Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Condition
2.2. Genome Sequencing
2.3. Genome Assembling
2.4. Gene Prediction and Annotation
2.5. Collinearity Analysis, Gene Family Construction, and Species Tree Construction
2.6. Protein Extraction and Peptide Digestion
2.7. Proteomic Analysis and Peptide Searching
3. Results and Discussion
3.1. Genome Assembly of S. rugosoannulata
3.2. Gene Prediction and Genome Comparisons
3.3. Annotation of the S. rugosoannulata Genome
3.4. Carbohydrate Active Enzymes (CAZymes)
3.5. Proteomic Analysis of Protein Expression Profiles in Different Parts of the S. rugosoannulata Fruiting Body
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, S.; Feng, X.; Huang, W.; Ibrahim, S.A.; Liu, Y. Effects of drying methods on non-volatile taste components of Stropharia rugoso-annulata mushrooms. LWT 2020, 127, 109428. [Google Scholar] [CrossRef]
- Song, Z.; Jia, L.; Xu, F.; Meng, F.; Deng, P.; Fan, K.; Liu, X. Characteristics of Se-enriched mycelia by Stropharia rugoso-annulata and its antioxidant activities in vivo. Biol. Trace. Elem. Res. 2009, 131, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Lai, P.F.; Shen, H.S.; Li, Y.B.; Zhou, X.H. Effect of spray drying technique on processing of Stropharia rugoso-annulata Farl: Murrill blanching liquid. Adv. J. Food Sci. Technol. 2014, 6, 512–516. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, C.F.; Feng, X.; Cheng, L.; Ibrahim, S.A.; Wang, C.T.; Huang, W. Isolation, characterization and antioxidant of polysaccharides from Stropharia rugosoannulata. Int. J. Biol. Macromol. 2020, 155, 883–889. [Google Scholar] [CrossRef]
- Jiang, L.; Hou, Y.; Ding, X. Structure identification and biological activities of a new polysaccharides from Stropharia rugosoannulata. Lat. Am. J. Pharm. 2020, 39, 1594–1604. [Google Scholar]
- Zhai, X.; Zhao, A.; Geng, L.; Xu, C. Fermentation characteristics and hypoglycemic activity of an exopolysaccharide produced by submerged culture of Stropharia rugosoannulata 2#. Ann. Microbiol. 2013, 63, 1013–1020. [Google Scholar]
