AoSsk1, a Response Regulator Required for Mycelial Growth and Development, Stress Responses, Trap Formation, and the Secondary Metabolism in Arthrobotrys oligospora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Sequence Analysis of AoSsk1
2.3. Disruption of Aossk1 and Its Verification
2.4. Analysis of Mycelial Growth, Colony Morphology, Conidiation, and Cell Nucleus
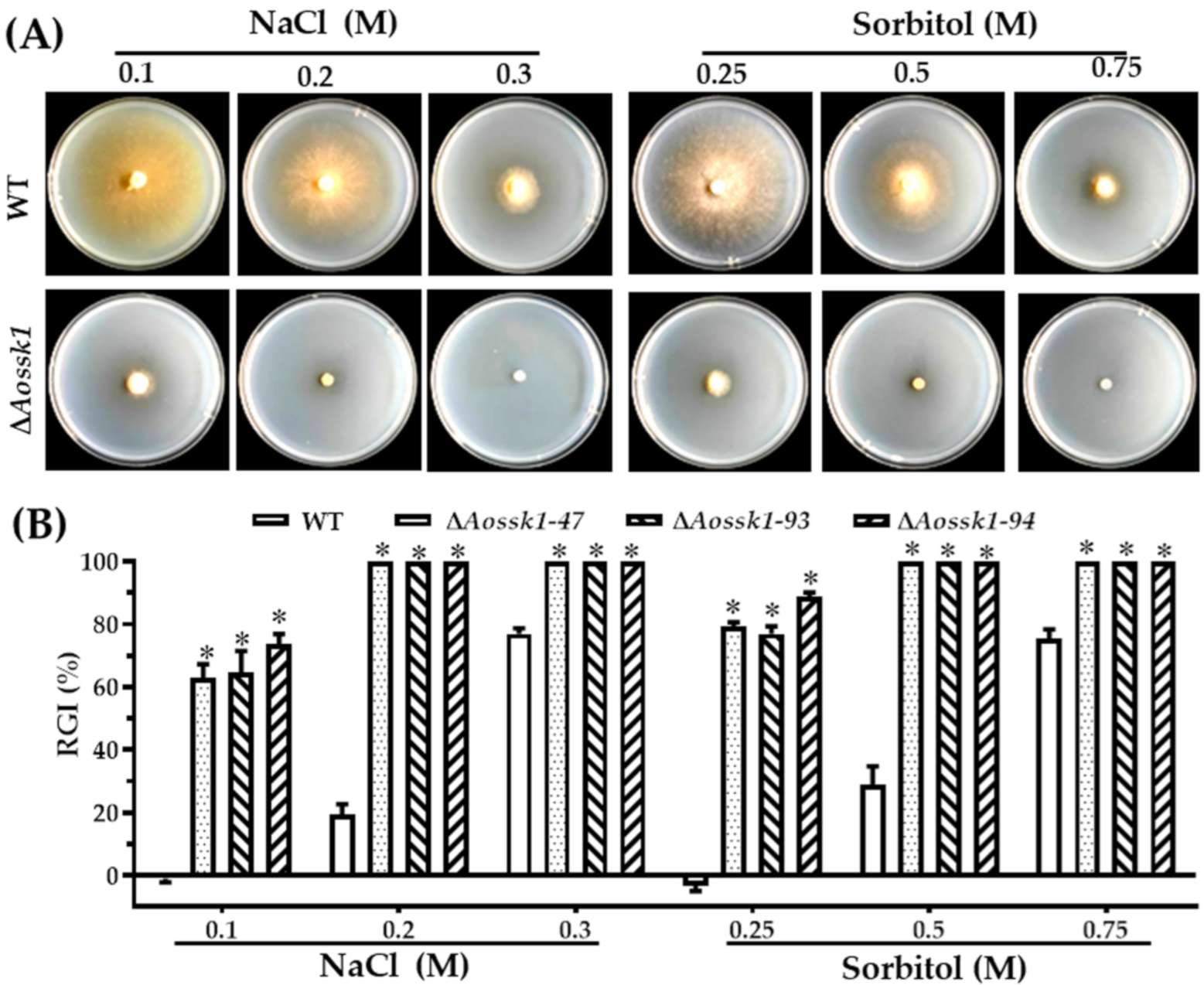
2.5. Comparison of Stress Resistance
2.6. Analysis of Autophagy, Lipid Droplets (LDs), and Endocytosis
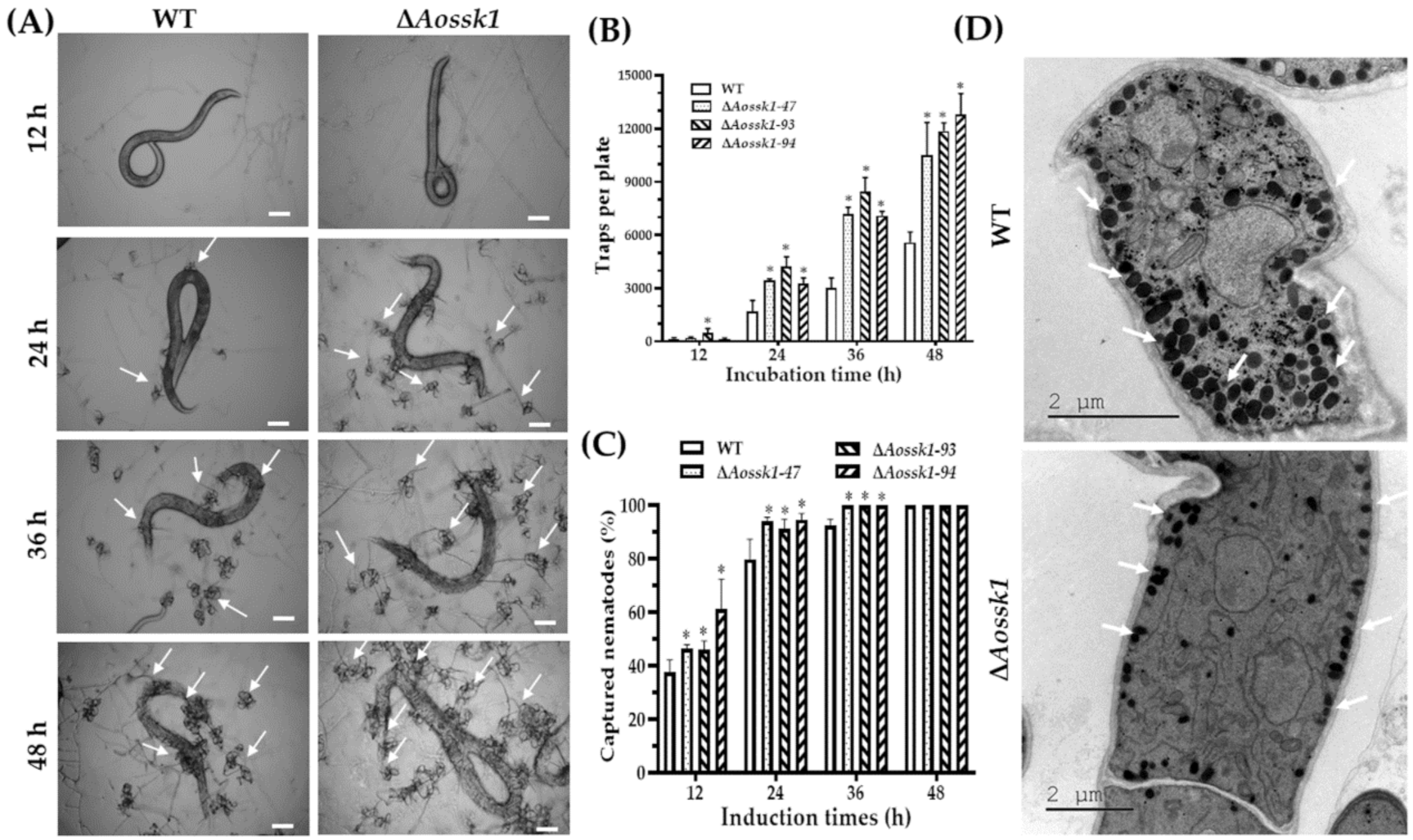
2.7. Analysis of Trap Formation and Nematode Predation
2.8. Reverse Transcription Quantitative PCR (RT-qPCR)
2.9. Metabolomics Analysis of Aossk1
2.10. Statistical Analysis
3. Results
3.1. Analysis of the AoSsk1 Sequence
3.2. Verification of Positive Transformants
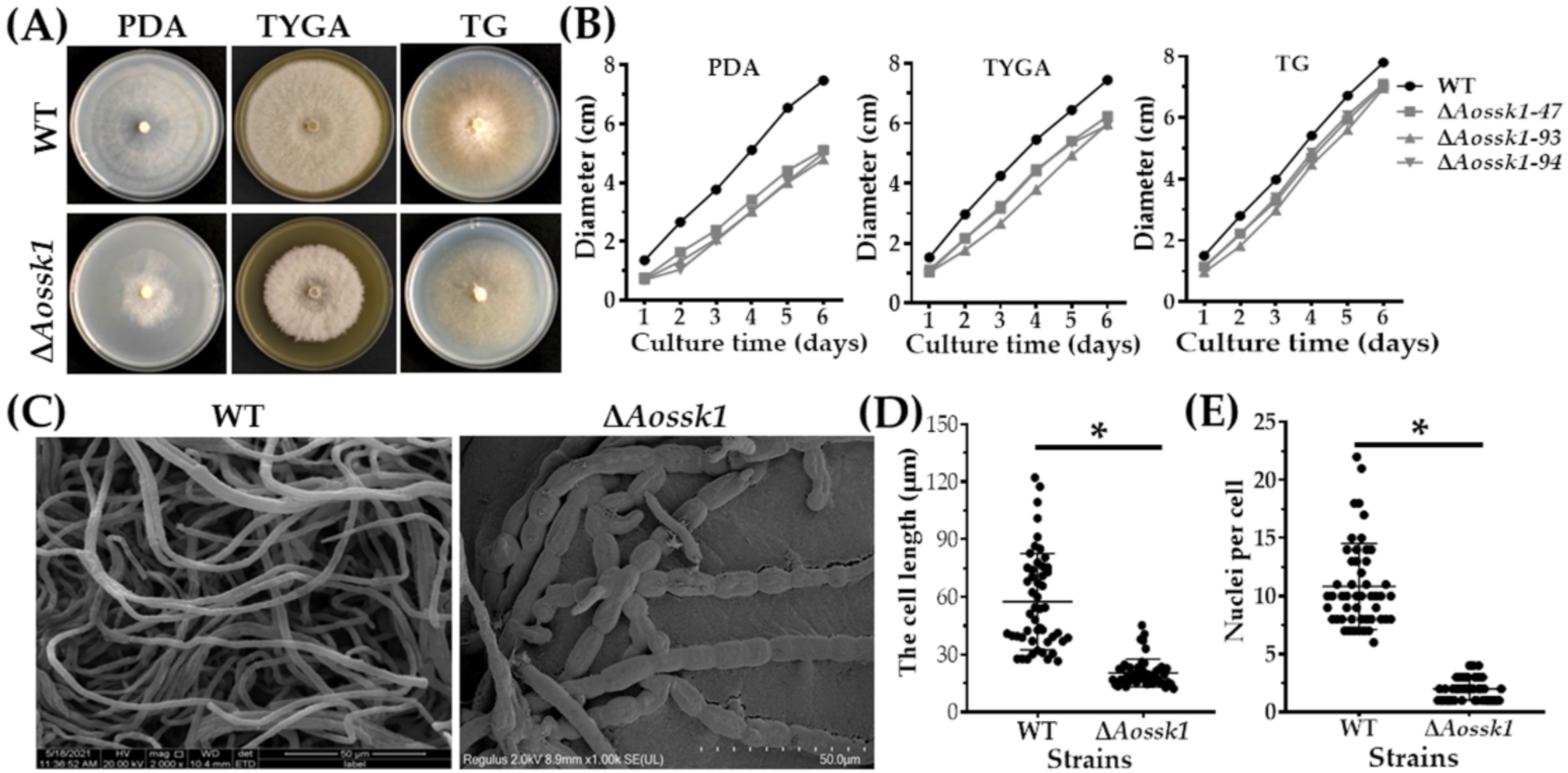
3.3. Aossk1 Plays a Vital Role in Mycelial Growth, Morphology, and Hyphal Cell Nuclei
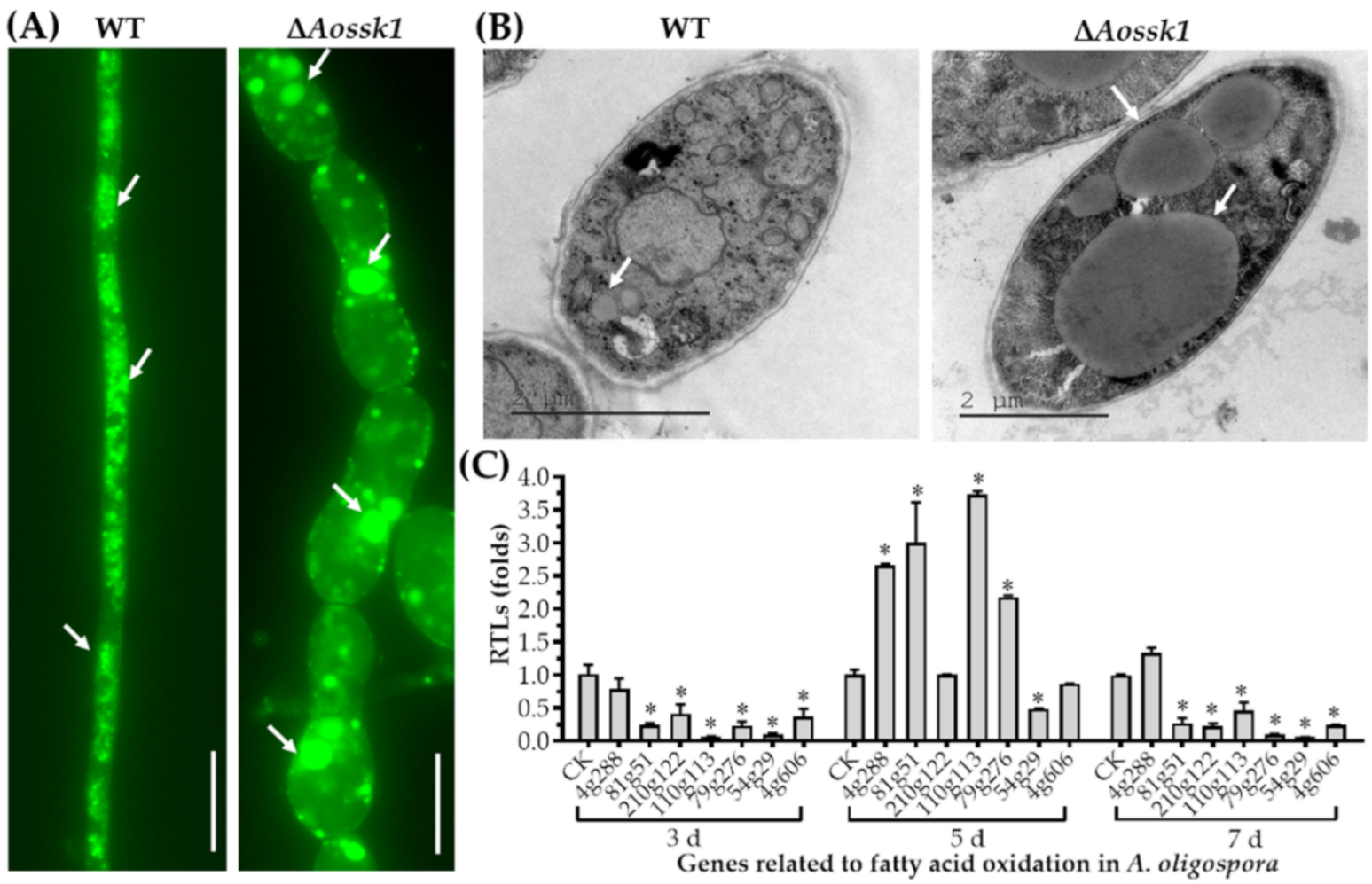
3.4. AoSsk1 Regulates LD Accumulation
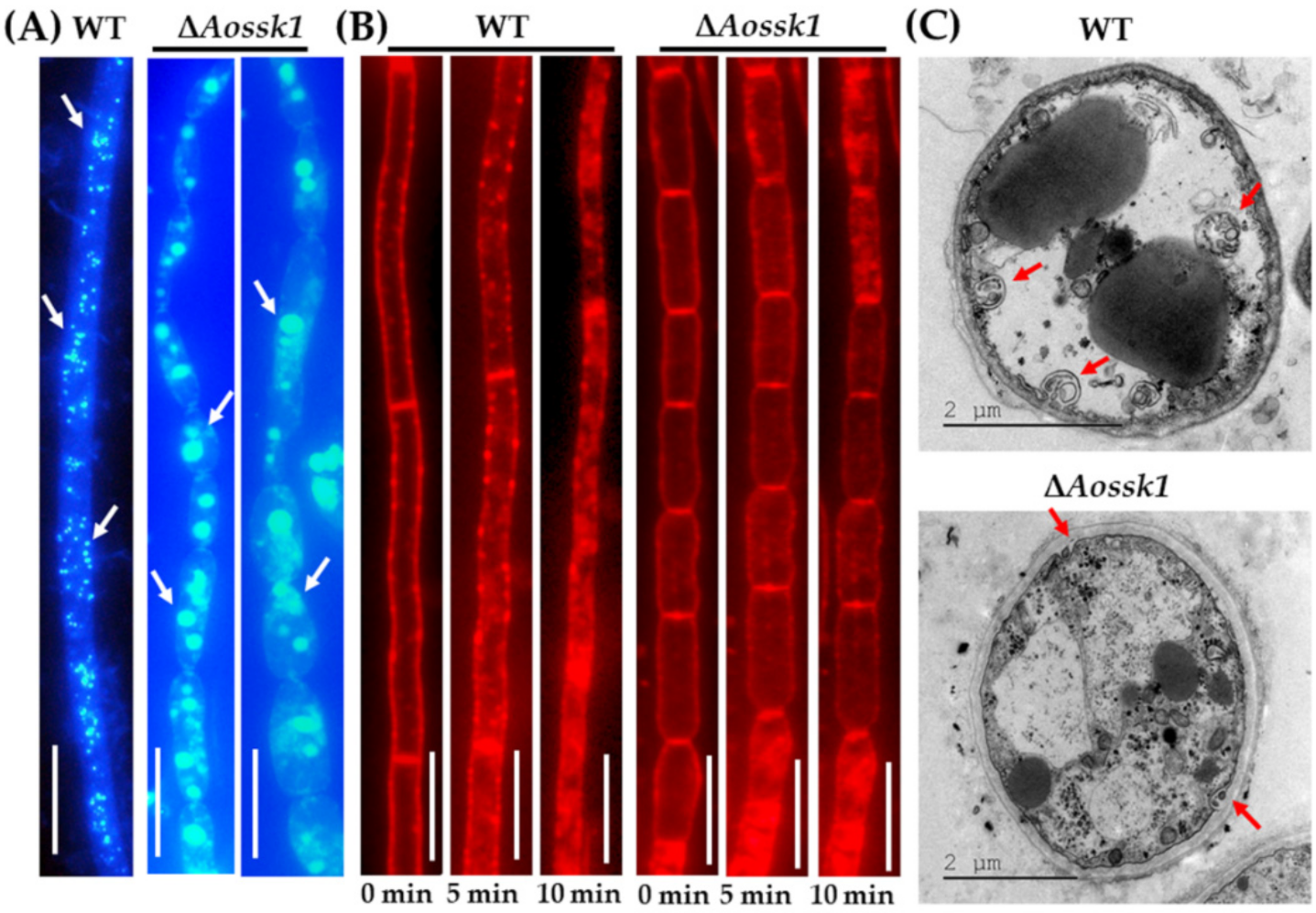
3.5. AoSsk1 Regulates Autophagy and Endocytosis
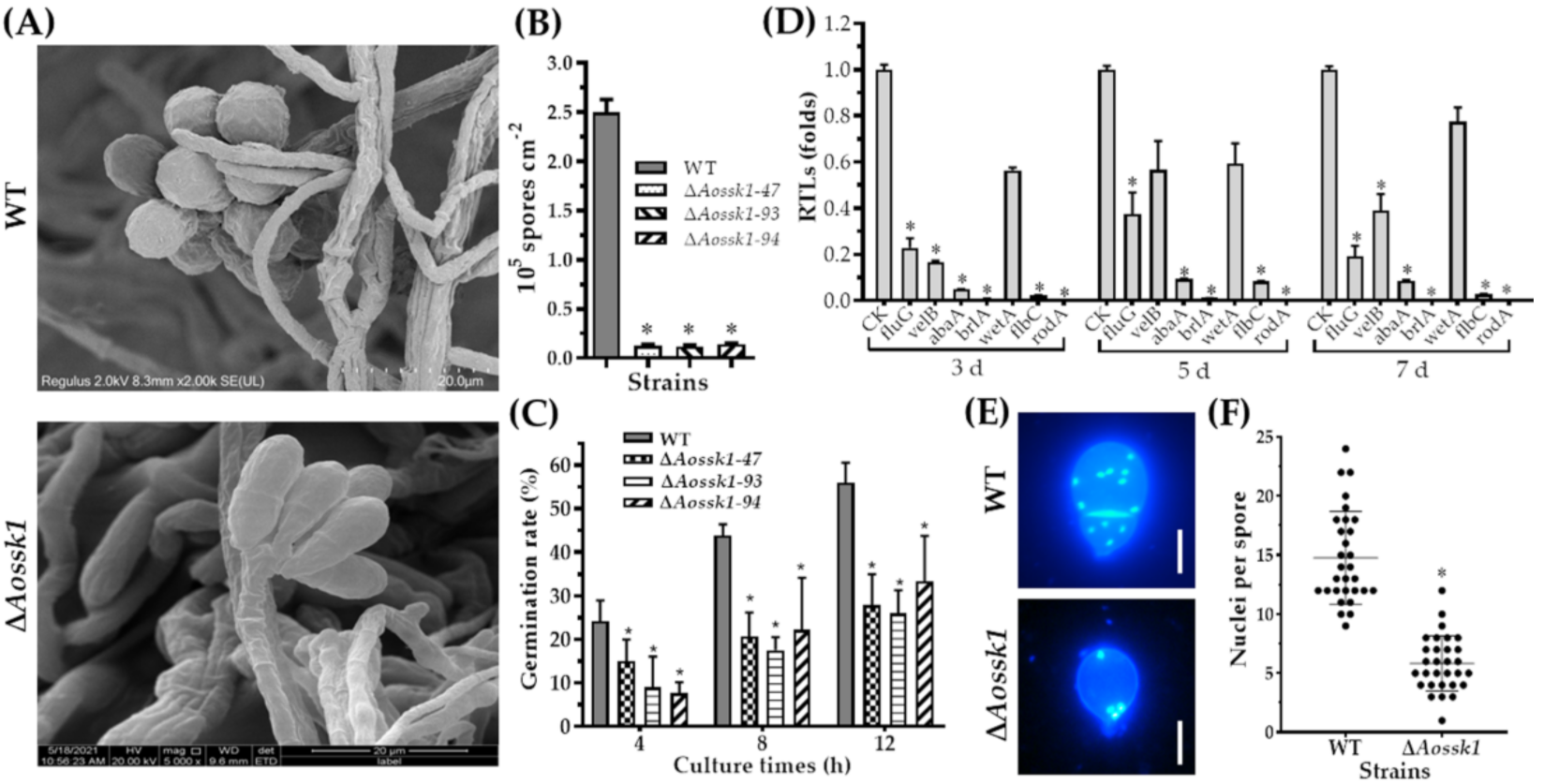
3.6. Aossk1 Plays a Crucial Role in Sporulation, Spore Germination, and Cell Nuclei
3.7. AoSsk1 Regulates Multiple Stress Responses
3.8. Aossk1 Regulates Trap Formation, Pathogenicity, and EDs
3.9. AoSsk1 Regulates the Secondary Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seger, R.; Krebs, E.G. The MAPK signaling cascade. FASEB J. 1995, 9, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, J.; Smith, D.A.; da Silva Dantas, A.; Doris, K.S.; Patterson, M.J.; Bruce, C.R.; Quinn, J. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 2007, 18, 4603–4614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamel, L.P.; Nicole, M.C.; Duplessis, S.; Ellis, B.E. Mitogen-activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. Plant. Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Rubio, G.; Fernández-Acero, T.; Martin, H.; Molina, M. Mitogen-activated protein kinase phosphatases (MKPs) in fungal signaling: Conservation, function, and regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.M.; Feng, M.G. Insights into regulatory roles of MAPK-cascaded pathways in multiple stress responses and life cycles of insect and nematode mycopathogens. Appl. Microbiol. Biotechnol. 2019, 103, 577–587. [Google Scholar] [CrossRef]
- Saito, H.; Tatebayashi, K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 2004, 136, 267–272. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Z.K.; Sun, H.H.; Ying, S.H.; Feng, M.G. Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2017, 19, 1808–1821. [Google Scholar] [CrossRef]
- Janiak-Spens, F.; Sparling, J.M.; Gurfinkel, M.; West, A.H. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J. Bacteriol. 1999, 181, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Tatebayashi, K.; Yamada, R.; Saito, H. Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 mitogen-activated protein kinase kinase kinase in the yeast high-osmolarity glycerol osmoregulatory pathway. Mol. Cell Biol. 2008, 28, 5172–5183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calera, J.A.; Zhao, X.J.; Calderone, R. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect. Immun. 2000, 68, 518–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, N.; Inglis, D.; Roman, E.; Pla, J.; Li, D.; Calera, J.A.; Calderone, R. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell 2003, 2, 1018–1024. [Google Scholar] [CrossRef] [Green Version]
- Shivarathri, R.; Jenull, S.; Stoiber, A.; Chauhan, M.; Mazumdar, R.; Singh, A.; Nogueira, F.; Kuchler, K.; Chowdhary, A.; Chauhan, N. The two-component response regulator Ssk1 and the mitogen-activated protein kinase Hog1 control antifungal drug resistance and cell wall architecture of Candida auris. mSphere 2020, 5, e00973-e20. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, F.; Li, C.; Feng, M.G. Bbssk1, a response regulator required for conidiation, multi-stress tolerance, and virulence of Beauveria bassiana. Appl. Microbiol. Biotechnol. 2014, 98, 5607–5618. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.L.; Chen, L.H.; Chung, K.R. How the pathogenic fungus Alternaria alternata copes with stress via the response regulators SSK1 and SHO1. PLoS ONE 2016, 11, e0149153. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Tang, C.; Deng, C.; Wang, Y. Involvement of a response regulator VdSsk1 in stress response, melanin biosynthesis and full virulence in Verticillium dahliae. Front. Microbiol. 2019, 10, 606. [Google Scholar] [CrossRef]
- Jiang, X.; Xiang, M.; Liu, X. Nematode-trapping fungi. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. 2017, 92, 357–368. [Google Scholar] [CrossRef]
