Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome
Abstract
1. Introduction
1.1. The Number of Kinesins in Eukaryotic Organisms
1.2. Kinesin Structure and Function
1.3. Schizophyllum Commune Kinesins
2. Cytoplasmic Transport Kinesins
2.1. Kinesin-1, Schcokin1
2.2. Kinesin3, Schcokin3
3. Mitotic Kinesins
3.1. Kinesin-5, Schcokin5
3.2. Kinesin-14, Schco14
3.3. Kinesin-4, Schcokin4
3.4. Kinesin-6, Rab6- Kinesin, Schcokin6
3.5. Kinesin 7, Schcokin7A, Schcokin7B
3.6. Kinesin-8, Schcokin8
3.7. Kinesin-10, Schcokin10
4. Are Kinesins Necessary for Growth and Nuclear Movements in Filamentous Basidiomycetes?
5. Summary and Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raper, J.R. Genetics of Sexuality in Higher Fungi; The Roland Press Company: New York, NY, USA, 1966; pp. 3–277. [Google Scholar]
- Ohm, R.A.; de Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; de Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 9, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Raudaskoski, M.; Kothe, E. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell 2010, 9, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Raudaskoski, M. Mating-type genes and hyphal fusions in filamentous basidiomycetes. Fungal Biol. Rev. 2015, 29, 179–193. [Google Scholar] [CrossRef]
- Wirth, S.; Freihorst, D.; Krause, K.; Kothe, E. What Role Might Non-Mating Receptors Play in Schizophyllum commune? J. Fungi 2021, 7, 399. [Google Scholar] [CrossRef]
- Salo, V.; Niini, S.S.; Virtanen, I.; Raudaskoski, M. Comparative immunocytochemistry of the cytoskeleton in filamentous fungi with dikaryotic and multinucleate hyphae. J. Cell Sci. 1989, 94, 11–24. [Google Scholar] [CrossRef]
- Raudaskoski, M.; Mao, W.-Z.; Yli-Mattila, T. Microtubule cytoskeleton in hyphal growth. Response to nocodazole in a sensitive and a tolerant strain of the homobasidiomycete Schizophyllum commune. Eur. J. Cell Biol. 1994, 64, 131–141. [Google Scholar]
- Rupeš, I.; Mao, W.-Z.; Åström, H.; Raudaskoski, M. Effects of nocodazole and brefeldin A on microtubule cytoskeleton and membrane organization in the homobasidiomycete Schizophyllum commune. Protoplasma 1995, 185, 212–221. [Google Scholar] [CrossRef]
- Jung, E.M.; Kothe, E.; Raudaskoski, M. The making of a mushroom: Mitosis, nuclear migration and the acton network. Fungal Genet. Biol. 2018, 111, 85–91. [Google Scholar] [CrossRef]
- Raudaskoski, M. The central role of septa in the basidiomycete Schizophyllum commune hyphal morphogenesis. Fungal Biol. 2019, 123, 638–649. [Google Scholar] [CrossRef]
- Runeberg, P.; Raudaskoski, M.; Virtanen, I. Cytoskeletal elements in the hyphae of the homobasidiomycete Schizophyllum commune visualized with indirect immunofluorescence and NBD-phallacidin. Eur. J. Cell Biol. 1986, 41, 25–32. [Google Scholar]
- Raudaskoski, M.; Rupes, I.; Timonen, S. Immunofluorescence microscopy of the cytoskeleton in filamentous fungi after quick-freezing and low-temperature fixation. Exp. Mycol. 1991, 15, 167–173. [Google Scholar] [CrossRef]
- Raudaskoski, M. The relationship between B-mating-type genes and nuclear migration in Schizophyllum commune. Fungal Genet. Biol. 1998, 24, 207–227. [Google Scholar] [CrossRef] [PubMed]
- Brunsch, M.; Schubert, D.; Gube, M.; Ring, C.; Hanish, L.; Linde, J.; Krause, K.; Kothe, E. Dynein heavy chain, encoded by two genes in Agaricomycetes, is required for nuclear migration in Schizophyllum commune. PLoS ONE 2015, 10, e0135616. [Google Scholar] [CrossRef] [PubMed]
- Straube, A.; Enard, W.; Berner, A.; Wedlich-Söldner, R.; Kahmann, R.; Steinberg, G. A split motor domain in cytoplasmic dynein. EMBO J. 2001, 20, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, E.R.; Hoyt, M.A. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2000, 1496, 99–116. [Google Scholar] [CrossRef]
- Kashiwazaki, J.; Yoneda, Y.; Mutoh, T.; Arai, R.; Yoshida, M.; Mabuchi, I. A unique kinesin-like protein, Klp8, is involved in mitosis and cell morphology through microtubule stabilization. Cytoskeleton 2019, 76, 355–367. [Google Scholar] [CrossRef]
