Differential Metabolomics Reveals Pathogenesis of Pestalotiopsis kenyana Causing Leaf Spot Disease of Zanthoxylum schinifolium
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Samples and Plant Samples
2.2. Instruments and Reagents
2.3. Sample Preparation before Processing
2.4. Metabolite Extraction
2.5. LC-MS/MS Analysis
2.5.1. Chromatographic Separation
2.5.2. Mass Spectrometry Acquisition
2.6. Pathogenicity Detection
2.7. Data Processing
3. Results
3.1. Experimental Quality Evaluation
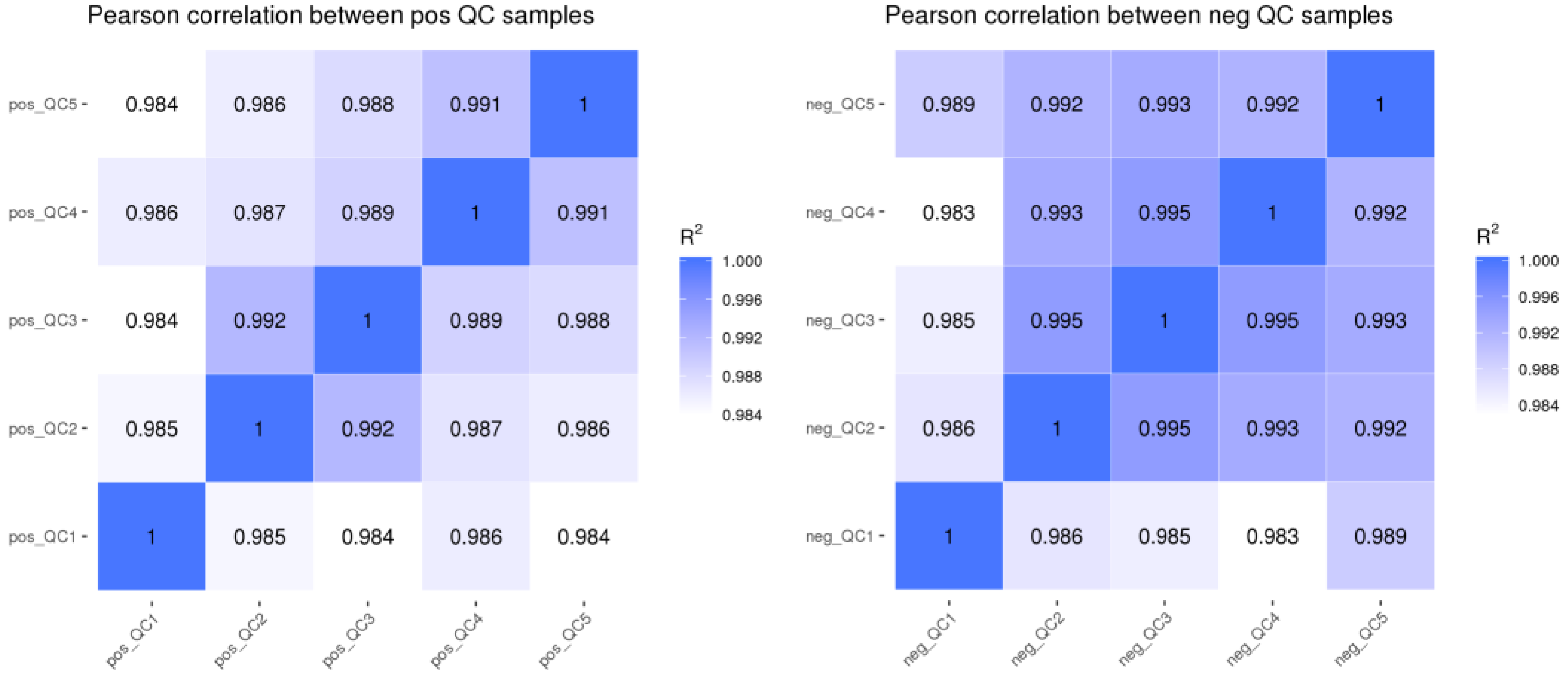