- He, P.; Geng, L.; Wang, J.; Xu, C. Production, purfication, molecular characterization and bioactivities of exopolysaccharides produced by the wine cap culinary-medicinal mushroom, Stropharia rugosoannulata 2# (higher basidiomycetes). Int. J. Med. Mushrooms 2012, 14, 365–376. [Google Scholar]
- Zhou, B.; Jia, L.; Meng, F.; Song, Z.; Liu, X.; Deng, P.; Fan, K. Statistical optimization of cultivation conditions for exopolysacchride production and mycelia growth by Stropharia rugosoannulata. Ann. Microbiol. 2010, 60, 89–96. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, Y.; Feng, X.; Ibrahim, S.A.; Huang, W.; Liu, Y. Effects of drying on the structural characteristics and antioxidant activities of polysaccharides from Stropharia rugosoannulata. Int. J. Food. Sci. Technol. 2021, 58, 3622–3631. [Google Scholar] [CrossRef]
- Wu, J.; Suzuki, T.; Choi, J.H.; Yasuda, N.; Noguchi, K.; Hirai, H.; Kawagishi, H. An unusual sterol from the mushroom Stropharia rugosoannulata. Tetrahedron Lett. 2013, 54, 4900–4902. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Kobori, H.; Kawaide, M.; Suzuki, T.; Choi, J.H.; Yasuda, N.; Noguchi, K.; Matsumoto, T.; Hirai, H.; Kawagishi, H. Isolation of bioactive steroids from the Stropharia rugosoannulata mushroom and absolute configuration of strophasterol B. Biosci. Biotechnol. Biochem. 2013, 77, 1779–1781. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Tokuyama, S.; Nagai, K.; Yasuda, N.; Noguchi, K.; Matsumoto, T.; Hirai, H.; Kawagishi, H. Strophasterols A to D with an unprecedented steroid skeleton: From the mushroom Stropharia rugosoannulata. Angew. Chem. 2012, 124, 10978–10980. [Google Scholar] [CrossRef]
