Four New Species of Aspergillus Subgenus Nidulantes from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Collection
2.2. Fungal Isolation and Preliminary Identification
2.3. Phylogenetic Analysis
2.4. Morphology
3. Results
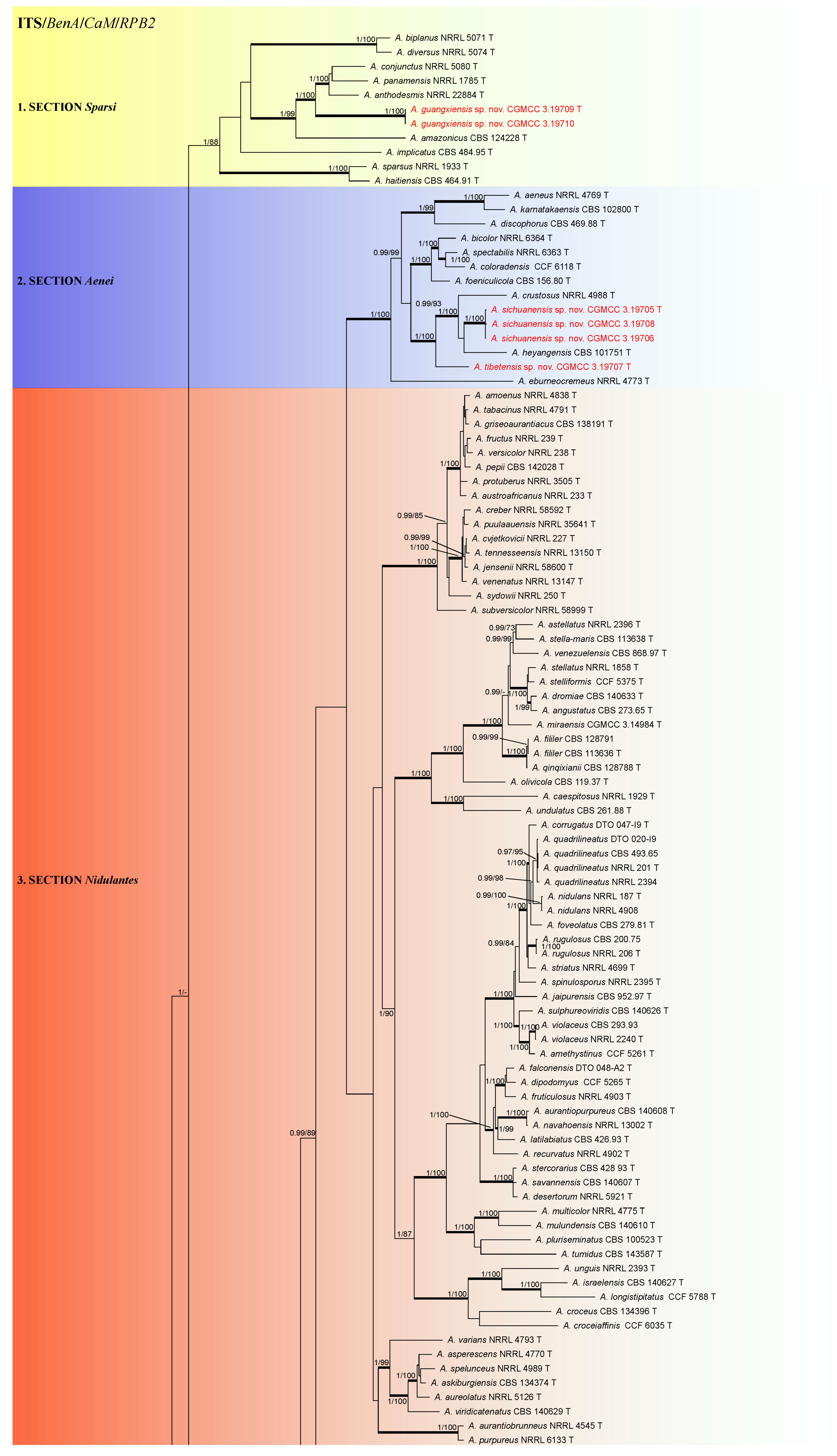
3.1. Phylogeny
3.2. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef]
- Samson, R.A.; Hubka, V.; Varga, J.; Houbraken, J.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Magistà, D.; Visagie, C.M.; et al. Response to Pitt & Taylor 2016: Conservation of Aspergillus with A. niger as the conserved type is unnecessary and potentially disruptive. TAXON 2017, 66, 1439–1446. [Google Scholar]
- Kocsubé, S.; Perrone, G.; Magistà, D.; Houbraken, J.; Varga, J.; Szigeti, G.; Hubka, V.; Hong, S.; Frisvad, J.C.; Samson, R.A. Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Stud. Mycol. 2016, 85, 199–213. [Google Scholar]
- Chen, A.J.; Frisvad, J.C.; Sun, B.D.; Varga, J.; Kocsubé, S.; Dijksterhuis, J.; Kim, D.H.; Hong, S.B.; Houbraken, J.; Samson, R.A. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84, 1–118. [Google Scholar] [CrossRef]
- Berkeley, M. Introduction to Cryptogamic Botany; Bailliere: London, UK, 1857; pp. 1–604. [Google Scholar]
- Frisvad, J.C.; Samson, R.A. Emericella venezuelensis, a new species with stellate ascospores producing sterigmatocystin and aflatoxin B1. Syst. Appl. Microbiol. 2004, 27, 672–680. [Google Scholar]
- Horie, Y. A new species of Emericella from Indian herbal drugs. Trans. Mycol. Soc. Japan 1978, 19, 313–317. [Google Scholar]
- Horie, Y. New or interesting Emericella (fungus) from herbal drugs. Trans. Mycol. Soc. Japan 1979, 20, 481–491. [Google Scholar]
- Horie, Y. Ascospore ornamentation and its application to the taxonomic re-evaluation in Emericella. Trans. Mycol. Soc. Japan 1980, 21, 483–493. [Google Scholar]
- Horie, Y.; Miyaji, M.; Nishimura, K.; Udagawa, S. Emericella falconensis, a new species from Venezuelan soil. Trans. Mycol. Soc. Japan 1989, 30, 257–263. [Google Scholar]
- Horie, Y.; Udagawa, S.; Abdullah, J.K.; Al-Bader, S.M. Emericella similis, a new species from Iraqi soil. Trans. Mycol. Soc. Japan 1990, 31, 425–430. [Google Scholar]
- Horie, Y.; Fukiharu, T.; Nishimura, K.; Taguchi, H.; Wang, D.; Li, R. New and interesting species of Emericella. Mycoscience 1996, 37, 323–329. [Google Scholar] [CrossRef]
- Horie, Y.; Li, D.M.; Fukiharu, T.; Li, R.; Abliz, P.; Nishimura, K.; Wang, D.L. Emericella appendiculata, a new species from Chinese soil. Mycoscience 1998, 39, 161–165. [Google Scholar]
- Horie, Y.; Miyaji, M.; Nishimura, K.; Franco, M.F.; Coelho, K.I.R. New and interesting species of Emericella from Brazilian soil. Mycoscience 1998, 37, 137–144. [Google Scholar] [CrossRef]
- Horie, Y.; Abliz, P.; Hui, Y.; Fukiharu, T.; Nishimura, K.; Li, D.M.; Li, R. Emericella qinqixianii, a new species from desert soil in China. Mycoscience 2000, 41, 183–187. [Google Scholar]
