia-like virus 1 | 99% | 0 | 72.70% |
| MfrcBOV2 | +ssRNA (2917) | RdRp (707) | Botrytis cinerea ourmia-like virus 13 | 100% | 0 | 73.38% |
| MfrcBOV3 | +ssRNA (2520) | RdRp (647) | Sclerotinia sclerotiorum ourmia-like virus 13 | 99% | 0 | 70.33% |
| MfrcBOV4 | +ssRNA (2617) | RdRp (730) | Botrytis cinerea ourmia-like virus 9 | 91% | 0 | 78.85% |
| MfrcBOV5 | +ssRNA (2792) | RdRp (656) | Monilinia ourmiavirus B | 100% | 0 | 99.70% |
| MfrcBOV6 | +ssRNA (3004) | RdRp (655) | Erysiphe necator associated ourmia-like virus 130 | 93% | 2 × 10−172 | 46.96% |
| MfrcBOV7 | +ssRNA (2382) | RdRp (645) | Plasmopara viticola associated ourmia-like virus 73 | 94% | 2 × 10−170 | 47.66% |
| MfrcBOV8 | +ssRNA (2695) | RdRp (668) | Oidiodendron maius ourmia-like virus 1 | 95% | 0 | 54.25% |
| MfrcBOV9 | +ssRNA (2638) | RdRp (780) | Monilinia ourmiavirus I | 69% | 0 | 86.64% |
| MfrcBOV10 | +ssRNA (2774) | RdRp (736) | Monilinia ourmiavirus A | 99% | 0 | 85.91% |
| MfrcBOV11 | +ssRNA (2438) | RdRp (667) | Pyricularia oryzae ourmia-like virus 1 | 97% | 0 | 54.71% |
| MfrcBOV12 | +ssRNA (2544) | RdRp (654) | Plasmopara viticola associated ourmia-like virus 47 | 97% | 0 | 50.54% |
| Hypoviridae | ||||||
| MfrcHV1 | +ssRNA (9338) | Polyprotein (2918) | Sclerotinia sclerotiorum hypovirus 7 | 100% | 0 | 97.74% |
| MfrcHV2 | +ssRNA (15,037) | Polyprotein (4784) | Monilinia hypovirus D | 99% | 0 | 93.24% |
| Fusariviridae | ||||||
| MfrcFV1 | +ssRNA (7425) | RdRp (1661) | Monilinia fusarivirus G | 99% | 0 | 81.24% |
| Protein ShFV1_gp2 (504) | Monilinia fusarivirus G | 98% | 0 | 71.54% | ||
| MfrcFV2 | +ssRNA (7330) | RdRp (1667) | Monilinia fusarivirus B | 100% | 0 | 97.36% |
| Protein BSB06_gp2 (509) | Monilinia fusarivirus B | 100% | 0 | 95.48% | ||
| MfrcFV3 | +ssRNA (5550) 1 | RdRp (1399) | Monilinia barnavirus J | 100% | 0 | 96.21% |
| Protein ShFV1_gp2 (434) | Sclerotinia sclerotiorum fusarivirus 2 | 99% | 0 | 70.00% | ||
| Barnaviridae | ||||||
| MfrcBV1 | +ssRNA (4993) | Peptidase (861) | Monilinia barnavirus A ORF1 | 99% | 0 | 94.52% |
| RdRp (691) | Monilinia barnavirus A ORF2 | 100% | 0 | 94.50% | ||
| Putative protein (238) | Monilinia barnavirus A ORF3 | 100% | 4 × 10−170 | 99.58% | ||
| Benyviridae | ||||||
| MfrcBeLV1 | +ssRNA (5761) | Replication-associated protein (1623) | Lentinula edodes ssRNA mycovirus | 100% | 0 | 47.38% |
| Hypothetical protein (214) | - | - | - | - | ||
| Parvoviridae | ||||||
| MfrcPV1 | ssDNA (1664) 1 | Non-structural protein NS3 (152) | Human CSF-associated densovirus | 94% | 3 × 10−64 | 67.13% |
| Structural protein VP (375) | Ciconia boyciana parvoviridae | 100% | 0 | 96.80% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Miccolis Angelini, R.M.; Raguseo, C.; Rotolo, C.; Gerin, D.; Faretra, F.; Pollastro, S. The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola. J. Fungi 2022, 8, 481. https://doi.org/10.3390/jof8050481
De Miccolis Angelini RM, Raguseo C, Rotolo C, Gerin D, Faretra F, Pollastro S. The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola. Journal of Fungi. 2022; 8(5):481. https://doi.org/10.3390/jof8050481
Chicago/Turabian StyleDe Miccolis Angelini, Rita Milvia, Celeste Raguseo, Caterina Rotolo, Donato Gerin, Francesco Faretra, and Stefania Pollastro. 2022. "The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola" Journal of Fungi 8, no. 5: 481. https://doi.org/10.3390/jof8050481
APA StyleDe Miccolis Angelini, R. M., Raguseo, C., Rotolo, C., Gerin, D., Faretra, F., & Pollastro, S. (2022). The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola. Journal of Fungi, 8(5), 481. https://doi.org/10.3390/jof8050481