- Zhang, W.; Tian, G.; Geng, X.; Zhao, Y.; Ng, T.B.; Zhao, L.; Wang, H. Isolation and characterization of a novel lectin from the edible mushroom Stropharia rugosoannulata. Molecules 2014, 19, 9880. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.X.; Huang, M.X.; Sun, P.; Cheng, S.X.; Zhang, Q.; Dai, H. Steroids, fatty acids and ceramide from the mushroom Stropharia rugosoannulata Farlow apud Murrill. Biochem. Syst. Ecol. 2020, 88, 103963. [Google Scholar] [CrossRef]
- Galletti, G.C. Detection of phenolics in wheat straw treated with white rot fungus Stropharia rugosoannulata by use of HPLC. J. Appl. Anim. Res. 1992, 2, 1–8. [Google Scholar] [CrossRef]
- Li, Y.B.; Lai, P.F.; Chen, J.C.; Shen, H.S.; Wu, L.; Tang, B.S. Physicochemical and Antioxidant Properties of Spray Drying Powders from “Strophari rugoso-annulata” and “Agaricus brunnescens” Blanching Liquid. Adv. Int. J. Food. Sci. Technol. 2015, 9, 372–378. [Google Scholar] [CrossRef]
- Luo, H.; Li, X.; Li, G.; Pan, Y.; Zhang, K. Acanthocytes of Stropharia rugosoannulata function as a nematode-attacking device. Appl. Environ. Microbiol. 2006, 72, 2982–2987. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Li, C.; Ni, S.; Zhang, H.; Dong, C. Ultrastructure and development of acanthocytes, specialized cells in Stropharia rugosoannulata, revealed by scanning electron microscopy (SEM) and cryo-SEM. Mycologia 2021, 113, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Zhang, H.; Meng, S.; Liu, Y. Chemotaxis of Caenorhabditis elegans toward volatile organic compounds from Stropharia rugosoannulata induced by amino acids. J. Nematol. 2018, 50, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Pozdnyakova, N.; Schlosser, D.; Dubrovskaya, E.; Balandina, S.; Sigida, E.; Grinev, V.; Turkovskaya, O. The degradative activity and adaptation potential of the litter-decomposing fungus Stropharia rugosoannulata. World J. Microbiol. Biotechnol. 2018, 34, 133. [Google Scholar] [CrossRef] [PubMed]
- Castellet-Rovira, F.; Lucas, D.; Villagrasa, M.; Rodríguez-Mozaz, S.; Barceló, D.; Sarrà, M. Stropharia rugosoannulata and Gymnopilus luteofolius: Promising fungal species for pharmaceutical biodegradation in contaminated water. J. Environ. Manag. 2018, 207, 396–404. [Google Scholar] [CrossRef]
