New Species of Mallocybe and Pseudosperma from North China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. DNA Extraction, PCR Amplification, Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
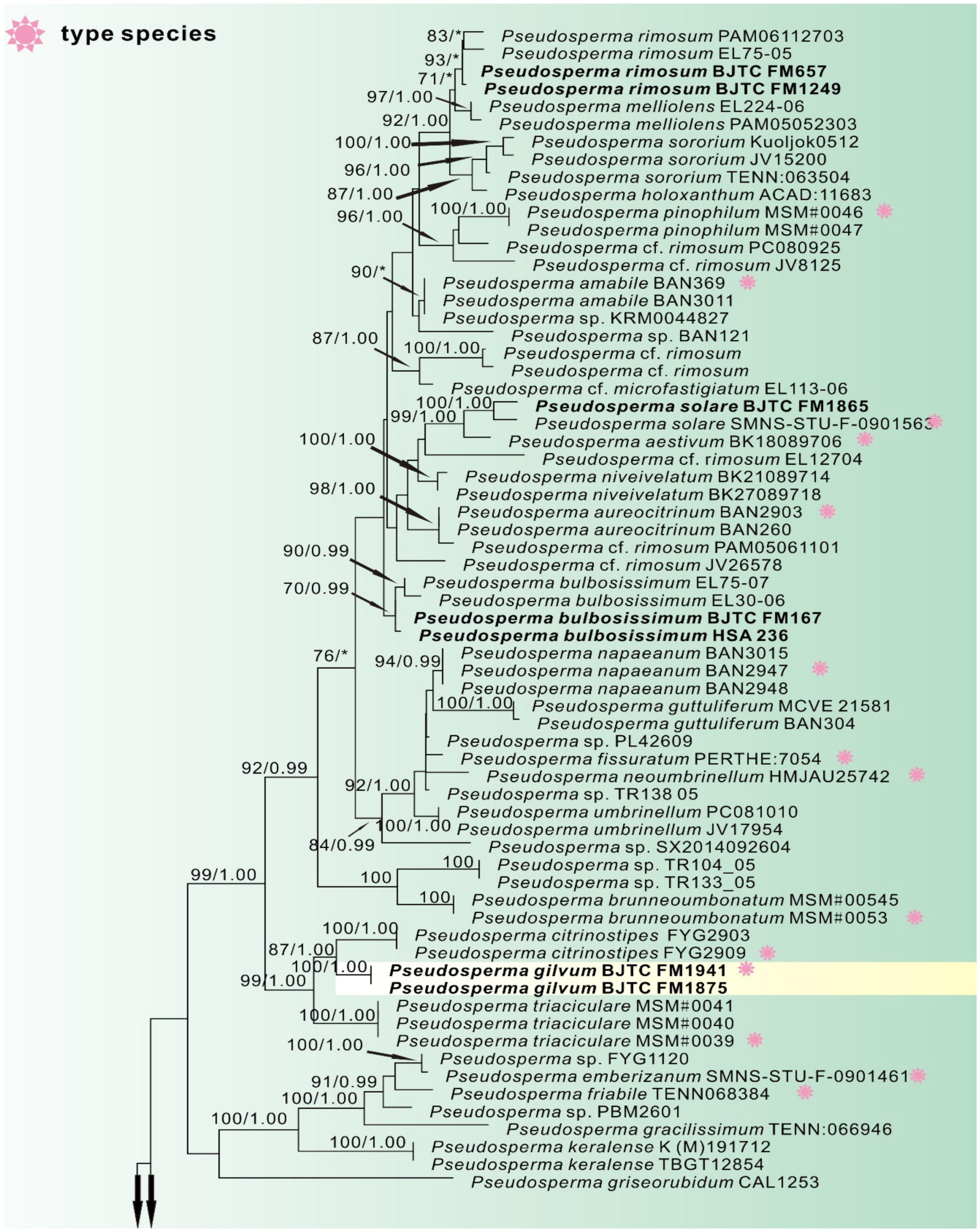
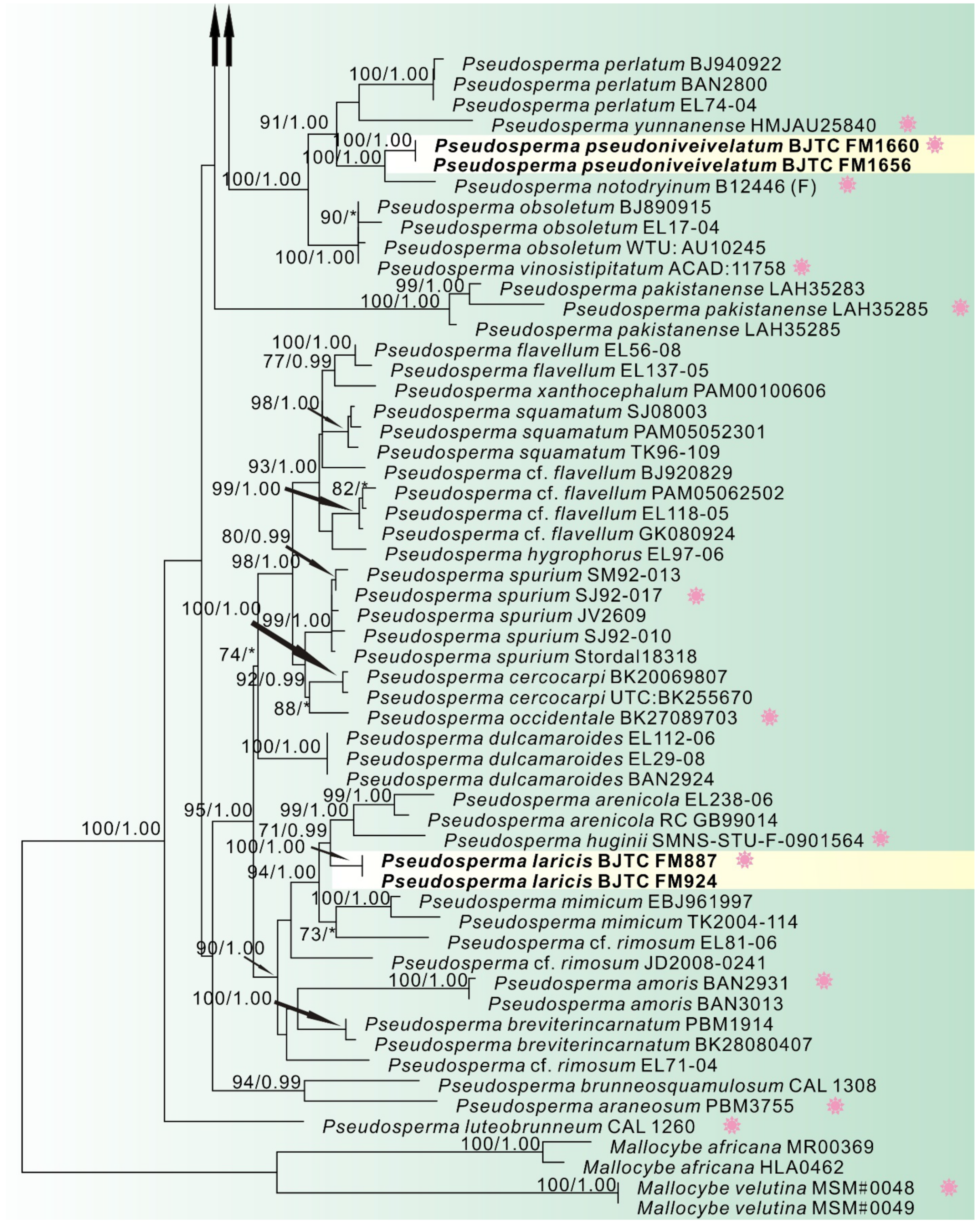
3. Results
3.1. Phylogenetic Analyses
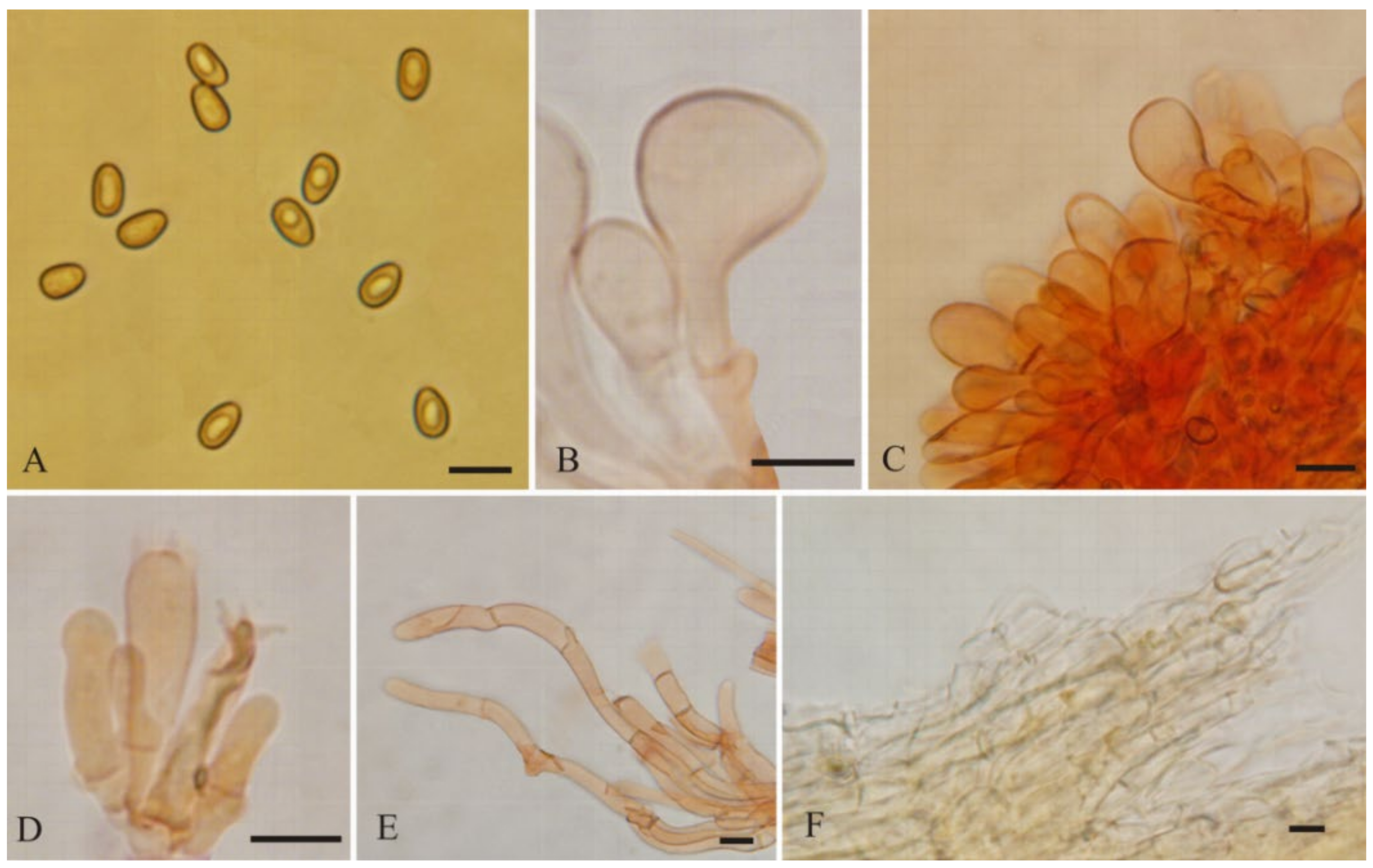
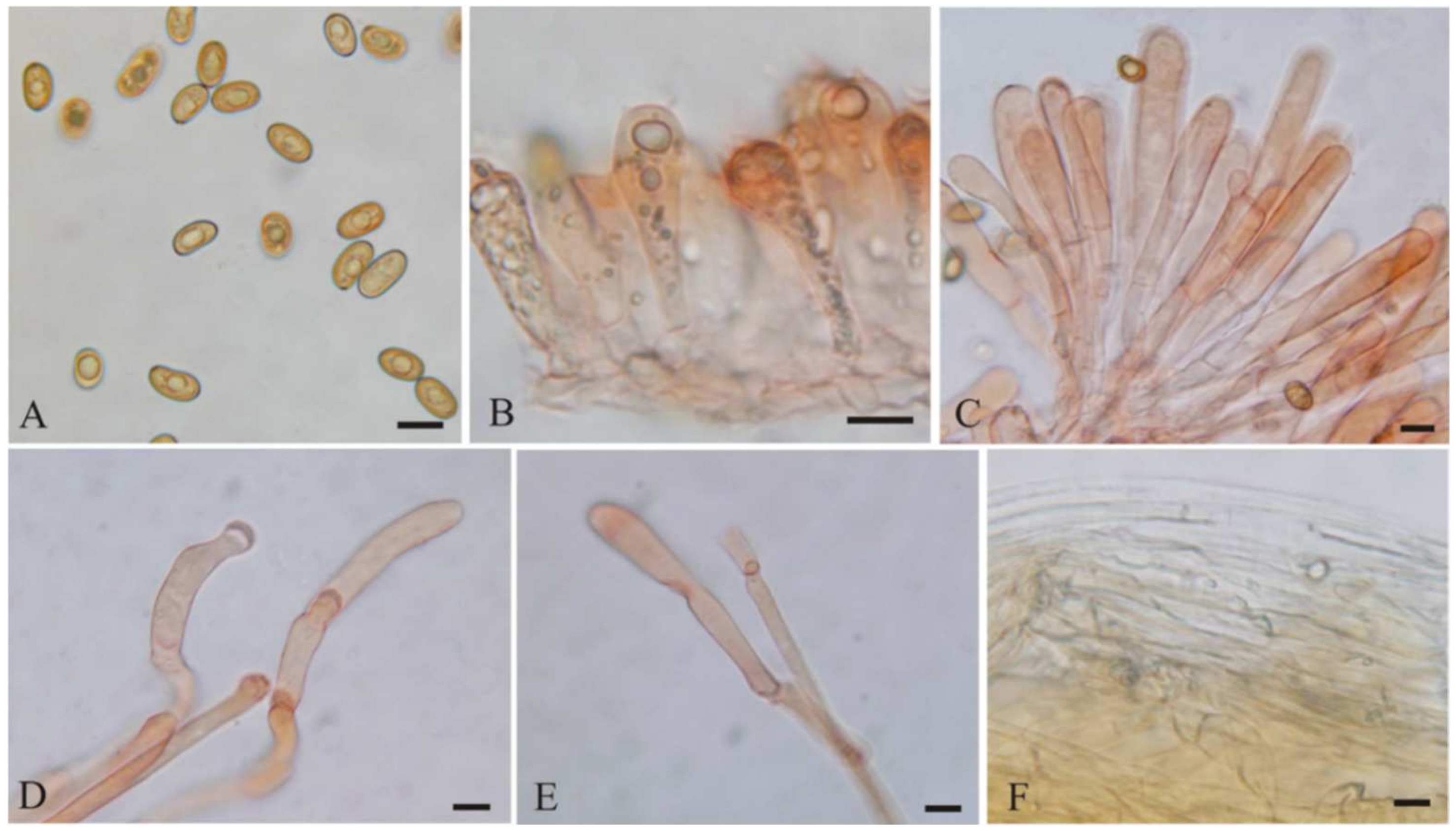
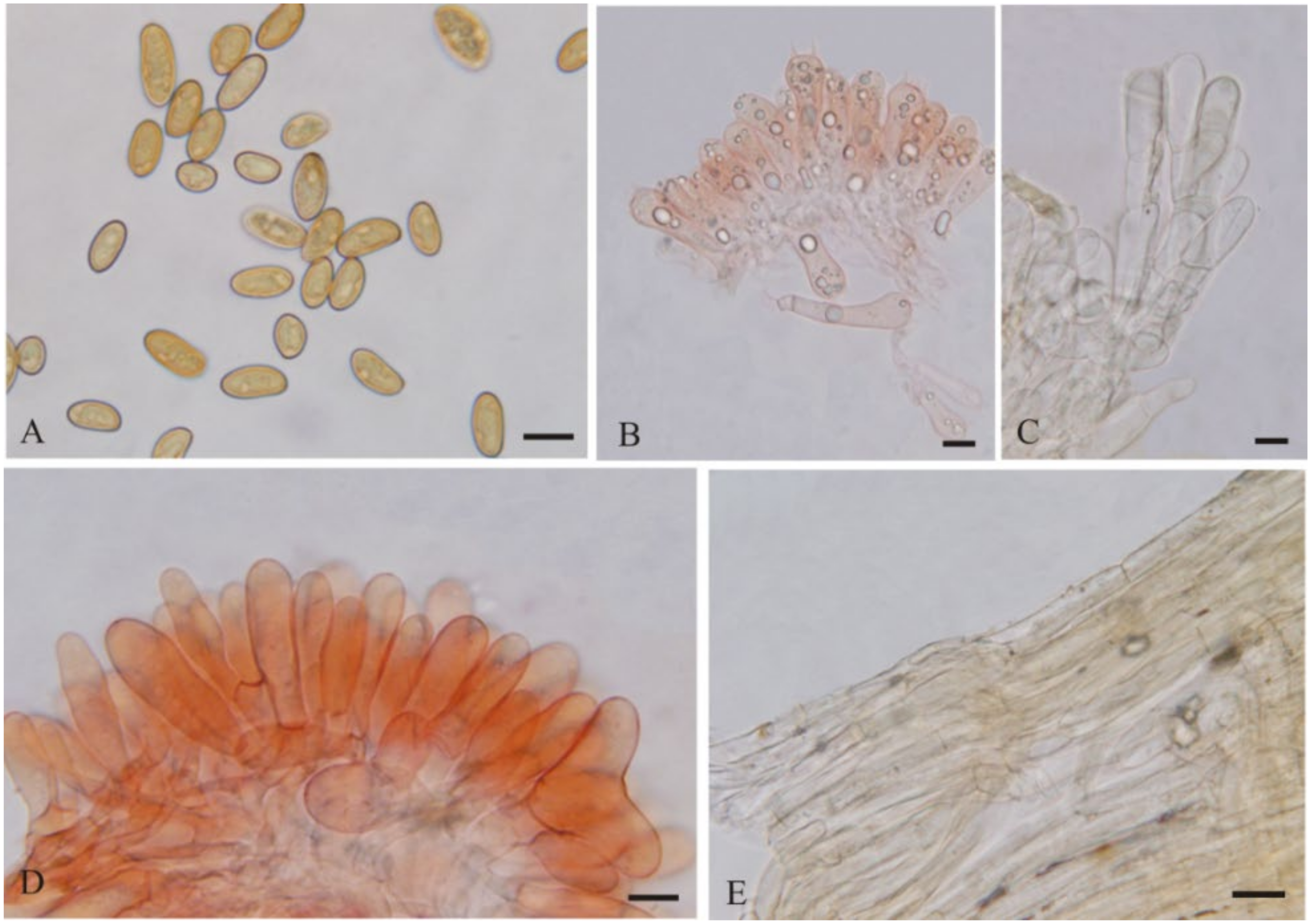
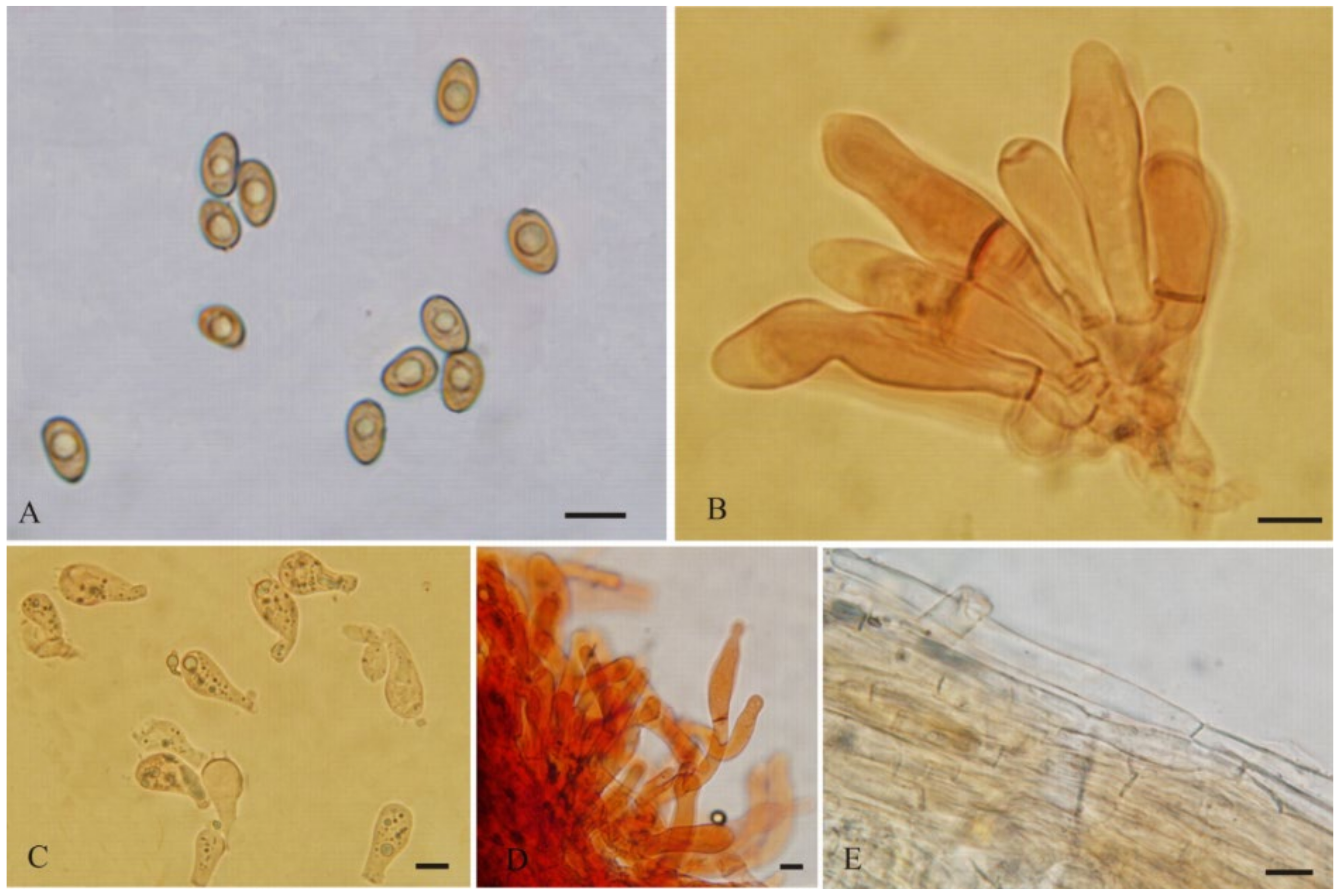
3.2. Taxonomy
4. Discussion
| Key to the species of Mallocybe from China | |
| 1. Annulus present | M. terrigena |
| 1. Annulus absent | 2 |
| 2. Pileus applanate to uplifted, with a shallow depression at the right | M. depressa |
| 2. Pileus plano-convex or applanate, with distinctly umbo or indistinctly umbo | 3 |
| 3. Habitat not associated with Picea | M. heimii |
| 3. Habitat associated with Picea | 4 |
| 4. Basidiospores subamygdaloid to subcylindrical, cylindrical, length > 9 μm | M. picea |
| 4. Basidiospores ellipsoid to subphaseoliform, cylindrical, length < 9 μm | M. leucoloma |
| Key to the species of Pseudosperma from China | |
| 1. Basidiomata uniformly brown (including pileus, lamellae, stipe) | P. neoumbrinellum |