- Nordbring-Hertz, B.; Jansson, H.B.; Tunlid, A. Nematophagous fungi. In Els; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar]
- Ji, X.; Li, H.; Zhang, W.; Wang, J.; Liang, L.; Zou, C.; Yu, Z.; Liu, S.; Zhang, K.Q. The lifestyle transition of Arthrobotrys oligospora is mediated by microRNA-like RNAs. Sci. China Life Sci. 2020, 63, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, N.; Zhang, G.; Wang, W.; Feng, H.; Yang, X.; Zheng, Y.; Yang, L.; Xie, M.; Zhang, K.Q.; Yang, J. Ric8 acts as a regulator of G-protein signalling required for nematode-trapping lifecycle of Arthrobotrys oligospora. Environ. Microbiol. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhao, Y.; Wang, Y.; Yang, L.; Li, D.; Yang, J.; Jiang, K.; Zhang, K.Q.; Yang, J. Functional analysis of seven regulators of G protein signaling (RGSs) in the nematode-trapping fungus Arthrobotrys oligospora. Virulence 2021, 12, 1825–1840. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.Y.; Chen, S.A.; Hsueh, Y.P. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J. Fungi 2020, 6, 191. [Google Scholar] [CrossRef]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.Q.; et al. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef]
- Park, G.; Colot, H.V.; Collopy, P.D.; Krystofova, S.; Crew, C.; Ringelberg, C.; Litvinkova, L.; Altamirano, L.; Li, L.; Curilla, S.; et al. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 2011, 722, 179–189. [Google Scholar]