- Schoch, C.L.; Aist, J.R.; Yoder, O.C.; Turgeon, G. A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet. Biol. 2003, 39, 1–15. [Google Scholar] [CrossRef]
- Rischitor, P.E.; Konzack, S.; Fischer, R. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell 2004, 3, 632–645. [Google Scholar] [CrossRef]
- Schuchardt, I.; Assmann, D.; Thines, E.; Schuberth, C.; Steinberg, G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 2005, 16, 5191–5201. [Google Scholar] [CrossRef]
- Welburn, J.P.I. The molecular basis for kinesin functional specificity during mitosis. Cytoskeleton 2013, 70, 476–493. [Google Scholar] [CrossRef]
- Lawrence, C.J.; Dawe, R.K.; Christie, K.R.; Cleveland, D.W.; Dawson, S.C.; Endow, S.A.; Goldstein, L.S.; Goodson, H.V.; Hirokawa, N.; Howard, J.; et al. A standardizied kinesin nomecclature. J. Cell Biol. 2004, 167, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Okada, Y.N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005, 9, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cao, L.; Wang, C.; Gigant, B.; Knossow, M. Kinesin, 30 years later: Recent insights from structural studies. Protein Sci. 2015, 7, 1047–1105. [Google Scholar] [CrossRef] [PubMed]
- Woehkle, G.; Ruby, A.K.; Hart, C.L.; Ly, B.; Hom-Booher, N.; Vale, R.D. Microtubule interaction site of the kinesin motor. Cell 1997, 90, 207–216. [Google Scholar]
- Cao, L.; Cantos-Fernandes, S.; Gigant, B. The structural switch of nucleotide-free kinesin. Sci Rep. 2017, 7, 42558. [Google Scholar] [CrossRef]
- Vale, R.D.; Fletterick, R.J. The design plan of kinesin motors. Ann. Rev. Cell Dev. Biol. 1997, 13, 745–777. [Google Scholar] [CrossRef]
- Guo, S.K.; Wang, P.Y.; Xie, P. Dynamics of dimeric kinesins: Limping, effect of longitudinal force, effects of neck linker extension and mutation, and comparison between kinesin-1 and kinesin-2. Int. J. Biol. Macromol. 2017, 105, 1126–1137. [Google Scholar] [CrossRef]
- Kojima, H.; Muto, E.; Higuchi, H.; Yanagida, T. Mechanics of single kinesin molecules measured by optical trapping nanometry. Biophys. J. 1997, 73, 2012–2022. [Google Scholar] [CrossRef]
- Yildiz, A.; Tomishige, M.; Vale, R.D.; Selvin, P.R. Kinesin walks hand-over-hand. Science 2004, 303, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Stepp, W.L.; Ökten, Z. Resolving kinesin stepping: One head at a time. Life Sci. Alliance 2019, 2, e201900456. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Romberg, L.; Pierce, D.W.; Vale, R.D. Role of the Kinesin neck region in processive microtubule-based motility. J. Cell Biol. 1998, 140, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Tripet, B.; Hodges, R.S. Helix capping interactions stabilize the N-terminus of the kinesin neck coiled-coil. J. Struct. Biol. 2002, 137, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, V.; Hancock, W.O. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell. Mol. Bioeng. 2009, 2, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.K.; Peter, L.G.; Gilbert, S.P.; Rayment, I. Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil. Biol. Chem. 2016, 291, 20372–20386. [Google Scholar] [CrossRef]
- Wade, R.H.; Kozielski, F. Structural links to kinesin directionality and movement. Nat. Struct. Biol. 2000, 6, 456–460. [Google Scholar] [CrossRef]
- Verhey, K.J.; Hammond, J.W. Traffic control: Regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 2009, 10, 765–777. [Google Scholar] [CrossRef]
- Hackney, D.D.; Stock, M.F. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2000, 2, 257–260. [Google Scholar] [CrossRef]
- Bathe, F.; Hahlen, K.; Dombi, R.; Driller, L.; Schliwa, M.; Woehlke, G. The Complex Interplay between the Neck and Hinge Domains in Kinesin-1 Dimerization and Motor Activity. Mol. Biol. Cell 2005, 8, 3529–3537. [Google Scholar] [CrossRef][Green Version]