- Kong, H.; Qi, Z. A new species of Emericella. Acta Mycol. Sin. 1986, 5, 211–214. [Google Scholar]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus, 1st ed.; Williams & Wilkins: Baltimore, MD, USA, 1965; pp. 1–686. [Google Scholar]
- Samson, R.A.; Mouchacca, J. Additional notes on species of Aspergillus, Eurotium and Emericella from Egyptian desert soil. Anton. Leeuw. 1975, 41, 343–351. [Google Scholar]
- Stchigel, A.M.; Guarro, J. A new species of Emericella from Indian soil. Mycologia 1997, 89, 937–941. [Google Scholar] [CrossRef]
- Thom, C.; Raper, K.B. The Aspergillus nidulans group. Mycologia 1939, 31, 653–669. [Google Scholar]
- Zalar, P.; Frisvad, J.C.; Gunde-Cimerman, N.; Varga, J.; Samson, R.A. Four new species of Emericella from the Mediterranean region of Europe. Mycologia 2008, 100, 779–795. [Google Scholar] [CrossRef]
- Samson, R.A.; Hounraken, J.; Frisvad, J.C.; Thrane, U.; Andersen, B. Food and Indoor Fungi; CBS-KNAW Fungal Biodiversity Center: Utrecht, The Netherlands, 2010; pp. 1–390. [Google Scholar]
- Samslon, R.A.; Varga, J.; Meijer, M.; Frisvad, J.C. New species in Aspergillus section Usti. Stud. Mycol. 2011, 69, 81–97. [Google Scholar] [CrossRef]
- Sklenář, F.; Jurjević, Ž.; Peterson, S.W.; Kolařík, M.; Nováková, A.; Flieger, M.; Stodůlková, E.; Kubátová, A.; Hubka, V. Increasing the species diversity in the Aspergillus section Nidulantes: Six novel species mainly from the indoor environment. Mycologia 2020, 112, 342–370. [Google Scholar] [PubMed]
- Gams, W.; Christensen, M.; Onions, A.H.S.; Pitt, J.I.; Samson, R.A. Infrageneric taxa of Aspergillus. In Advances in Penicillium and Aspergillus Systematics; Samson, R.A., Pitt, J.I., Eds.; NATO ASI Series. Ser. A; Life Sciences Plenum Press: New York, NY, USA, 1985; Volume 102, pp. 55–62. [Google Scholar]
- Peterson, S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008, 100, 205–226. [Google Scholar]
- Peterson, S.W.; Varga, J.; Frisvad, J.C.; Samson, R.A. Phylogeny and subgeneric taxonomy of Aspergillus. In Aspergillus in the Genomic Era; Varga, J., Samson, R.A., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2008; pp. 33–56. [Google Scholar]
- Varga, J.; Frisvad, J.C.; Samson, R.A. Aspergillus sect. Aenei sect. nov., a new section of the genus for A. karnatakaensis sp. nov. and some allied fungi. IMA Fungus 2010, 1, 197–205. [Google Scholar] [PubMed]
- Despot, J.D.; Sándor, K.; Bencsik, O.; Kecskeméti, A.; Szekeres, A.; Vágvölgyi, C.; Varga, J.; Klarić, M.Š. New sterigmatocystin-producing species of Aspergillus section Versicolores from indoor air in Croatia. Mycol. Prog. 2017, 16, 63–72. [Google Scholar]