- Yang, X.; Ma, N.; Yang, L.; Zheng, Y.; Zhen, Z.; Li, Q.; Xie, M.; Li, J.; Zhang, K.Q.; Yang, J. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2018, 102, 4601–4613. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Tunlid, A.; Ahman, J.; Oliver, R.P. Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 1999, 173, 111–116. [Google Scholar] [CrossRef]
- Xie, M.; Wang, Y.; Tang, L.; Yang, L.; Zhou, D.; Li, Q.; Niu, X.; Zhang, K.Q.; Yang, J. AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 2019, 21, 4648–4661. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhang, K.Q.; Yang, J. Pleiotropic roles of Ras GTPases in the nematode-trapping fungus Arthrobotrys oligospora identified through multi-omics analyses. iScience 2021, 24, 102820. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhou, D.; Zhao, Y.; Li, D.; Niu, X.; Zhang, K.Q.; Yang, J. Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 2020, 10, 3065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Yang, J.; Jiang, K.; Bai, N.; Zhu, M.; Zhu, Y.; Zhang, K.Q.; Yang, J. AoBck1 and AoMkk1 are necessary to maintain cell wall integrity, vegetative growth, conidiation, stress resistance, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 2021, 12, 649582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zheng, Y.; Ma, Y.; Yang, L.; Xie, M.; Zhou, D.; Niu, X.; Zhang, K.Q.; Yang, J. The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 2019, 10, 1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Zhu, Y.; Bai, N.; Yang, L.; Xie, M.; Yang, J.; Zhu, M.; Zhang, K.Q.; Yang, J. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2021, 65, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, X.; Bai, N.; Yang, X.; Zhang, K.Q.; Yang, J. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Spectr. 2022, 10, e0175921. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, X.; Xie, M.; Zhang, G.; Yang, L.; Bai, N.; Zhao, Y.; Li, D.; Zhang, K.Q.; Yang, J. The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet. Biol. 2020, 138, 103352. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xie, M.; Ma, N.; Bai, N.; Zhu, M.; Zhang, K.Q.; Yang, J. Phospholipase C (AoPLC2) regulates mycelial development, trap morphogenesis, and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. J. Appl. Microbiol. 2022, 132, 2144–2156. [Google Scholar] [CrossRef]
- Lee, Y.M.; Kim, E.; An, J.; Lee, Y.; Choi, E.; Choi, W.; Moon, E.; Kim, W. Dissection of the HOG pathway activated by hydrogen peroxide in Saccharomyces cerevisiae. Environ. Microbiol. 2017, 19, 584–597. [Google Scholar] [CrossRef]
- Ruprich-Robert, G.; Chapeland-Leclerc, F.; Boisnard, S.; Florent, M.; Bories, G.; Papon, N. Contributions of the response regulators Ssk1p and Skn7p in the pseudohyphal development, stress adaptation, and drug sensitivity of the opportunistic yeast Candida lusitaniae. Eukaryot. Cell 2008, 7, 1071–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Xie, M.; Bai, N.; Yang, L.; Zhang, K.Q.; Yang, J. The autophagy-related gene Aolatg4 regulates hyphal growth, sporulation, autophagosome formation, and pathogenicity in Arthrobotrys oligospora. Front. Microbiol. 2020, 11, 592524. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhu, Y.; Bai, N.; Xie, M.; Zhang, K.-Q.; Yang, J. Aolatg1 and Aolatg13 regulate autophagy and play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Cell Infect. Microbiol. 2022, 11, 824407. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef]