- Cai, D.; Hoppe, A.D.; Swanson, J.A.; Verhey, K.J. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J. Cell Biol. 2007, 176, 51–63. [Google Scholar] [CrossRef]
- Hoyt, M.A.; Le, L.; Loo, K.K.; Saunders, V.S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992, 118, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, E.R.; Gheber, L.; Kingsbury, T.; Hoyt, M.A. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 2006, 281, 26004–26013. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, M.A.; He, L.; Totis, L.; Saunders, W.S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 1993, 135, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Fridman, V.; Gerson-Gurwitz, A.; Shapira, O.; Movshovich, N.; Lakämper, S.; Schmidt, C.F.; Gheber, L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 2013, 126, 4147–4159. [Google Scholar] [CrossRef]
- Augustine, B.; Chen, F.C.; Yeong, F.M. Role of Kip2 during early mitosis—Impact on spindle pole body separation and chromosome capture. J. Cell Sci. 2018, 131, jcs211425. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.; Gupta, M.L., Jr.; Hoyt, M.A.; Pellman, D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell 2004, 6, 815–829. [Google Scholar] [CrossRef]
- Roberts, A.J.; Goodman, B.S.; Reck-Peterson, S.L. Reconstitution of dynein transport to the microtubule plus end by kinesin. eLife 2014, 3, e02641. [Google Scholar] [CrossRef]
- Gupta, M.L., Jr.; Carvalho, P.; Roof, D.M.; Pellman, D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006, 8, 913–923. [Google Scholar] [CrossRef]
- Meluh, P.M.; Rose, M.D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 1990, 60, 1029–1041. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 1992, 356, 74–76. [Google Scholar] [CrossRef]
- Edamatsu, M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem. Biophys. Res. Commun. 2014, 446, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, M.; Okazaki, M.; Teratani, Y.; Furuta, K.; Toda, T. Kinesin-6 Klp9 plays motor-dependent and -independent roles in collaboration with Kinesin-5 Cut7 and the microtubule crosslinker Ase1 in fission yeast. Sci. Rep. 2019, 9, 7336. [Google Scholar] [CrossRef]
- Browning, H.; Hayles, J.; Mata, J.; Aveline, L.; Nurse, P.; McIntosh, J.R. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 2000, 151, 15–28. [Google Scholar] [CrossRef]
- Unsworth, A.; Masuda, H.; Dhut, S.; Toda, T. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol. Biol. Cell 2008, 19, 5104–5115. [Google Scholar] [CrossRef]
- Lin, Y.; Wei, Y.-L.; She, Z.-Y. Kinesin-8 motors: Regulation of microtubule dynamics and chromosome movements. Chromosoma 2020, 129, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Troxell, C.L.; Sweezy, M.A.; West, R.R.; Reed, K.D.; Carson, B.D.; Pidoux, A.L.; Cande, W.Z.; McIntosh, J.R. pkl1+ and klp2+: Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol. Biol. Cell 2001, 12, 3476–3488. [Google Scholar] [CrossRef] [PubMed]
- Requena, N.; Alberti-Segui, C.; Winzenburg, E.; Horn, C.; Schliwa, M.; Philippsen, P.; Liese, L.; Fischer, R. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 2001, 42, 121–132. [Google Scholar] [CrossRef]
- Takeshita, N.; Wernet, V.; Tsuizaki, M.; Grün, N.; Hoshi, H.O.; Ohta, A.; Fischer, R.; Horiuchi, H. Transportation of Aspergillus nidulans class III and V chitin synthases to the hyphal tips depends on conventional kinesin. PLoS ONE 2015, 10, e0125937. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Fischer, R.; Xiang, X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 2003, 14, 1479–1488. [Google Scholar] [CrossRef]
- Zekert, N.; Fischer, R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol. Biol. Cell 2009, 20, 673–684. [Google Scholar] [CrossRef]
- Enos, A.P.; Morris, R.N. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 1990, 60, 1019–1027. [Google Scholar] [CrossRef]