- Xiao, K.; Liu, H.; Dong, S.; Fan, X.; Chen, Y.; Xu, H. Interfacial effect of Stropharia rugoso-annulata in liquid medium: Interaction of exudates and nickel-quintozene. RSC Adv. 2016, 6, 86068–86081. [Google Scholar] [CrossRef]
- Yan, P.S.; Jiang, J.H.; Li, G.F.; Deng, C.L. Mating system and DNA polymorphism of monokaryons with different mating type of Stropharia rugoso-annulata. World J. Microbiol. Biotechnol. 2003, 19, 737–740. [Google Scholar] [CrossRef]
- Yan, P.S.; Jiang, J.H. Preliminary research of the RAPD molecular marker-assisted breeding of the edible basidiomycete Stropharia rugoso-annulata. World J. Microbiol. Biotechnol. 2005, 21, 559–563. [Google Scholar] [CrossRef]
- Yan, P.S.; Jiang, J.H.; Cui, W.S. Characterization of protoplasts prepared from the edible fungus, Stropharia rugoso-annulata. World J. Microbiol. Biotechnol. 2004, 20, 173–177. [Google Scholar] [CrossRef]
- Xie, Y.; Zhong, Y.; Chang, J.; Kwan, H.S. Chromosome-level de novo assembly of Coprinopsis cinerea A43mut B43mut pab1-1# 326 and genetic variant identification of mutants using Nanopore MinION sequencing. Fungal Genet. Biol. 2021, 146, 103485. [Google Scholar] [PubMed]
- Chen, J.; Li, J.M.; Tang, Y.J.; Ma, K.; Li, B.; Zeng, X.; Liu, X.B.; Li, Y.; Yang, Z.L.; Xu, W.N.; et al. Genome-wide analysis and prediction of genes involved in the biosynthesis of polysaccharides and bioactive secondary metabolites in high-temperature-tolerant wild Flammulina filiformis. BMC Genom. 2020, 21, 719. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, C.S.; Kong, W.S. Genomic Insights into the Fungal Lignocellulolytic Machinery of Flammulina rossica. Microorganisms 2019, 7, 421. [Google Scholar] [CrossRef] [Green Version]