- Guo, C.T.; Peng, H.; Tong, S.M.; Ying, S.H.; Feng, M.G. Distinctive role of fluG in the adaptation of Beauveria bassiana to insect-pathogenic lifecycle and environmental stresses. Environ. Microbiol. 2021, 23, 5184–5199. [Google Scholar] [CrossRef]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and AbaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 2019, 4, e00140-19. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Yang, Q.; Jiang, J.; Michailides, T.J.; Ma, Z. Involvement of a putative response regulator Brrg-1 in the regulation of sporulation, sensitivity to fungicides, and osmotic stress in Botrytis cinerea. Appl. Microbiol. Biotechnol. 2011, 90, 215–226. [Google Scholar] [CrossRef]
- Kuo, T.H.; Yang, C.T.; Chang, H.Y.; Hsueh, Y.P.; Hsu, C.C. Nematode-trapping fungi produce diverse metabolites during predator-prey interaction. Metabolites 2020, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.X.; Tan, J.L.; Wei, L.X.; Wang, Y.L.; Zhang, C.P.; Wu, D.K.; Zhu, C.Y.; Zhang, Y.; Zhang, K.Q.; Niu, X.M. Morphology regulatory metabolites from Arthrobotrys oligospora. J. Nat. Prod. 2012, 75, 1419–1423. [Google Scholar] [CrossRef]
- Xu, Z.F.; Wang, B.L.; Sun, H.K.; Yan, N.; Zeng, Z.J.; Zhang, K.Q.; Niu, X.M. High trap formation and low metabolite production by disruption of the polyketide synthase gene involved in the biosynthesis of arthrosporols from nematode-trapping fungus Arthrobotrys oligospora. J. Agric. Food Chem. 2015, 63, 9076–9082. [Google Scholar] [CrossRef]