- Stock, M.F.; Chu, J.; Hackney, D.D. The kinesin family member BimC contains a second microtubule binding region attached to the N terminus of the motor domain. J. Biol. Chem. 2003, 278, 52315–52322. [Google Scholar] [CrossRef] [PubMed]
- Gerson-Gurwitz, A.; Thiede, C.; Movshovich, N.; Fridman, V.; Podolskaya, M.; Danieli, T.; Lakämper, S.; Klopfenstein, D.R.; Schmidt, C.F.; Gheber, L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011, 30, 4942–4954. [Google Scholar] [CrossRef]
- Konzack, S.; Rischitor, P.E.; Enke, C.; Fischer, R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 2005, 16, 497–506. [Google Scholar] [CrossRef]
- Herrero, S.; Takeshita, N.; Fischer, R. The Aspergillus nidulans CENP-E kinesin motor KipA interacts with the fungal homologue of the centromere-associated protein CENP-H at the kinetochore. Mol. Microbiol. 2011, 80, 981–994. [Google Scholar] [CrossRef]
- Efimov, V.P.; Zhang, J.; Xiang, X. CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol. Biol. Cell 2006, 17, 2021–2034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garcia, M.A.; Koonrugsa, N.; Toda, T. Spindle–kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002, 21, 6015–6024. [Google Scholar] [CrossRef]
- O’Connell, M.J.; Meluh, P.B.; Rose, M.D.; Morris, N.R. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J. Cell Biol. 1993, 120, 153–162. [Google Scholar] [CrossRef]
- Prigozhina, N.L.; Walker, R.A.; Oakley, E.C.; Oakley, B.R. γ-Tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 2001, 12, 3161–3174. [Google Scholar] [CrossRef]
- Mouriño-Pérez, R.R.; Riquelme, M.; Callejas-Negrete, O.A. Microtubules and associated molecular motors in Neurospora crassa. Mycologia 2016, 108, 515–527. [Google Scholar] [CrossRef]
- Vale, R.D.; Reese, T.S.; Sheetz, M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 1985, 42, 39–50. [Google Scholar] [CrossRef]
- Steinberg, G.; Schliwa, M. The Neurospora organelle motor: A distant relative of conventional kinesin with unconventional properties. Mol. Biol. Cell 1995, 11, 1605–1618. [Google Scholar] [CrossRef] [PubMed]
- Lehmler, C.; Steinberg, G.; Snetselaar, K.M.; Schliwa, M.; Kahmann, R.; Bölker, M. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 1997, 16, 3464–3473. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.; Schliwa, M. Characterization of the biophysical and motility properties of kinesin from the fungus Neurospora crassa. J. Biol. Chem. 1996, 271, 7516–7521. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G. A kinesin-like mechanoenzyme from the zygomycete Syncephalastrum racemosum shares biochemical similarities with conventional kinesin from Neurospora crassa. Eur. J. Cell Biol. 1997, 73, 124–131. [Google Scholar]
- Kirchner, J.; Woehlke, G.; Schliwa, M. Universal and unique features of kinesin motors: Insights from a comparison of fungal and animal conventional kinesins. Biol. Chem. 1999, 380, 915–921. [Google Scholar] [CrossRef][Green Version]
- Song, Y.H.; Marx, A.; Muller, J.; Woehlke, G.; Schliwa, M.; Krebs, A.; Hoenger, A.; Mandelkow, E. Structure of fast kinesin: Implications for ATPase mechanism and interactions with microtubules. EMBO J. 2001, 20, 6213–6225. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Zhangb, J.; Xiangb, X.; Pantazopouloua, A. Transport of fungal RAB11 secretory vesicles involves myosin-5, dynein/dynactin/p25, and kinesin-1 and is independent of kinesin-3. Mol. Biol. Cell 2017, 28, 947–961. [Google Scholar] [CrossRef]
- Seidel, C.; Moreno-Velásquez, S.D.; Riquelme, M.; Fischer, R. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot. Cell 2013, 12, 1020–1032. [Google Scholar] [CrossRef][Green Version]
- Schuster, M.; Martin-Urdiroz, M.; Higuchi, Y.; Hacker, C.; Kilaru, S.; Gurr, S.J.; Steinberg, G. Co-delivery of cell-wall-forming enzymes in the same vesicle for coordinated fungal cell wall formation. Nat. Microbiol. 2016, 1, 16149. [Google Scholar] [CrossRef]