- Fang, M.; Wang, X.; Chen, Y.; Wang, P.; Lu, L.; Lu, J.; Yao, F.; Zhang, Y. Genome sequence analysis of Auricularia heimuer combined with genetic linkage map. J. Fungi 2020, 6, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Wu, F.; Si, J.; Zhao, Y.F.; Dai, Y.C. Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2019, 111, 50–58. [Google Scholar] [CrossRef]
- Bao, D.; Gong, M.; Zheng, H.; Chen, M.; Zhang, L.; Wang, H.; Jiang, J.; Wu, L.; Zhu, Y.; Zhu, G.; et al. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS ONE 2013, 8, e58294. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Lu, D.; Wang, S.; Zhao, Y.; Gao, S.; Han, R.; Yu, J.; Zheng, W.; Geng, J.; Hu, S. Genome assembly and pathway analysis of edible mushroom Agrocybe cylindracea. Genom. Proteom. Bioinform. 2020, 18, 341–351. [Google Scholar] [CrossRef]
- Zhang, J.; Shen, N.; Li, C.; Xiang, X.; Liu, G.; Gui, Y.; Patev, S.; Hibbett, D.S.; Barry, K.; Andreopoulos, W.; et al. Population genomics provides insights into the genetic basis of adaptive evolution in the mushroom-forming fungus Lentinula edodes. J. Adv. Res. 2021, in press. [Google Scholar] [CrossRef]
- Wang, G.; Chen, L.; Tang, W.; Wang, Y.; Zhang, Q.; Wang, H.; Zhou, X.; Wu, H.; Guo, L.; Dou, M.; et al. Identifying a melanogenesis-related candidate gene by a high-quality genome assembly and population diversity analysis in Hypsizygus marmoreus. J. Genet. Genomics 2021, 48, 75–87. [Google Scholar] [CrossRef]
- Xu, L.; Guo, L.; Yu, H. Label-free comparative proteomics analysis revealed heat stress responsive mechanism in Hypsizygus marmoreus. Front. Microbiol. 2021, 11, 3359. [Google Scholar] [CrossRef]
- Chen, Y.; Nie, F.; Xie, S.Q.; Zheng, Y.F.; Dai, Q.; Bray, T.; Wang, Y.X.; Xing, J.F.; Huang, Z.J.; Wang, D.P.; et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021, 12, 60. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- Isaacson, T.; Damasceno, C.M.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Li, H.; Wu, S.; Ma, X.; Chen, W.; Zhang, J.; Duan, S.; Gao, Y.; Kui, L.; Huang, W.; Wu, P.; et al. The genome sequences of 90 mushrooms. Sci. Rep. 2018, 8, 9982. [Google Scholar] [CrossRef] [PubMed]