| 1. Basidiomata not uniformly brown | 2 |
| 2. Pileus color with pinkish tinges | 3 |
| 2. Pileus color without pinkish tinges | 4 |
| 3. Pileus yellowish buff, grayish brown to pale pinkish beige, basidiospores 10–15 × 5.5–7.5 μm | P. sororium |
| 3. Pileus gray brown to pinkish gray, basidiospores 9–13 × 5–6 μm | P. obsoletum |
| 4. Pileus surface with a distinct pale white or gray white velipellis | 5 |
| 4. Pileus surface without velipellis | 7 |
| 5. Stipe surface fibrillose with densely squamules | P. yunnanense |
| 5. Stipe surface fibrillose without squamules | 6 |
| 6. Basidiospores mostly subphaseoliform, subcylindrical to cylindrical, Lm × Wm = 11.40 × 6.34 μm | P. gilvum |
| 6. Basidiospores mostly ellipsoid, Lm × Wm = 10.19 × 6.22 μm | P. pseudoniveivelatum |
| 7. Pileus color with yellow tinges | 8 |
| 7. Pileus color without yellow tinges | 9 |
| 8. Habitat associated with Larix | P. laricis |
| 8. Habitat not associated with Larix | 10 |
| 9. Pileus brownish to dark brown | P. perlatum |
| 9. Pileus white or pallid ivory | P. bulbosissimum |
| 10. Basidiospores small, length < 10 μm | P. avellaneum |
| 10. Basidiospores large, length > 10 μm | 11 |
| 11. Cheilocystidia missing (sub)capitate | P. rimosum |