- Wedlich-Söldner, R.; Schulz, I.; Straube, A.; Steinberg, G. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 2002, 13, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Wedlich-Söldner, R.; Straube, A.; Friedrich, M.W.; Steinberg, G. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 2002, 21, 2946–2957. [Google Scholar] [CrossRef] [PubMed]
- Lenz, J.H.; Schuchardt, I.; Straube, A.; Steinberg, G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006, 25, 2275–2286. [Google Scholar] [CrossRef]
- Schuster, M.; Assmann, M.A.; Lenz, P.; Lipowsky, R.; Steinberg, G. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA 2011, 108, 3618–3623. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Kilarua, S.; Fink, G.; Collemarec, J.; Rogera, Y.; Steinberg, G. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell 2011, 22, 3645–3657. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.; Schuster, M. The dynamic fungal cell. Fungal Biol. Rev. 2011, 25, 14–37. [Google Scholar] [CrossRef]
- Steinberg, G. Endocytosis and early endosome motility in filamentous fungi. Curr. Opin. Microbiol. 2014, 20, 10–18. [Google Scholar] [CrossRef]
- Steinberg, G. Kinesin-3 in the basidiomycete Ustilago maydis transports organelles along the entire microtubule array. Fungal Genet. Biol. 2015, 74, 59–61. [Google Scholar] [CrossRef]
- Baumann, S.; Pohlmann, T.; Jungbluth, M.; Brachmann, A.; Feldbrügge, M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 2012, 125, 2740–2752. [Google Scholar] [CrossRef]
- Slogiannis, J.; Reck-Peterson, S.L. Hitchhiking: A non-canonical mode of microtubule-based transport. Trends Cell Biol. 2017, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- von Loeffelholz, O.; Moores, C.A. Cryo-EM structure of the Ustilago maydis kinesin-5 motor domain bound to microtubules. J. Struct. Biol. 2019, 207, 312–316. [Google Scholar] [CrossRef]
- Simm, D.; Hatje, K.; Kollmar, M. Waggawagga: Comparative visualization of coiled-coil predictions and detection of stable single α-helices (SAH domains). Bioinformatics 2015, 31, 767–769. [Google Scholar] [CrossRef]
- Kallipolitou, A.; Deluca, D.; Majdic, U.; Lakämper, S.; Cross, R.; Meyhöfer, E.; Moroder, L.; Schliwa, M.; Woehlke, G. Unusual properties of the fungal conventional kinesin neck domain from Neurospora crassa. EMBO J. 2001, 20, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, F.; Deluca, D.; Majdic, U.; Kirchner, J.; Schliwa, M.; Moroder, L.; Woehlke, G. A conserved tyrosine in the neck of a fungal kinesin regulates the catalytic motor core. EMBO J. 2003, 22, 450–458. [Google Scholar] [CrossRef]
- Hahlen, K.; Ebbing, B.; Reinders, J.; Mergler, J.; Sickmann, A.; Woehlke, G. Feedback of the kinesin-1 neck-linker position on the catalytic site. J. Biol. Chem. 2006, 281, 18868–18877. [Google Scholar] [CrossRef] [PubMed]
- Adio, S.; Reth, J.; Bathe, F.; Woehlke, G. Review: Regulation mechanisms of Kinesin-1. J. Muscle Res. Cell Mot. 2006, 27, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Seiler, S.; Kirchner, J.; Horn, C.; Kallipolitou, A.; Woehlke, G.; Schliwa, M. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2000, 2, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Seiler, S.; Nargang, F.E.; Steinberg, G.; Schliwa, M. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 1997, 16, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, A.J.; Jeyaprakash, A.; García-Añoveros, J.; Tang, L.Z.; Fisk, G.; Hartshorne, T.; Franco, R.; Born, T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron 1991, 1, 113–122. [Google Scholar] [CrossRef]
- Okada, Y.; Yamazaki, H.; Sekine-Aizawa, Y.; Hirokawa, N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 1995, 81, 769–780. [Google Scholar] [CrossRef]
- Becht, P.; Vollmeister, E.; Feldbrügge, M. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot. Cell 2005, 4, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Becht, P.; König, J.; Feldbrügge, M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 2006, 119, 4964–4973. [Google Scholar] [CrossRef] [PubMed]