- Pallister, E.; Gray, C.J.; Flitsch, S.L. Enzyme promiscuity of carbohydrate active enzymes and their applications in biocatalysis. Curr. Opin. Struct. Biol. 2020, 65, 184–192. [Google Scholar] [CrossRef]
- Garron, M.L.; Henrissat, B. The continuing expansion of CAZymes and their families. Curr. Opin. Chem. Biol. 2019, 53, 82–87. [Google Scholar] [CrossRef]
- Zhang, J.; Siika-Aho, M.; Tenkanen, M.; Viikari, L. The role of acetyl xylan esterase in the solubilization of xylan and enzymatic hydrolysis of wheat straw and giant reed. Biotechnol. Biofuels 2011, 4, 60. [Google Scholar] [CrossRef] [Green Version]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. CSH Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [Green Version]
- Carrocci, T.J.; Hoskins, A.A. RNA processing: Fungal spliceosomes break the mold. Curr. Biol. 2021, 31, R1482–R1484. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akinyi, M.V.; Frilander, M.J. At the intersection of major and minor spliceosomes: Crosstalk mechanisms and their impact on gene expression. Front. Genet. 2021, 12, 700744. [Google Scholar] [CrossRef]
- Xiong, F.; Li, S. Spliceosome component JANUS fulfills a role of mediator in transcriptional regulation during Arabidopsis development. Plant Signal. Behav. 2021, 16, 1841974. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Jung, E.S.; Singh, D.; Lee, D.E.; Kim, S.; Lee, Y.W.; Kim, J.G.; Lee, C.H. Spatial (cap & stipe) metabolomic variations affect functional components between brown and white beech mushrooms. Food Res. Int. 2017, 102, 544–552. [Google Scholar]
- Nasiri, F.; Tarzi, B.G.; Bassiri, A.R.; Hoseini, S.E.; Aminafshar, M. Comparative study on the main chemical composition of button mushroom’s (Agaricus bisporus) cap and stipe. J. Food Biosci. Technol. 2013, 3, 41–48. [Google Scholar]