- Sun, X.; Wang, F.; Lan, N.; Liu, B.; Hu, C.; Xue, W.; Zhang, Z.; Li, S. The Zn(II)2Cys6-type transcription factor ADA-6 regulates conidiation, sexual development, and oxidative stress response in Neurospora crassa. Front. Microbiol. 2019, 10, 750. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, K.-X.; Liu, Q.-Q.; Bai, N.; Zhu, M.-C.; Zhang, K.-Q.; Yang, J.-K. AoSsk1, a Response Regulator Required for Mycelial Growth and Development, Stress Responses, Trap Formation, and the Secondary Metabolism in Arthrobotrys oligospora. J. Fungi 2022, 8, 260. https://doi.org/10.3390/jof8030260
Jiang K-X, Liu Q-Q, Bai N, Zhu M-C, Zhang K-Q, Yang J-K. AoSsk1, a Response Regulator Required for Mycelial Growth and Development, Stress Responses, Trap Formation, and the Secondary Metabolism in Arthrobotrys oligospora. Journal of Fungi. 2022; 8(3):260. https://doi.org/10.3390/jof8030260
Chicago/Turabian StyleJiang, Ke-Xin, Qian-Qian Liu, Na Bai, Mei-Chen Zhu, Ke-Qin Zhang, and Jin-Kui Yang. 2022. "AoSsk1, a Response Regulator Required for Mycelial Growth and Development, Stress Responses, Trap Formation, and the Secondary Metabolism in Arthrobotrys oligospora" Journal of Fungi 8, no. 3: 260. https://doi.org/10.3390/jof8030260
APA StyleJiang, K.-X., Liu, Q.-Q., Bai, N., Zhu, M.-C., Zhang, K.-Q., & Yang, J.-K. (2022). AoSsk1, a Response Regulator Required for Mycelial Growth and Development, Stress Responses, Trap Formation, and the Secondary Metabolism in Arthrobotrys oligospora. Journal of Fungi, 8(3), 260. https://doi.org/10.3390/jof8030260