3.1.1. QC Sample Correlation Analysis
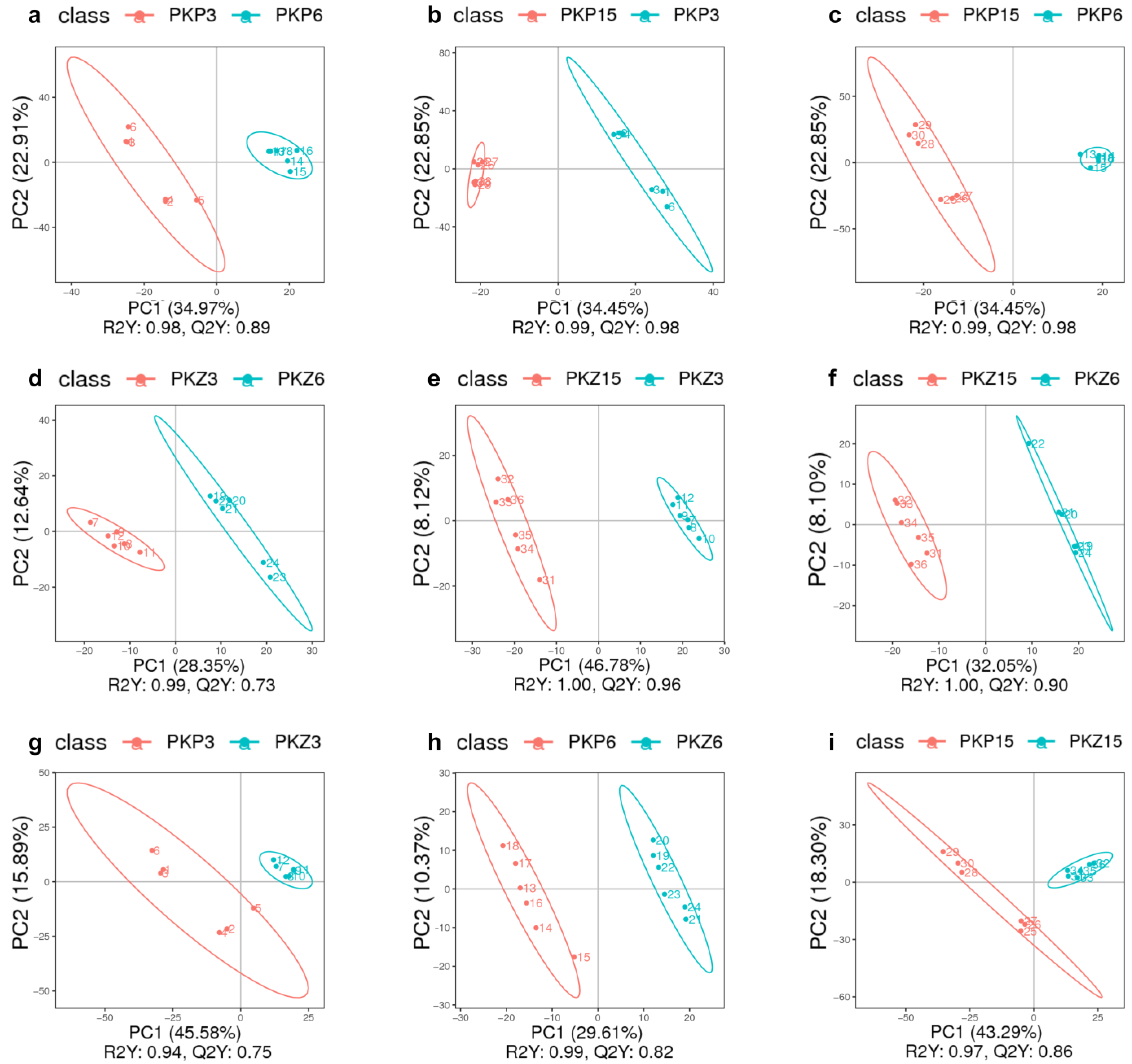
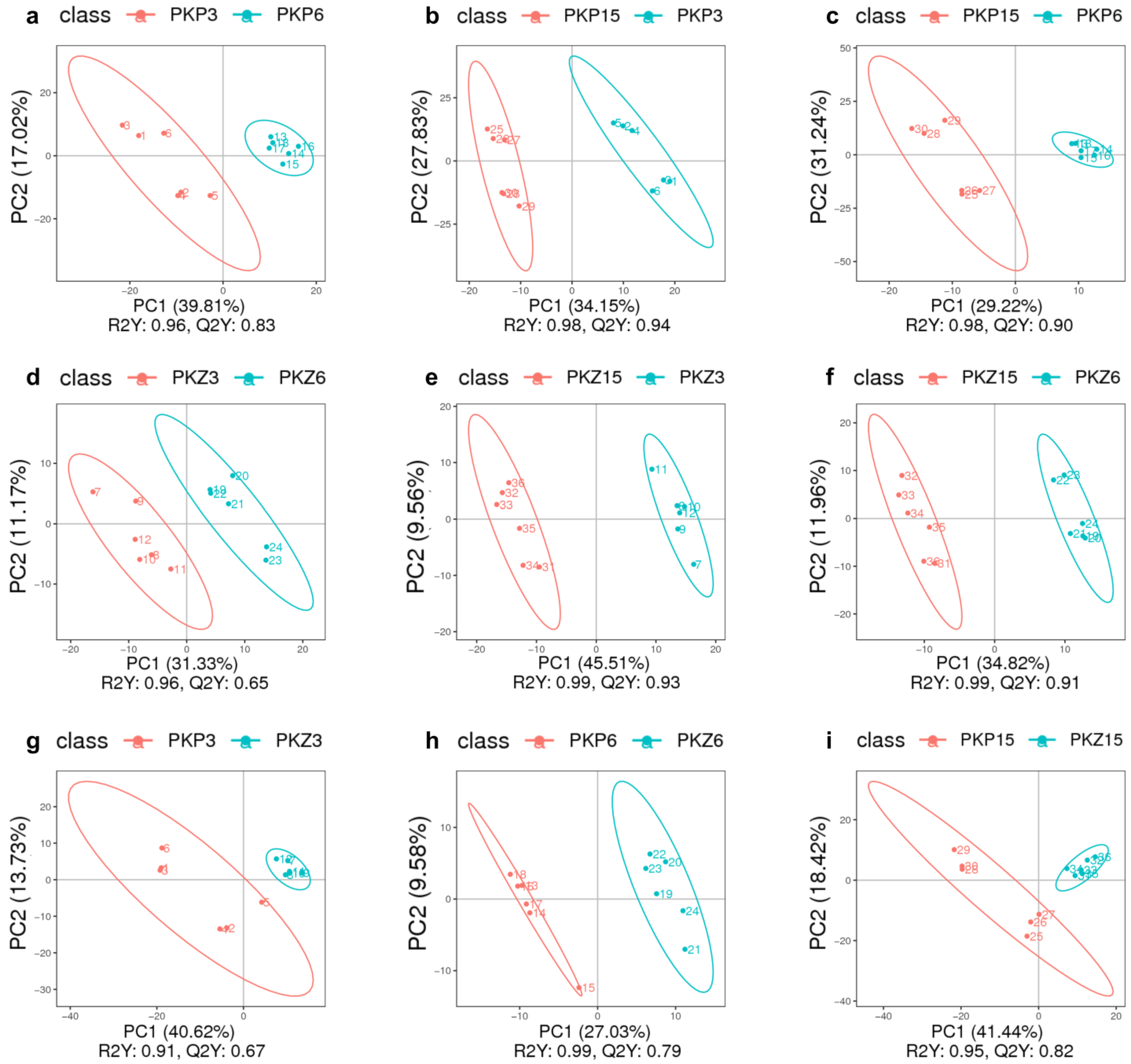
3.1.2. Overall Sample PLS-DA
3.2. PLS-DA for Each Comparison Group
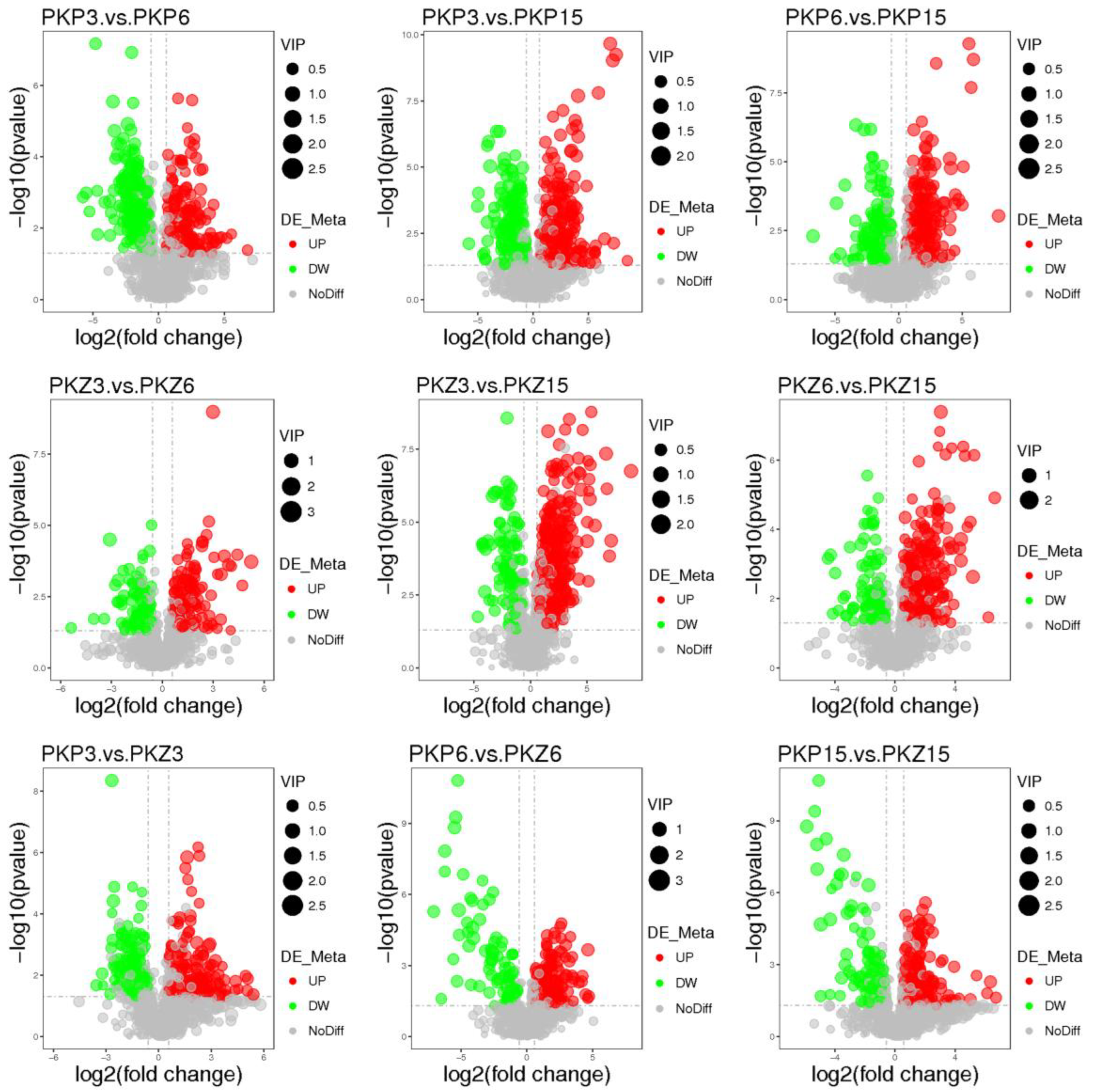
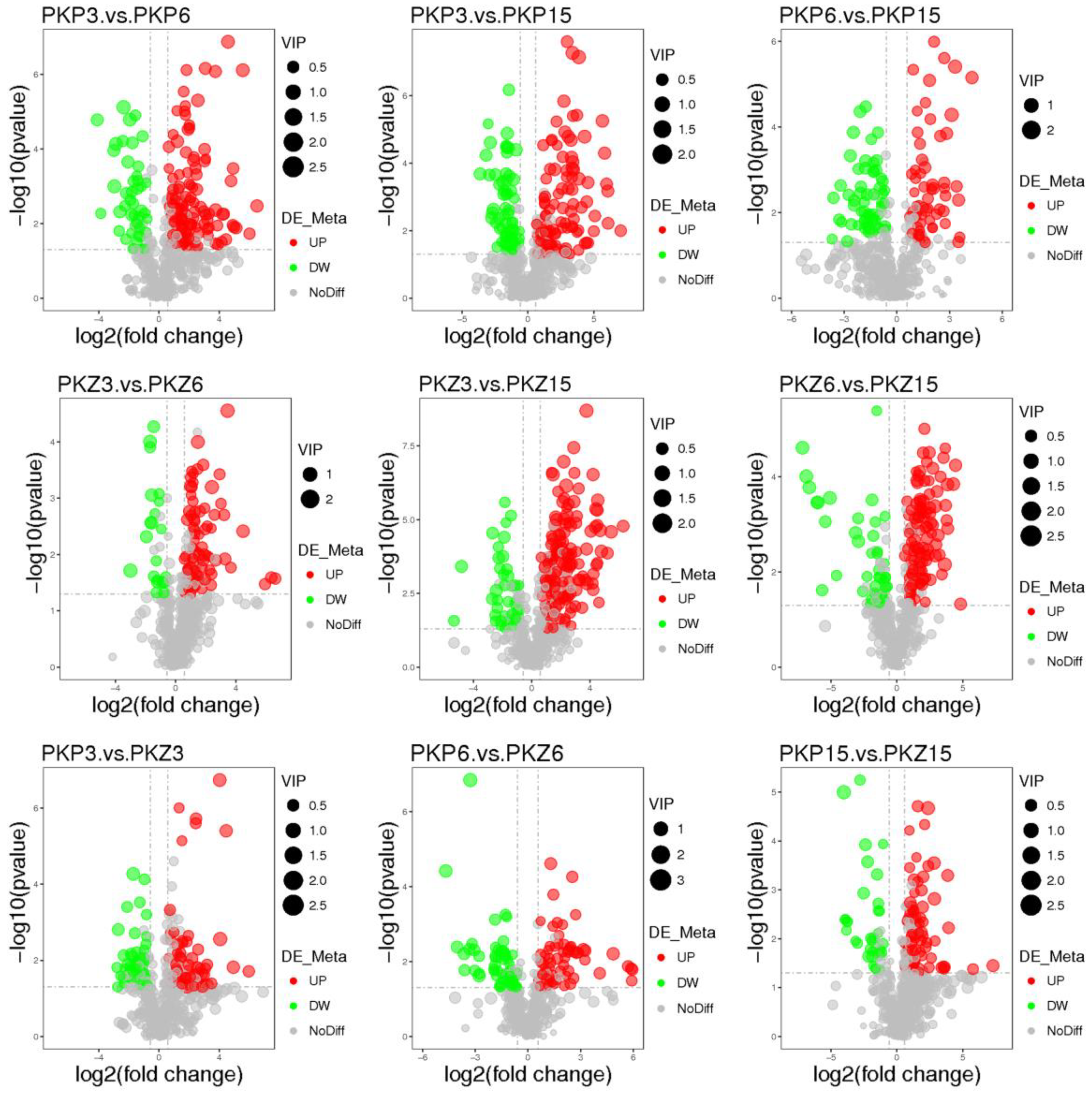
3.3. Univariate Statistical Analysis
3.4. Significantly Different Metabolites
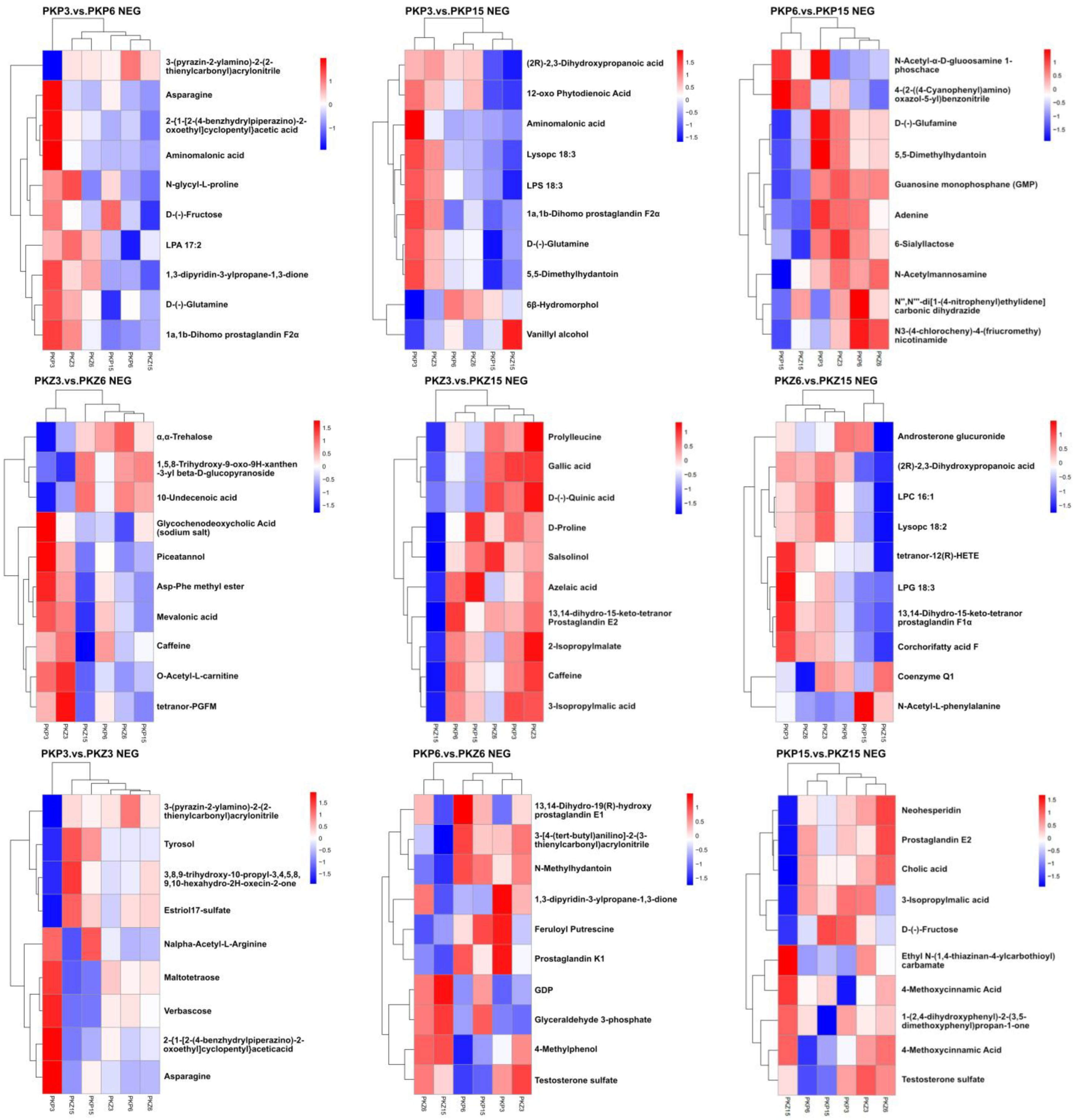
3.5. Hierarchical Cluster Analysis
3.6. KEGG Metabolic Pathway Analysis