| 11. Cheilocystidia (sub)capitate | 12 |
| 12. Stipe surface with lemon yellow fibrils | P. citrinostipes |
| 12. Stipe surface with whitish to dingy whitish rough fibres or glabrous | P. solare |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matheny, P.B.; Kudzma, L.V. New species of Inocybe (Agaricales) from eastern North America. J. Torrey Bot. Soc. 2019, 146, 213–235. [Google Scholar] [CrossRef]
- Matheny, P.B.; Hobbs, A.M.; Esteve-Raventós, F. Genera of Inocybaceae: New skin for the old ceremony. Mycologia 2019, 112, 83–120. [Google Scholar] [CrossRef] [PubMed]
- Massee, G. A monograph of the genus Inocybe, Karsten. Ann. Bot. 1904, 18, 459–504. [Google Scholar] [CrossRef]
- Heim, R. Le Genre Inocybe, Encylopédie Mycologique 1; Paul Lechevalier & Fils: Paris, France, 1931; 429p. [Google Scholar]
- Kühner, R.; Romagnesi, H. Flore Analytique des Champignos Supérieurs; Masson et Cie: Paris, France, 1953; 556p. [Google Scholar]
- Horak, E. Synopsis generum Agaricalium (Die Gattungstypen der Agaricales). Beiträge Zur Kryptogamenflora Der Schweiz 1967, 13, 1–741. [Google Scholar]
- Kuyper, T.W. A revision of the genus Inocybe in Europe 1. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia 1986, 3, 1–247. [Google Scholar]