- Zarnack, K.; Feldbrügge, M. Microtubule-Dependent mRNA Transport in Fungi. Eukaryot. Cell 2010, 9, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Pohlmann, T.; Feldbrügge, M. Core components of endosomal mRNA transport are evolutionarily conserved in fungi. Fungal Genet. Biol. 2019, 126, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Blangy, A.; Heidi, A.; Lane, H.A.; d’Herin, P.; Harper, M.; Kress, M.S.; Nigg, E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 1995, 63, 1159–1169. [Google Scholar] [CrossRef]
- Bannigan, A.; Scheible, W.-R.; Lukowitz, W.; Fagerstrom, C.; Wadsworth, P.; Somerville, C.; Baskin, T.I. A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 2007, 120, 2819–2827. [Google Scholar] [CrossRef]
- Roof, D.M.; Meluh, P.B.; Rose, M.D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992, 118, 95–108. [Google Scholar] [CrossRef]
- Acar, S.; Carlson, D.B.; Budamagunta, M.S.; Yarov-Yarovoy, V.; Correia, J.J.; Niñonuevo, M.R.; Jia, W.; Tao, L.; Leary, J.A.; Voss, J.C.; et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 2013, 4, 1343. [Google Scholar] [CrossRef]
- Scholey, J.E.; Nithianantham, S.; Scholey, J.M.; Al-Bassam, J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. eLife 2014, 3, e02217. [Google Scholar] [CrossRef]
- Hoyt, M.A. Cellular roles of kinesin and related proteins. Curr. Opin. Cell Biol. 1994, 6, 63–68. [Google Scholar] [CrossRef]
- Tao, L.; Mogilner, A.; Civelekoglu-Scholey, G.; Wollman, R.; Evans, J.; Stahlberg, H.; Scholey, J.M. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 2006, 16, 2293–2302. [Google Scholar] [CrossRef] [PubMed]
- Rincon, S.A.; Lamson, A.; Blackwell, R.; Syrovatkina, V.; Fraisier, V.; Paoletti, A.; Betterton, M.D.; Tran, P.T. Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat. Commun. 2017, 8, 15286. [Google Scholar] [CrossRef] [PubMed]
- Britto, M.; Goulet, A.; Rizvi, S.; von Loeffelholz, O.; Moores, C.A.; Cross, R.A. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 7483–7489. [Google Scholar] [CrossRef] [PubMed]
- McDonald, H.B.; Stewart, R.J.; Goldstein, L.S. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 1990, 63, 1159–1165. [Google Scholar] [CrossRef]
- Manning, B.D.; Barrett, J.G.; Wallace, J.A.; Granok, H.; Snyder, M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 1999, 144, 1219–1233. [Google Scholar] [CrossRef]
- Endow, S.A.; Higuchi, H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature 2000, 406, 9013–9916. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Shigematsu, H.; Yokoyama, T.; Kikkawa, M.; Sugawa, M.; Aoki, M.; Shirouzu, M.; Yajima, J.; Nitta, R. Structural basis of backwards motion in kinesin-1-kinesin-14 chimera: Implication for kinesin-14 motility. Structure 2016, 24, 1322–1334. [Google Scholar] [CrossRef] [PubMed]
- Sekine, Y.; Okada, Y.; Noda, Y.; Kondo, S.; Aizawa, H.; Takemura, R.; Hirokawa, N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J. Cell Biol. 1994, 127, 187–201. [Google Scholar] [CrossRef]
- Sheng, L.; Hao, S.-L.; Yang, W.-X.; Sun, Y. The multiple functions of kinesin-4 family motor protein KIF4 and its clinical potential. Gene 2018, 678, 90–99. [Google Scholar] [CrossRef]
- Almeida, A.C.; Maiato, H. 2018: Chromokinesins. Curr. Biol. 2018, 28, R1131–R1135. [Google Scholar] [CrossRef]
- Mazumdar, M.; Sung, M.-H.; Misteli, T. Chromatin maintenance by a molecular motor protein. Nucleus 2011, 2, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Heintz, T.G.; Heller, J.P.; Zhao, R.; Caceres, A.; Eva, R.; Fawcett, J.W. Kinesin KIF4A transports integrin β1 in developing axons of cortical neurons. Mol. Cell Neurosci. 2014, 63, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.J.; Nader, G.P.; Ramalingam, N.; Bartolini, F.; Gundersen, G.G. Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS ONE 2014, 9, e91568. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fu, C.; Ward, J.J.; Loiodice, I.; Velve-Casquillas, G.; Nedelec, F.J.; Tran, P.T. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell 2009, 17, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Krüger, L.K.; Sanchez, J.-L.; Paoletti, A.; Tran, P.T. Kinesin-6 regulates cell-size-dependent spindle elongation velocity to keep mitosis duration constant in fission yeast. eLife 2019, 8, e42182. [Google Scholar] [CrossRef]