3.7. Pathogenicity Test Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Paik, S.K.; Koh, K.H.; Beak, S.M.; Paek, S.H.; Kim, J.A. The essential oils from Zanthoxylum schinifolium pericarp induce apoptosis of HepG2 human hepatoma cells through increased production of reactive oxygen species. Biol. Pharm. Bull. 2005, 28, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.W.; Peng, W.Y.; Ma, Q.Q.; Wang, Z.; Wu, P. Research Progress on Nutrient Composition of Green Pepper. E3S Web Conf. 2021, 251, 2064. [Google Scholar] [CrossRef]
- Gong, Y.; Sun, W.H.; Xu, T.T.; Zhang, L.; Huang, X.Y.; Tan, Z.H.; Di, D.L. Chemical constituents from the pericarps of Zanthoxylum bungeanum and their chemotaxonomic significance. Biochem. Syst. Ecol. 2021, 95, 104213. [Google Scholar] [CrossRef]
- Li, P.Q.; Wu, Z.; Liu, T.; Wang, Y.N. Biodiversity, Phylogeny, and Antifungal Functions of Endophytic Fungi Associated with Zanthoxylum bungeanum. Int. J. Mol. Sci. 2016, 17, 1541. [Google Scholar] [CrossRef]
- Zhang, M.M.; Wang, J.L.; Zhu, L.; Li, T.; Jiang, W.D.; Zhou, J.; Peng, W.; Wu, C.J. Zanthoxylum