- Singer, R. The Agaricales in Modern Taxonomy, 4th ed.; Koeltz Scientific Books: Koenigsten, Germany, 1986; 981p. [Google Scholar]
- Horak, E. Röhrlinge und Blätterpilze in Europa; Spektrum Akademischer Verlag: Heildelberg, Germany, 2005; 575p. [Google Scholar]
- Stangl, J. Die Gattung Inocybe in Bayern. Hoppea 1989, 46, 5–388. [Google Scholar]
- Jacobsson, S. Key to Inocybe. In Funga Nordica: Agaricoid, Boletoid and Cyphelloid Genera; Knudsen, H., Vesterholt, J., Eds.; Nordsvamp: Copenhagen, Denmark, 2008; pp. 868–906. [Google Scholar]
- Cripps, C.L.; Larsson, E.; Horak, E. Subgenus Mallocybe (Inocybe) in the Rocky Mountain alpine zone with molecular reference to European arctic-alpine material. N. Am. Fungi 2010, 5, 97–126. [Google Scholar]
- Saba, M.; Khalid, A.N. Mallocybe velutina (Agaricales, Inocybaceae), a new species from Pakistan. Mycoscience 2020, 61, 348–352. [Google Scholar] [CrossRef]
- Aïgnon, H.; Naseer, A.; Matheny, P.B.; Yorou, N.S.; Ryberg, M. Mallocybe africana (Inocybaceae, Fungi), the first species of Mallocybe described from Africa. Phytotaxa 2021, 478, 49–60. [Google Scholar] [CrossRef]