- Fontijn, R.D.; Goud, B.; Echard, A.; Jollivet, F.; van Marle, J.; HPannekoek, H.; Horrevoets, A.J. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell Biol. 2001, 8, 2944–2955. [Google Scholar] [CrossRef] [PubMed]
- Janisch, K.M.; McNeely, K.C.; Dardick, J.M.; Lim, S.M.; Dwyer, N.D. 2 Kinesin-6 KIF20B is required for efficient cytokinetic furrowing and timely abscission in human cells. Mol. Biol. Cell 2018, 29, 166–179. [Google Scholar] [CrossRef]
- Craske, B.; Welburn, J.P.I. Leaving no-one behind: How CENP-E facilitates chromosome alignment. Essays Biochem. 2020, 64, 313–324. [Google Scholar]
- Cottingham, F.R.; Hoyt, M.A. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 1997, 138, 1041–1053. [Google Scholar] [CrossRef]
- Fischer, R.; Zekert, N.; Takeshita, N.N. Polarized growth in fungi—Interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 2008, 68, 813–826. [Google Scholar] [CrossRef]
- Schunck, T.; Herrero, S.; Fischer, R. The Aspergillus nidulans CENP_E kinesin KipA is able to dimerize and to move processively along microtubules. Curr. Genet. 2011, 57, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Manck, R.; Ishitsuka, Y.; Herrero, S.; Takeshita, N.; Nienhaus, G.U.; Fischer, R. Genetic evidence for a microtubule-capture mechanism during polarised growth of Aspergillus nidulans. J. Cell Sci. 2015, 128, 3569–3582. [Google Scholar] [CrossRef] [PubMed]
- Messin, L.J.; Millar, J.B.A. Role and regulation of kinesin-8 motors through the cell cycle. Syst. Syn. Biol. 2014, 8, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Qiu, W.; Gupta, M.L., Jr.; Pereira-Leal, J.B.; Reck-Peterson, S.L.; Pellman, D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell 2011, 43, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Weaver, L.N.; EmsMcClung, S.C.; Stout, J.R.; LeBlanc, C.; Shaw, S.L.; Gardner, M.K.; Walczak, C.E. Kif18A Uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr. Biol. 2011, 21, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Tokai, N.; Fujimoto-Nishiyama, A.; Toyoshima, Y.; Yonemura, S.; Tsukita, S.; Inoue, J.; Yamamota, T. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 1996, 15, 457–467. [Google Scholar] [CrossRef]
- Afshar, K.; Scholey, J.; Hawley, R.S. Identification of the chromosome localization domain of the Drosophila nod kinesin-like protein. J. Cell Biol. 1995, 131, 833–843. [Google Scholar] [CrossRef]
- Cane, S.; Ye, A.A.; Luks-Morgan, S.J.; Maresca, T.J. Elevated polar ejection forces stabilize kinetochore-microtubule attachments. J. Cell Biol. 2013, 200, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Tokai-Nishizumi, N.; Ohsugi, M.; Suzuki, E.; Yamamoto, T. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol. Biol. Cell 2005, 16, 5455–5463. [Google Scholar] [CrossRef]
- Cross, R.A.; McAinsh, A. Prime movers: The mechanochemistry of mitotic kinesins. Nat. Rev. Mol. Cell Biol. 2014, 15, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Kurgan, L.A. DRNApred, fast sequence-based method that accurately predicts and discriminates DNA- and RNA-binding residues. Nucleic Acids Res. 2017, 45, e84. [Google Scholar] [CrossRef]
- Haag, C.; Steuten, B.; Feldbrϋgge, M. Membrane-coupled mRNA trafficking in fungi. Ann. Rev. Microbiol. 2015, 69, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Girbardt, M. Ultrastructure of the fungal nucleus I. Z. Allg. Mikrobiol. 1970, 10, 451–468. [Google Scholar] [CrossRef]