- Hubka, V.; Nováková, A.; Peterson, S.W.; Frisvad, J.C.; Sklenář, F.; Matsuzawa, T.; Kubátová, A.; Kolařík, M. A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin-producing species. Plant Syst. Evol. 2016, 302, 1267–1299. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.S.J.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.F.P.; Souza-Motta, C.M.; et al. Fungal planet description sheets: 716–784. Persoonia 2018, 40, 240–393. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.J.; Smith, D.; Summerell, B.A.; Cano-Lira, J.F.; Guarro, J.; Houbraken, J.; et al. Fungal planet description sheets: 625–715. Persoonia 2017, 39, 270–467. [Google Scholar] [CrossRef]
- Sun, B.D.; Houbraken, J.; Frisvad, J.C.; Jiang, X.Z.; Chen, A.J.; Samson, R.A. New species in Aspergillus section Usti and an overview of Aspergillus section Cavernicolarum. Int. J. Syst. Evol. Microbiol. 2020, 70, 5401–5416. [Google Scholar]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar]
- Sklenář, F.; Glässnerová, K.; Jurjević, Ž.; Houbraken, J.; Samson, R.A.; Visagie, C.M.; Yilmaz, N.; Gené, J.; Cano, J.; Chen, A.J.; et al. Taxonomy of Aspergillus series Versicolores: Species reduction and lessons learned about intraspecific variability. Stud. Mycol. 2022, 102. [Google Scholar] [CrossRef]
- Bartoli, A.; Maggi, O. Four new species of Aspergillus from Ivory coast soil. Trans. Br. Mycol. Soc. 1978, 71, 383–394. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Skouboe, P.; Samson, R.A. Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Syst. Appl. Microbiol. 2005, 28, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus section Sparsi. IMA Fungus 2010, 1, 187–195. [Google Scholar]
- Horie, Y.; Udagawa, S. Emericeila omanensis, a new species from Oman soil. Mycoscience 1995, 36, 391–394. [Google Scholar] [CrossRef]
- Klich, M.; Mendoza, C.; Mullaney, E.; Keller, N.; Bennett, J.W. A new sterigmatocystin-producing Emericella variant from agricultural desert soils. Syst. Appl. Microbiol. 2001, 24, 131–138. [Google Scholar]
- Polacheck, I.; Nagler, A.; Okon, E.; Drakos, P.; Plaskowitz, J.; Kwon-Chung, K.J. Aspergillus quadrilineatus, a new causative agent of fungal sinusitis. J. Clin. Microbiol. 1992, 30, 3290–3293. [Google Scholar] [PubMed]
- de Hoog, G.S.; Guarro, J.; Gene, J.; Figueras, M.J. Atlas of Clinical Fungi, 3rd ed.; CBS-KNAW Fungal Biodiversity Center: Utrecht, The Netherlands, 2014; pp. 1–1126. [Google Scholar]
- Verweij, P.E.; Varga, J.; Houbraken, J.; Rijs, A.J.M.M.; Antonius, J.M.M.; VerdyunLunel, L.M.; Blijlevens, N.M.A.; Shen, Y.R.; Holland, S.M.; Warris, A.; et al. Emericella quadrilineata as cause of invasive aspergillosis. Emerg. Infect. Dis. 2008, 14, 566–572. [Google Scholar] [CrossRef]
- Yu, J.; Mu, X.; Li, R. Invasive pulmonary aspergillosis due to Emericella nidulans var. echinulata, successfully cured by voriconazol and micafungin. J. Clin. Microbiol. 2013, 51, 1327–1329. [Google Scholar]