- Strange, R.C.; Jones, P.W.; Fryer, A.A. Glutathione S-transferase: Genetics and role in toxicology. Toxicol. Lett. 2000, 112, 357–363. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, L.; Zhang, Q.; Zhuansun, X.; Li, H.; Chen, X.; Krugman, T.; Sun, Q.; Xie, C. Exogenous sodium diethyldithiocarbamate, a Jasmonic acid biosynthesis inhibitor, induced resistance to powdery mildew in wheat. Plant Direct 2020, 4, e00212. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Z.; Xu, W.; Zeng, J.; Li, L.; Li, S.; Gao, Z. Bacillus pumilus LZP02 promotes rice root growth by improving carbohydrate metabolism and phenylpropanoid biosynthesis. Mol. Plant Microbe. Interact. 2020, 33, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
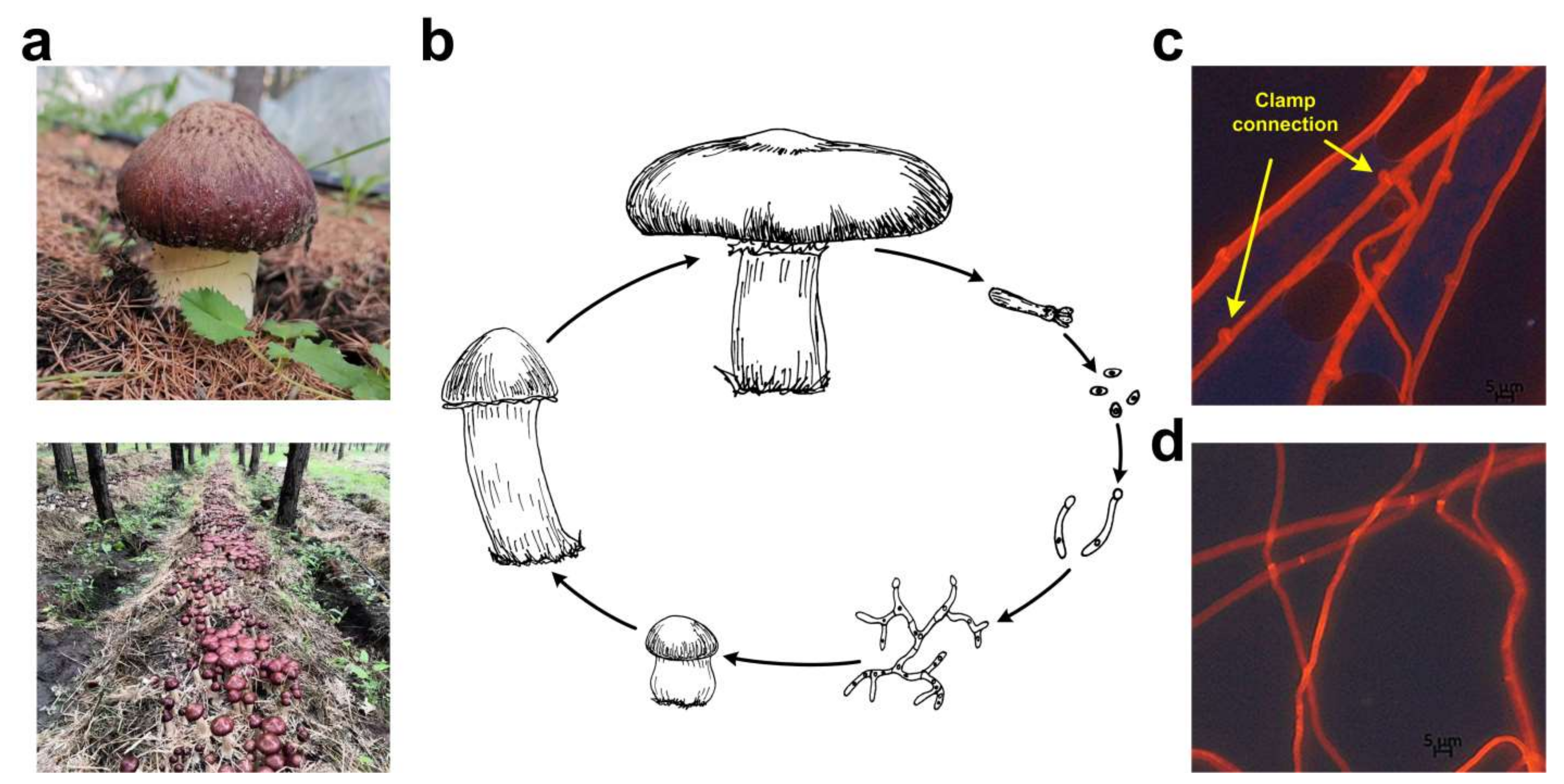
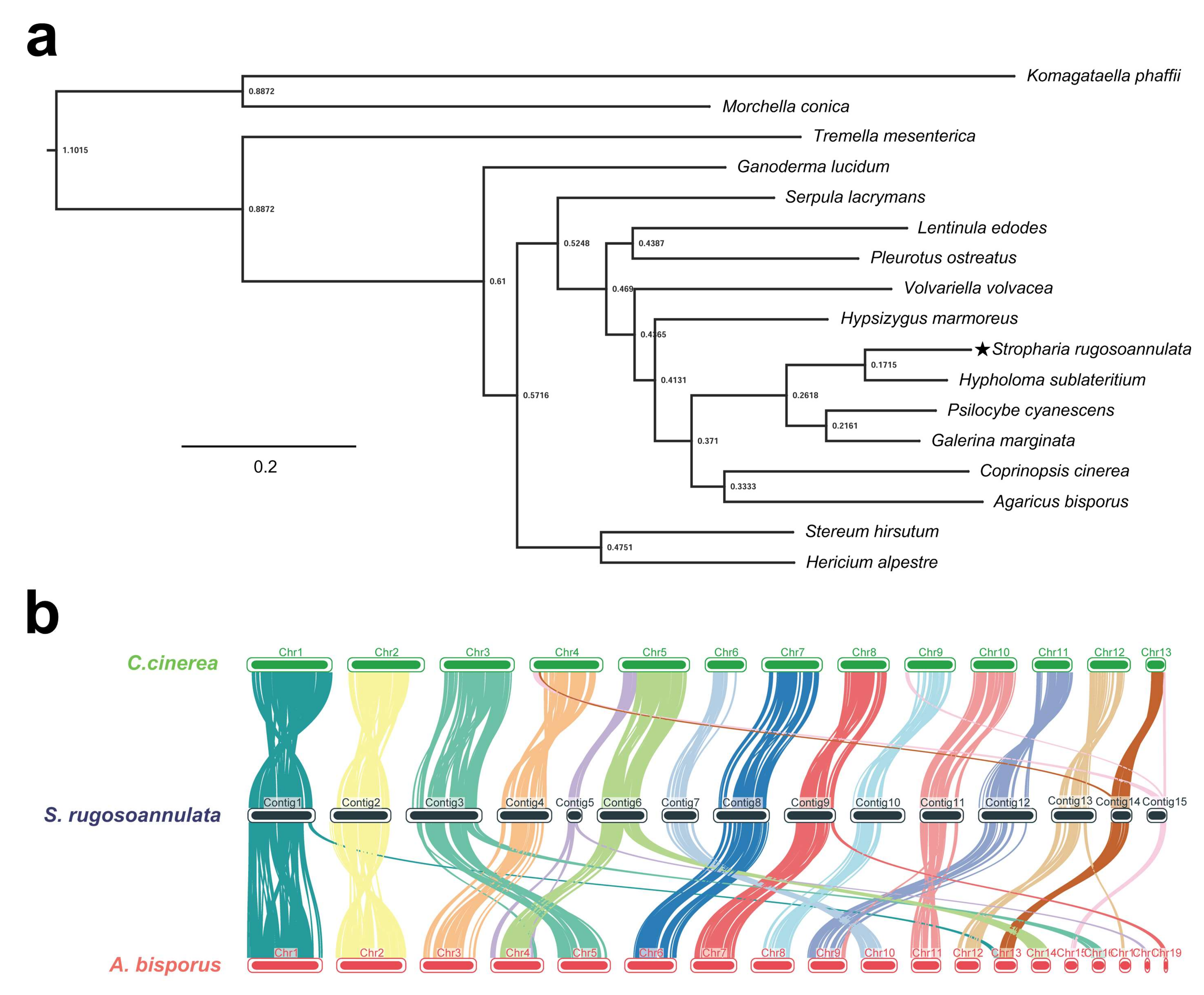


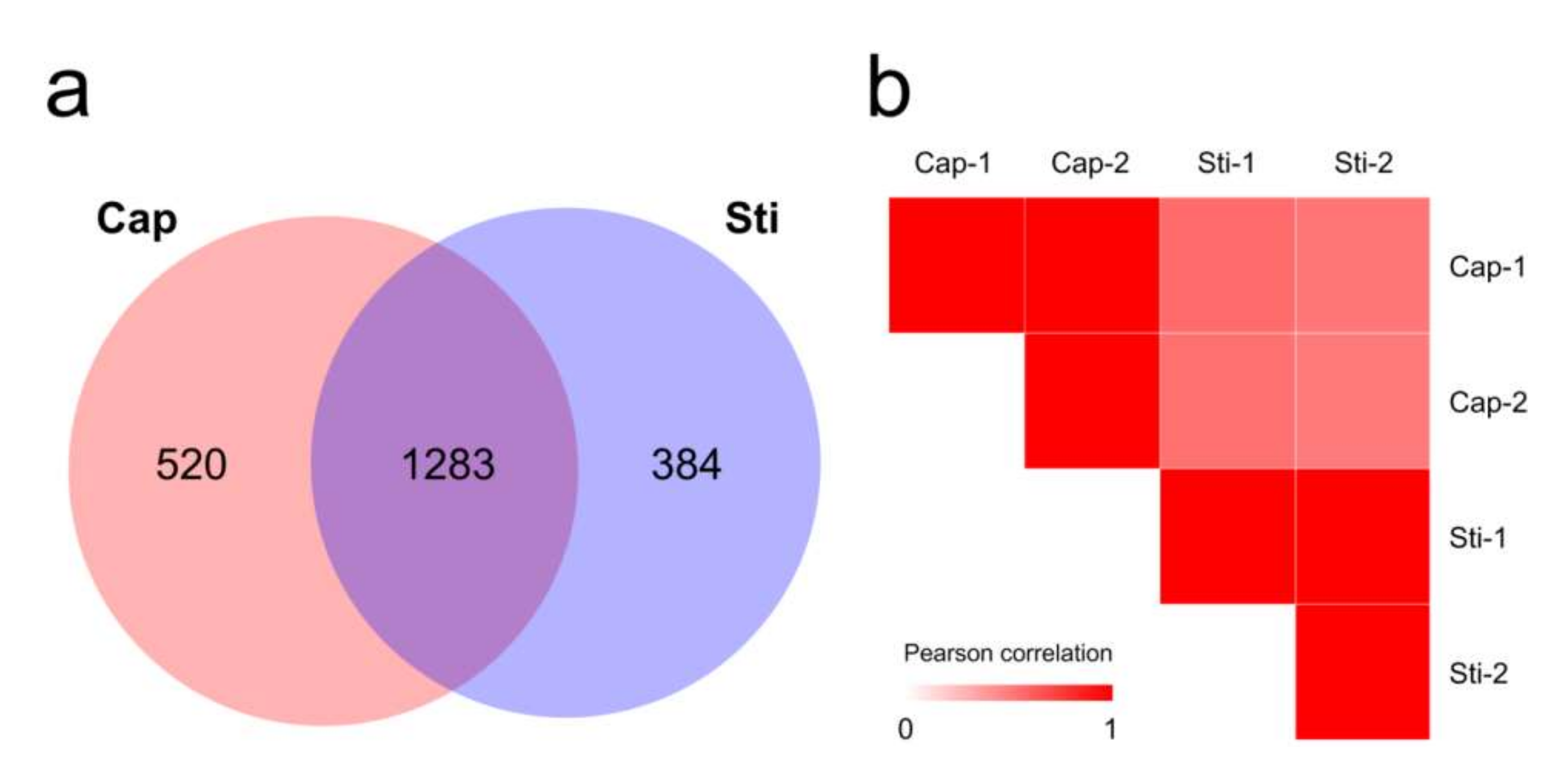

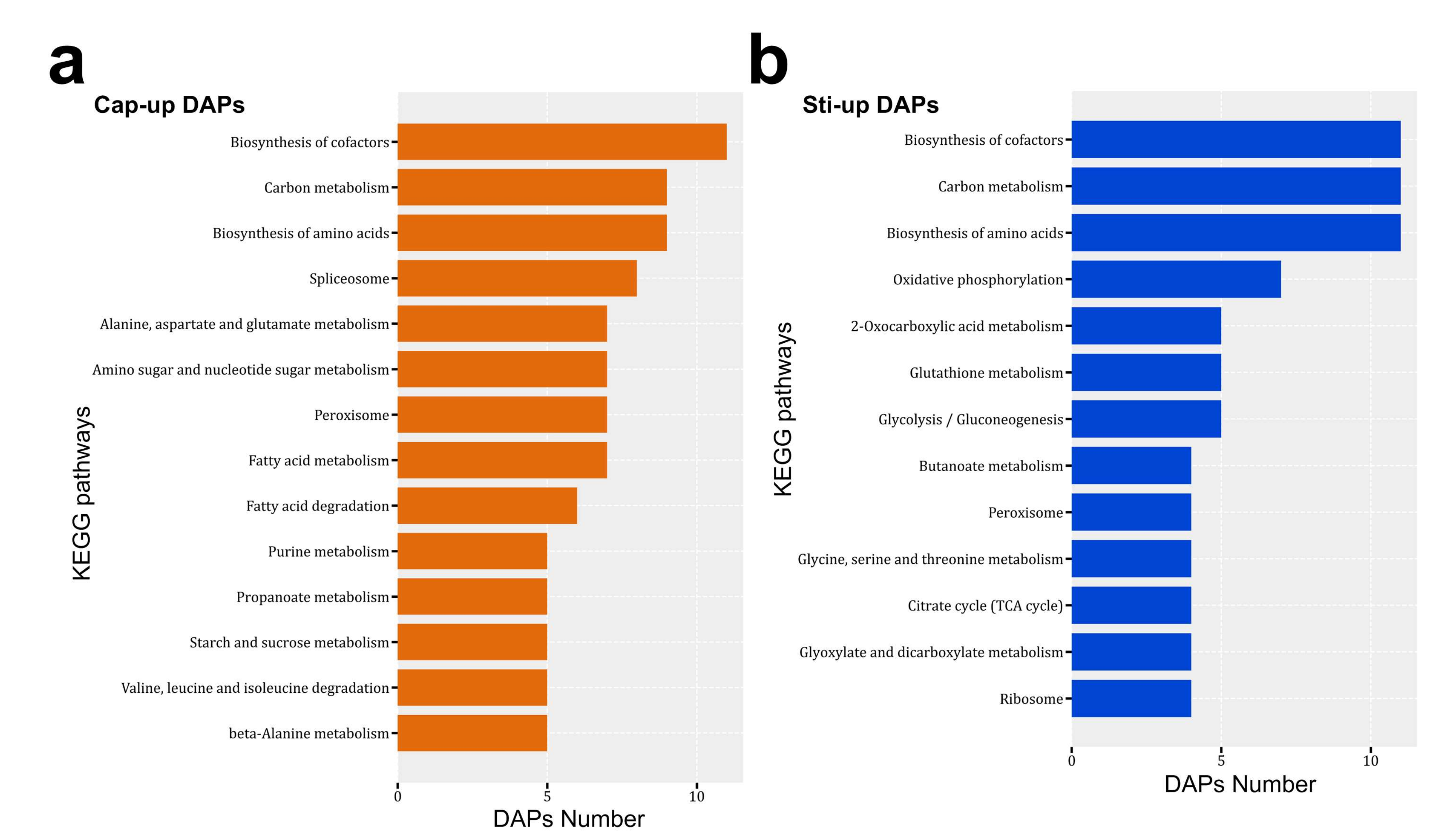
| Species | Strain | Sequencing Technology | Genome Size (Mbp) | GC Content (%) | Sequencing Read Coverage Depth | Contigs | Scaffold | Scaffold N50 (Mbp) |
|---|---|---|---|---|---|---|---|---|
| S. rugosoannulata | A15 | Nanopore + Illumina | 47.89 | 47.94 | 165.4 | 20 | 20 | 3.64 |
| Content | Number/Length |
|---|---|
| Gene number | 12,752 |
| Total concatenated gene length | 26,992,228 bp (56.4% of the genome) |
| Average gene length | 2116.71 bp |
| Average exon length | 258.19 bp |
| Average exon number | 6.55 |
| Average intron length | 76.75 bp |
| Fungal Species | Genes | Genes in Orthogroups (%) | Unassigned Genes (%) | Orthogroups Containing Species (%) | Number of Species-Specific Orthogroups | Genes in Species-Specific Orthogroups |
|---|---|---|---|---|---|---|
| S.rugosoannulata | 12,752 | 12,342 (96.8%) | 410 (3.2%) | 7766 (46.6%) | 197 | 881 |
| A. bisporus | 11,280 | 10,430 (92.5%) | 850 (7.5%) | 6543 (39.2%) | 216 | 1792 |
| C. cinerea | 13,356 | 11,751 (88%) | 1605 (12%) | 7382 (44.3%) | 371 | 1699 |
| Ganoderma lucidum | 16,495 | 14,002 (84.9%) | 2493 (15.1%) | 7274 (43.6%) | 618 | 2796 |