- Xiang, X. Nuclear movement in fungi. Semin. Cell Dev. Biol. 2018, 82, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Girbardt, M. Ultrastructure and dynamics of the moving nucleus. Symp. Soc. Exp. Biol. 1968, 22, 249–259. [Google Scholar] [PubMed]
- Snider, P.I.; Raper, J.R. Nuclear migration in the basidiomycete Schizophyllum commune. Am. J. Bot. 1958, 45, 538–546. [Google Scholar] [CrossRef]
- Niederpruem, D.J. Direct studies of dikaryotization in Schizophyllum commune. I. Live inter-cellular nuclear migration patterns. Arch. Microbiol. 1980, 128, 172–178. [Google Scholar] [CrossRef]
- Makino, R.; Kamada, T. Isolation and characterization of mutations that affect nuclear migration for dikaryosis in Coprinus cinereus. Curr. Genet. 2004, 45, 149–156. [Google Scholar] [CrossRef]
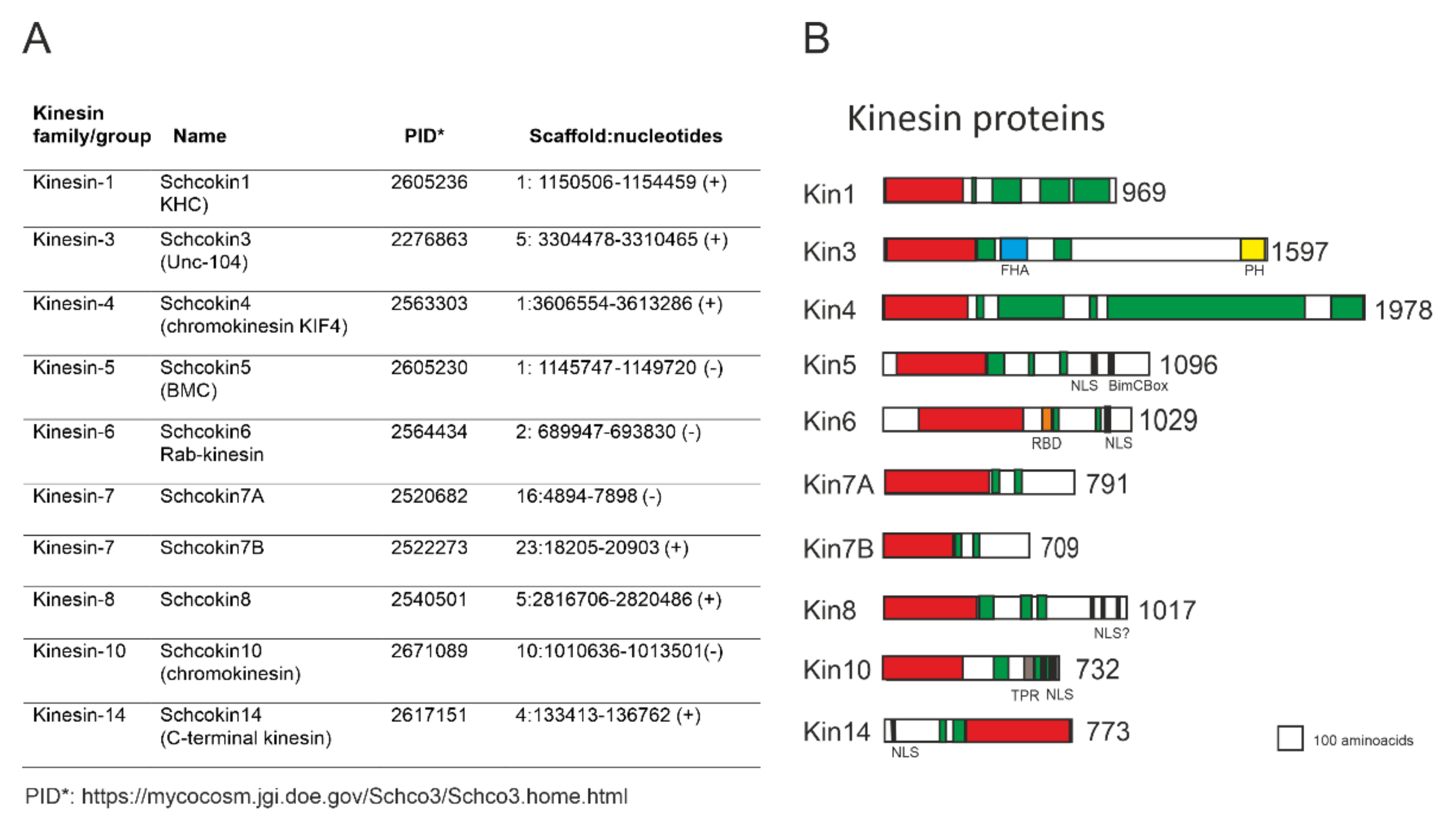
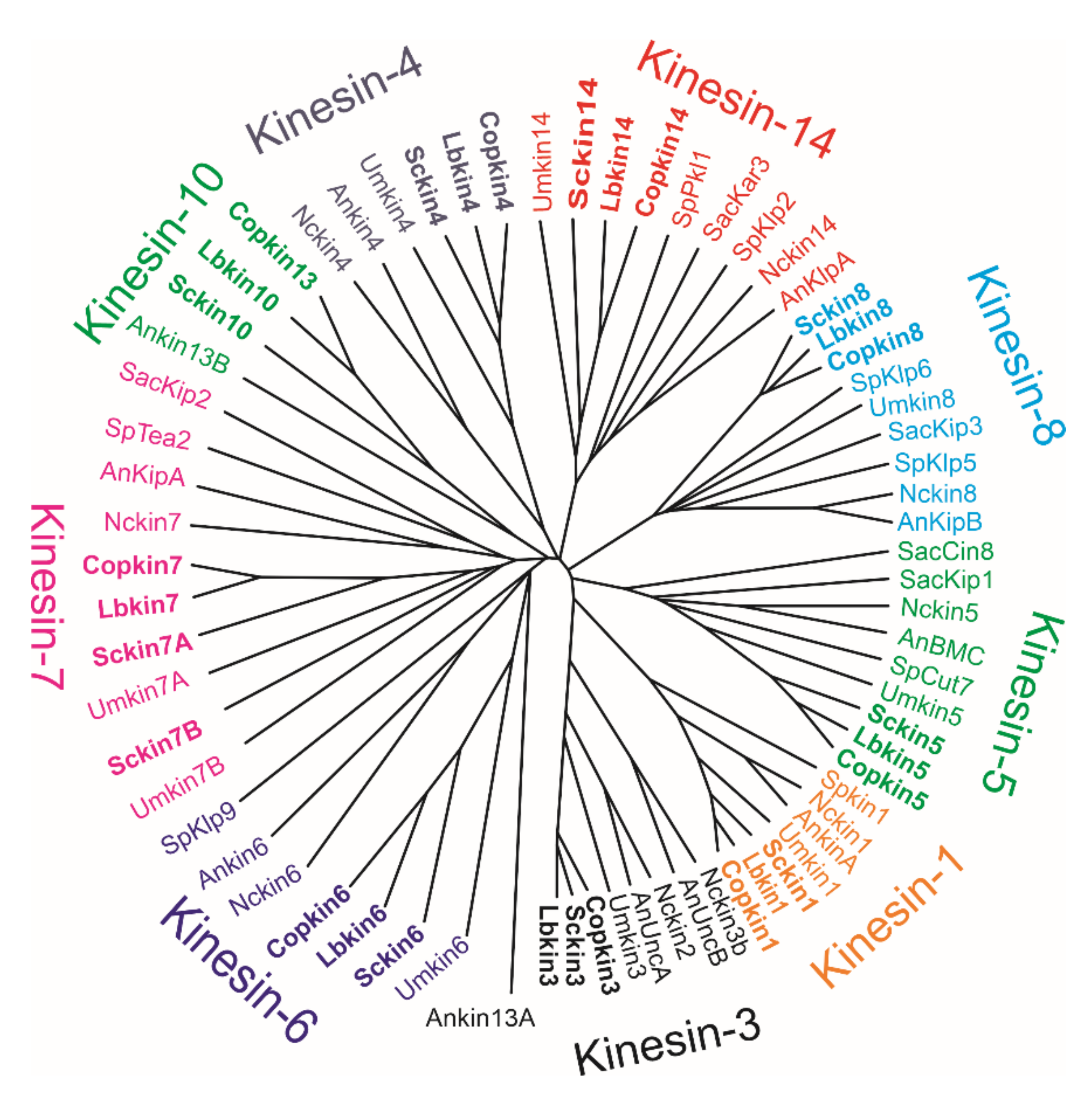
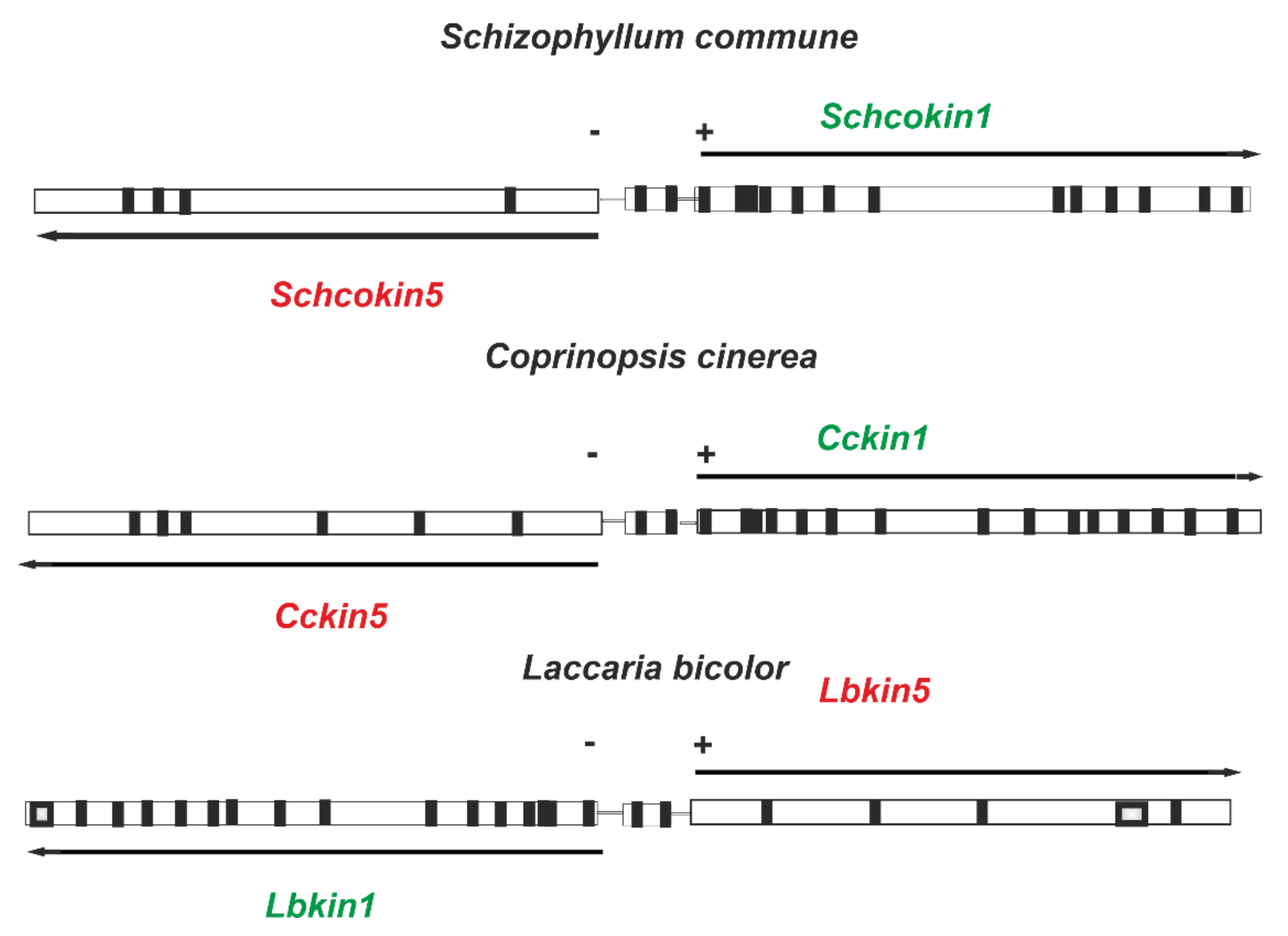
| GeneBank Accession | References | GeneBank Assecion Number | References |
|---|---|---|---|
| SacCin8 NP_010853 | [16,18,42,43,44] | Copkin1 XP_001833251.1 | |
| SacKip1 NP_009490.1 | [16,18,42,45] | Copkin3 XP_001841223.1 | |
| Sac Kip2 NP_015170.1 | [16,18,46,47,48] | Copkin4 XP_001833035.1 | |
| SacKip3 NP_011299.1 | [16,18,49] | Copkin5 XP_001833249.1 | |
| Sack Kar3 NP_015467.1 | [16,18,44,50] | Copkin6 XP_001828624.1 | |
| Copkin7 XP_001835410.1 | |||
| SpKlp3 CAB75775.1 | [17,18] | Copkin8 XP_001836899.1 | |
| SpCut7 CAA94636.1 | [17,18,51,52] | Copkin13 XP_001832695.1 | |
| Spkin6 Klp9 CAA21179.2 | [17,18,53] | Copkin14 XP_001831857.1 | |
| SpkinTea2 CAA22353.1 | [17,18,54] | ||
| SpkinKlp5 CAB10160.1 | [17,18,55,56] | Lbkin1 XP_001875399.1 | |
| SpkinKlp6 CAA20063.2 | [17,18,55] | Lbkin3 XP_001884395.1 | |
| SpkinKlp2 CAB65811.1 | [17,18,57] | Lbkin4 XP_001875444.1 | |
| SpkinPkl1 CAB16597.1 | [17,18,57] | Lbkin5 XP_001874862.1 | |
| Lbkin6 XP_001873613.1 | |||
| AnkinA XP_662947.1 | [18,19,58,59,60] | Lbkin7 XP_001875606.1 | |
| AnUncA XP_680816.1 | [19,61] | Lbkin8 XP_001874733.1 | |
| AnUncB XP_664467.1 | [19,61] | LbkinTPR XP_001881344.1 | |
| Ankin4 XP_664479.1 | [19] | Lbkin14 PID985042 | |
| AnBMC XP_660967.1 | [19,62,63,64] | ||
| Ankin6 XP_660728.1 | Schcokin1 ACG58879.1 | ||
| AnKipA XP_681555.1 | [19,65,66,67] | Schcokin3 XP_003036969.1 | |
| AnKipB XP_662117.1 | [19,68] | Schcokin4 EU860363 | |
| Ankin13A XP_661574.1 | Schcokin5 EU850808.1 | [14] | |
| Ankin13B XP_661325.1 | [19] | Schcokin6 PID2564434 | |
| AnKlpA XP_663944.1 | [18,19,69,70] | Schcokin7A XP_003026378.1 | |
| Schcokin7B XP_003025887.1 | |||
| Nckin1 KHC XP_964432.2 | [18,19,50,51,52,53,54,55,71,72,73,74,75,76,77,78,79] | Schcokin8 XP_003036868.1 | |
| Nckin2 XP_960661.2 | [19,80] | Schcokin10 XP_003028679.1 | |
| Nckin3b XP_961491.1 | [19] | Schcokin14 XP_003032452.1 | [14] |
| Nckin4 XP_963673.1 | [19] | ||
| Nckin5 XP_964753.1 | [19] | Umkin1 XP_760365.1 | [18,20,74,81] |
| Nckin6 XP_961843.1 | [19] | Umkin3 XP_762398.1 | [20,82,83,84,85,86,87,88,89,90,91] |
| Nckin7 XP_964051.1 | [19] | Umkin4 XP_759304.1 | [18,20,74] |
| Nckin8 XP_960006.2 | [19] | Umkin5 XP_760872.1 | [20,92] |
| Nckin14 XP_958282.1 | [19] | Umkin6 XP_760874.1 | [20] |
| Umkin7A XP_760671.1 | [20] | ||
| Umkin7B XP_757043.1 | [20] | ||
| Umkin8 XP_757707.1 | [20] | ||
| Umkin14 XP_760654.1 | [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raudaskoski, M. Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome. J. Fungi 2022, 8, 294. https://doi.org/10.3390/jof8030294
Raudaskoski M. Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome. Journal of Fungi. 2022; 8(3):294. https://doi.org/10.3390/jof8030294
Chicago/Turabian StyleRaudaskoski, Marjatta. 2022. "Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome" Journal of Fungi 8, no. 3: 294. https://doi.org/10.3390/jof8030294
APA StyleRaudaskoski, M. (2022). Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome. Journal of Fungi, 8(3), 294. https://doi.org/10.3390/jof8030294