- de Fontbrune, F.S.; Denis, B.; Meunier, M.; Garcia-Hermoso, D.; Bretagne, S.; Alanio, A. Iterative breakthrough invasive aspergillosis due to TR (34)/298H azole-resistant Aspergillus fumigatus and Emericella sublata in a single hematopoietic stem cell transplant patient. Transpl. Infect. Dis. 2014, 16, 687–691. [Google Scholar] [CrossRef]
- Sabino, R.; Verissimo, C.; Parada, H.; Brandao, J.; Viegas, C.; Corolino, E.; Clemons, K.V.; Stevens, D.A. Molecular screening of 246 Portuguese Aspergillus isolates among different clinical and environmental sources. Med. Mycol. 2014, 52, 517–527. [Google Scholar]
- Tsang, C.C.; Hui, T.W.S.; Lee, K.C.; Chen, J.H.K.; Ngan, A.H.Y.; Tam, E.W.T.; Chan, J.F.W.; Wu, A.L.; Cheung, M.; Tse, B.P.H.; et al. Genetic diversity of Aspergillus species isolated from onychomycosis and Aspergillus hongkongensis sp. nov., with implications to antifungal susceptibility testing. Diagn. Microbiol. Infect. Dis. 2016, 84, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Al-Hatmi, A.M.S.; Castro, M.A.; de Hoog, G.S. Epidemiology of Aspergillus species causing keratitis in Mexico. Mycoses 2019, 62, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Chen, M.; Mo, X.; Liu, J.; Yan, F.; Li, Z.; Xie, S.; Chen, D. The first case report of kerion-type scalp mycosis caused by Aspergillus protuberus. BMC Infect. Dis. 2019, 19, 506. [Google Scholar]
- de Vries, R.P.; Robert, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar]
- Pontecorvo, G.; Gloor, E.T.; Forbes, E. Analysis of mitotic recombination in Aspergillus nidulans. J. Genet. 1954, 52, 226–237. [Google Scholar]
- Herbert, N.; Arst, J.R. Integrator gene in Aspergillus nidulans. Nature 1976, 262, 231–234. [Google Scholar]
- Dean, R.A.; Timberlake, W.E. Production of cell wall-degrading enzymes by Aspergillus nidulans: A model system for fungal pathogenesis of plants. Plant Cell 1989, 1, 265–273. [Google Scholar]
- Schoustra, S.E.; Debets, A.J.M.; Slakhorst, M.; Hoekstra, R.F. Reducing the cost of resistance; experimental evolution in the filamentous fungus Aspergillus nidulans. J. Evol. Biol. 2006, 19, 1115–1127. [Google Scholar]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. [Google Scholar] [CrossRef]
- Yaegashi, J.; Oakley, B.R.; Wang, C.C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotechnol. 2014, 41, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Wang, B.T.; Wang, Z.D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar]
- Andersen, M.R.; Nielsen, J.B.; Klitgaard, A.; Petersen, L.M.; Zachariasen, M.; Hansen, T.J.; Blicher, L.H.; Gotfredsen, C.H.; Larsen, T.O.; Nielsen, K.F.; et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. PNAS 2013, 110, E99–E107. [Google Scholar] [PubMed]
- Vazquez, J.A.; Sobel, J.D. Anidulafungin: A novel echinocandin. Clin. Infect. Dis. 2006, 43, 215–222. [Google Scholar]