- Larsson, E.; Ryberg, M.; Moreau, P.A.; Mathiesen, Å.D.; Jacobsson, S. Taxonomy and evolutionary relationships within species of section Rimosae (Inocybe) based on ITS, LSU and mtSSU sequence data. Persoonia 2009, 23, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Saba, M.; Haelewaters, D.; Pfister, D.H.; Khalid, A.N. New species of Pseudosperma (Agaricales, Inocybaceae) from Pakistan revealed by morphology and multi-locus phylogenetic reconstruction. MycoKeys 2020, 69, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Bandini, D.; Oertel, B. Three new species of the genus Pseudosperma (Inocybaceae). Czech Mycol. 2020, 72, 221–250. [Google Scholar] [CrossRef]
- Yu, W.J.; Chang, C.; Qin, L.W.; Zeng, N.K.; Wang, S.X.; Fan, G.Y. Pseudosperma citrinostipes (Inocybaceae), a new species associated with Keteleeria from southwestern China. Phytotaxa 2020, 450, 8–16. [Google Scholar] [CrossRef]
- Dring, D.M. Techniques for microscopic preparation. In Methods in Microbiology; Booth, C., Ed.; Academic Press: New York, NY, USA, 1971; Volume 4. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4239–4246. [Google Scholar] [CrossRef] [Green Version]
- Matheny, P.B.; Wang, Z.; Binder, M.; Curtis, J.M.; Lim, Y.W.; Nilsson, R.H.; Hughes, K.W.; Hofstetter, V.; Ammirati, J.F.; Schoch, C.L.; et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 2007, 43, 430–451. [Google Scholar] [CrossRef]