| G. marginata | 21,391 | 18,671 (87.3%) | 2720 (12.7%) | 9196 (55.1%) | 788 | 2960 |
| Hericium alpestre | 11,007 | 10,182 (92.5%) | 825 (7.5%) | 6040 (36.2%) | 293 | 1120 |
| Hypsizygus marmoreus | 16,627 | 14,218 (85.5%) | 2409 (14.5%) | 8164 (48.9%) | 548 | 2154 |
| H. sublateritium | 17,771 | 15,233 (85.7%) | 2538 (14.3%) | 8489 (50.9%) | 466 | 1575 |
| K. phaffii | 5040 | 4226 (83.8%) | 814 (16.2%) | 3700 (22.2%) | 33 | 92 |
| Lentinula edodes | 12,051 | 10,676 (88.6%) | 1375 (11.4%) | 6737 (40.4%) | 415 | 1284 |
| M. conica | 11,593 | 7893 (68.1%) | 3700 (31.9%) | 5420 (32.5%) | 339 | 1225 |
| P. cyanescens | 15,936 | 14,415 (90.5%) | 1521 (9.5%) | 8089 (48.5%) | 395 | 1853 |
| Pleurotus ostreatus | 12,296 | 11,377 (92.5%) | 919 (7.5%) | 7233 (43.4%) | 284 | 1077 |
| Stereum hirsutum | 14,066 | 12,588 (89.5%) | 1478 (10.5%) | 7510 (45%) | 351 | 1251 |
| Serpula lacrymans | 14,481 | 12,377 (85.5%) | 2104 (14.5%) | 6852 (41.1%) | 288 | 1804 |
| Tremella mesenterica | 8074 | 6264 (77.6%) | 1810 (22.4%) | 5079 (30.4%) | 217 | 772 |
| Volvariella volvacea | 11,448 | 10,929 (95.5%) | 519 (4.5%) | 6634 (39.8%) | 215 | 1451 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhao, S.; Hu, C.; Mao, C.; Guo, L.; Yu, H.; Yu, H. Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu). J. Fungi 2022, 8, 99. https://doi.org/10.3390/jof8020099
Li S, Zhao S, Hu C, Mao C, Guo L, Yu H, Yu H. Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu). Journal of Fungi. 2022; 8(2):99. https://doi.org/10.3390/jof8020099
Chicago/Turabian StyleLi, Shuwen, Shuxue Zhao, Chunhui Hu, Chengzhi Mao, Lizhong Guo, Hailong Yu, and Hao Yu. 2022. "Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu)" Journal of Fungi 8, no. 2: 99. https://doi.org/10.3390/jof8020099
APA StyleLi, S., Zhao, S., Hu, C., Mao, C., Guo, L., Yu, H., & Yu, H. (2022). Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu). Journal of Fungi, 8(2), 99. https://doi.org/10.3390/jof8020099