- Sofjan, A.K.; Mitchell, A.; Shah, D.N.; Nguyen, T.; Sim, M.; Trojcak, A.; Beyda, N.D.; Garey, K.W. Rezafungin (CD101), a next-generation echinocandin: A systematic literature review and assessment of possible place in therapy. J. Glob. Antimicrob. Resist. 2018, 14, 58–64. [Google Scholar]
- Houbraken, J.; Samson, R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011, 70, 1–51. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute: London, UK, 1970; pp. 1–34. [Google Scholar]
- Sun, Z.M.; Qi, Z.T. New taxa and a new record of Aspergillus and Eurotium. Acta Mycol. Sin. 1994, 13, 81–87. [Google Scholar]
- Christensen, M.; Raper, K.B.; States, J.S. Two new Aspergillus nidulans group members from Wyoming soils. Mycologia 1978, 70, 332–342. [Google Scholar] [CrossRef]
- Raper, K.B.; Thom, C. New Aspergilli from soil. Mycologia 1944, 36, 555–557. [Google Scholar]
- Maggi, O.; Persiani, A.M. Aspergillus implicatus, a new species isolated from Ivory coast forest soil. Mycol. Res. 1994, 98, 869–873. [Google Scholar] [CrossRef]
- Mares, D.; Andreotti, E.; Maldonado, M.E.; Pedrini, P.; Colalongo, C.; Romagnoli, C. Three new species of Aspergillus from amazonian forest soil (Ecuador). Curr. Microbiol. 2008, 57, 222–229. [Google Scholar] [CrossRef]
- Smith, G. Some new species of soil moulds. Trans. Br. Mycol. Soc. 1956, 39, 111–114. [Google Scholar] [CrossRef]
- Shumi, W.; Hossain, M.T.; Anwar, M.N. Isolation and purification of fungus Aspergillus funiculosus G. Smith and its enzyme protease. Pak. J. Biol. Sci. 2004, 7, 312–317. [Google Scholar]
- Qi, Z.T. Flora Fungorum Sinicorum. Aspergillus et Teleomorphi Cognati; Science Press: Beijing, China, 1997; Volume 5, pp. 1–198. (In Chinese) [Google Scholar]
- Sun, B.D.; Ding, G.; Zhang, Y.S.; Zhao, G.Z.; Zhou, Y.G.; Chen, A.J. Current taxonomy of Aspergillus subgenus Nidulantes and re-identification of several strains. Mycosystema 2017, 36, 1192–1209. [Google Scholar]
- Li, D.M.; Wang, D.L.; Li, R.Y.; Wang, X.H.; Horie, Y.; Fukiharu, T. Emericella spp. in soils of north China. Mycosystema 1998, 17, 130–136. [Google Scholar]
- Wang, L. Aspergillus keveioides, a new species of Aspergillus sect. Usti from Shandong Province, China. Mycosystema 2013, 32, 136–144. [Google Scholar]
- Zhang, L.C.; Chen, J.; Guo, W.H.; Guo, S.X. A new species of Emericella from China. Mycotaxon 2013, 125, 131–138. [Google Scholar] [CrossRef]





| Species | Origin | Strain No. | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|
| Aspergillus guangdongensis | Farmland soil, Guangdong | CGMCC 3.19704 T | MN640760 | MN635246 | MN635257 | MN635269 |
| A. guangxiensis | Farmland soil, Guangxi | CGMCC 3.19709 T | MN640765 | MN635251 | MN635262 | MN635274 |
| A. guangxiensis | Farmland soil, Guangxi | CGMCC 3.19710 | MN640766 | MN635252 | MN635263 | MN635275 |
| A. sichuanensis | Farmland soil, Sichuan | CGMCC 3.19705 T | MN640761 | MN635247 | MN635258 | MN635270 |
| A. sichuanensis | Farmland soil, Sichuan | CGMCC 3.19706 | MN640762 | MN635248 | MN635259 | MN635271 |