- Ryberg, M.; Nilsson, R.H.; Kristiansson, E.; Töpel, M.; Jacobsson, S.; Larsson, E. Mining metadata from unidentified ITS sequences in GenBank: A case study in Inocybe (Basidiomycota). BMC Evol. Biol. 2008, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Matheny, P.B. A phylogenetic classification of the Inocybaceae. McIlvainea 2009, 18, 11–21. [Google Scholar]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Ryberg, M.; Larsson, E.; Jacobsson, S. An evolutionary perspective on morphological and ecological characters in the mushroom family Inocybaceae (Agaricomycotina, Fungi). Mol. Phylogenet. Evol. 2010, 55, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Kropp, B.R.; Matheny, P.B.; Hutchison, L.J. Inocybe section Rimosae in Utah: Phylogenteic affinities and new species. Mycologia 2013, 105, 728–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horak, E.; Matheny, P.B.; Desjardin, D.E.; Soytong, K. The genus Inocybe (Inocybaceae, Agaricales, Basidiomycota) in Thailand and Malaysia. Phytotaxa 2015, 230, 201–238. [Google Scholar] [CrossRef]
- Pradeep, C.K.; Vrinda, K.B.; Varghese, S.P.; Korotkin, H.B.; Matheny, P.B. New and noteworthy species of Inocybe (Agaricales) from tropical India. Mycol. Prog. 2016, 15, 24. [Google Scholar] [CrossRef]
- Brugaletta, E.; Consiglio, G.; Marchetti, M. Inocybe siciliana, una nuova specie del Sottogenere Mallocybe. Riv. Micol. 2018, 60, 195–209. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Phylogenet. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J. MrModeltest, Version 2.2; Computer Software; Evolutionary Biology Centre, University of Uppsala: Uppsala, Sweden, 2004.
- Page, R.D. TreeView; Glasgow University: Glasgow, UK, 2001. [Google Scholar]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Phylogenet. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Du, F.; Liu, B.; Li, Z.Y. Investigation Report on Wild Eating Mushrooms and Poisonous Mushrooms in Shanxi (Part 2). J. Shanxi Univ. 1980, 71–78. [Google Scholar] [CrossRef]
- Fan, G.Y.; Tolgor, B. Taxonomy in Inocybe subgen. Mallocybe from China. J. Fungal Res. 2016, 14, 129–141. [Google Scholar]
- Matheny, P.B. Key to Species of Inocybaceae from Eastern North America—v7; University of Tennessee: Knoxville, TN, USA, 2020; pp. 1–23. [Google Scholar]
- Tolgor, B.; Fan, G.Y. Three new species of Inocybe sect. Rimosae from China. Mycosystema 2018, 37, 693–702. [Google Scholar]