| A. sichuanensis | Farmland soil, Sichuan | CGMCC 3.19708 | MN640764 | MN635250 | MN635261 | MN635273 |
| A. tibetensis | Farmland soil, Tibet | CGMCC 3.19707 T | MN640763 | MN635249 | MN635260 | MN635272 |
| Macromorphology Colony Diam 25 °C, 7 d (mm) | Micromorphology (μm) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | CYA | CYA 37 °C | MEA | Conidial Head | Vesicle | Stipe Length | Conidia Ornamentation | Conidia Shape and Size | Ascomata | Hülle Cells | References |
| Aspergillus guangdongensis | 30–33 | 3–4 | 43–44 | Uniseriate | 25–35 | 230–450 | Smooth | Subglobose to ellipsoidal, 3–4 × 2–3 | NOB | NOB | This study |
| A. funiculosus | * 30–35 after 10–14 d on CA | - | - | Uniseriate | 8–35 | 400–600 | Smooth to coarsely rugulose | Elliptical to globose, 3–3.5 × 2–2.5 | NOB | NOB | [17] |
| A. guangxiensis | 11–13 | NG | 9–11 | Biseriate | 17–28 | 350–600 | Finely roughened | Subglobose to globose, 3–4 | NOB | NOB | This study |
| A. conjunctus | * 30 after 10 d on CA | - | 40 after 10 d | Biseriate | 15–30 | 500–700 | Smooth to finely roughened | Subglobose to globose, 2.5–4 | NOB | elongate | [17] |
| A. haitiensis | 30–35 after 14 d | - | 50–60 after 14 d | Biseriate | 10–25 | 200–500 | Smooth | Globose to ellipsoidal, 4–5.6 × 5–6 | NOB | NOB | [38] |
| A. sichuanensis | 14–23 | 5–6 | 13–16 | Biseriate | 10–17 | 200–500 | Echinulate | Globose, 3–4 | NOB | globose to ovoid | This study |
| A. heyangensis | 19–23 | NG | - | Biseriate | 8–12 | 60–150–(200) | Roughened | Subglobose to globose, 2.5–3.5 | NOB | NOB | [67] |
| A. crustosus | * 20–25 after 14 d on CA | - | - | Biseriate | (5.5)–7–10– (12) | 150–250 | Echinulate | Granular to echinulate, 2.5–3 | NOB | globose to irregular | [17] |
| A. tibetensis | 13–17 | NG | 20–22 | Biseriate | 20–26 | 125–300 | Smooth | Globose, 2.5–3 | NOB | globose to ovoid | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Luo, C.; Bills, G.F.; Li, J.; Huang, P.; Wang, L.; Jiang, X.; Chen, A.J. Four New Species of Aspergillus Subgenus Nidulantes from China. J. Fungi 2022, 8, 1205. https://doi.org/10.3390/jof8111205
Sun B, Luo C, Bills GF, Li J, Huang P, Wang L, Jiang X, Chen AJ. Four New Species of Aspergillus Subgenus Nidulantes from China. Journal of Fungi. 2022; 8(11):1205. https://doi.org/10.3390/jof8111205
Chicago/Turabian StyleSun, Bingda, Chunling Luo, Gerald F. Bills, Jibing Li, Panpan Huang, Lin Wang, Xianzhi Jiang, and Amanda Juan Chen. 2022. "Four New Species of Aspergillus Subgenus Nidulantes from China" Journal of Fungi 8, no. 11: 1205. https://doi.org/10.3390/jof8111205
APA StyleSun, B., Luo, C., Bills, G. F., Li, J., Huang, P., Wang, L., Jiang, X., & Chen, A. J. (2022). Four New Species of Aspergillus Subgenus Nidulantes from China. Journal of Fungi, 8(11), 1205. https://doi.org/10.3390/jof8111205