- Zheng, G.Y.; Bi, Z.S.; Li, C.; Li, T.H. A preliminary report on the Taxonomy of Inocybe in Guangdong Province. J. Shanxi Univ. 1985, 3, 65–75. [Google Scholar]
- Song, G.; Wang, L.Y.; Yang, W.S. Taxonomy of higher fungi in Helanshan Mountains II. Yinshan Acad. J. 1994, 12, 30–44. [Google Scholar]
- Tolgor, B. List of Agarics and Boletoid Fungi from Eastern Inner Mongolia. J. Fungal Res. 2012, 10, 20–30. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, N.; Xu, Y.-Y.; Zhao, T.-Y.; Lv, J.-C.; Fan, L. New Species of Mallocybe and Pseudosperma from North China. J. Fungi 2022, 8, 256. https://doi.org/10.3390/jof8030256
Mao N, Xu Y-Y, Zhao T-Y, Lv J-C, Fan L. New Species of Mallocybe and Pseudosperma from North China. Journal of Fungi. 2022; 8(3):256. https://doi.org/10.3390/jof8030256
Chicago/Turabian StyleMao, Ning, Yu-Yan Xu, Tao-Yu Zhao, Jing-Chong Lv, and Li Fan. 2022. "New Species of Mallocybe and Pseudosperma from North China" Journal of Fungi 8, no. 3: 256. https://doi.org/10.3390/jof8030256
APA StyleMao, N., Xu, Y.-Y., Zhao, T.-Y., Lv, J.-C., & Fan, L. (2022). New Species of Mallocybe and Pseudosperma from North China. Journal of Fungi, 8(3), 256. https://doi.org/10.3390/jof8030256