bungeanum Maxim. (Rutaceae): A Systematic Review of Its Traditional Uses, Botany, Phytochemistry, Pharmacology, Pharmacokinetics, and Toxicology. Int. J. Mol. Sci. 2017, 18, 2172. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Liu, Y.; Xie, C.X.; Li, X.W.; Yu, Y.D.; Ye, M.; Chen, S.L. The Chemical and Genetic Characteristics of Szechuan Pepper (Zanthoxylum bungeanum and Z. armatum) Cultivars and Their Suitable Habitat. Front. Plant Sci. 2016, 7, 467. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Lu, Y.H.; Mo, R.; He, Q. Antioxidant and nitrite-scavenging activities of Zanthoxylum bungeanum maxim. and Capsicum annuum L.: A synergistic, additive or antagonistic effect of the extracts? Eur. Food Res. Technol. 2021, 247, 2877–2885. [Google Scholar] [CrossRef]
- Patil, K.; Mallya, R. Genus Zanthoxylum as Sources of Drugs for Treatment of Tropical Parasitic Diseases. Curr. Drug Discov. Technol. 2022, 19, e040322201773. [Google Scholar] [CrossRef]
- Yang, Q.Q.; Yang, Q.Q.; Mei, X.F.; Wang, Z.R.; Chen, X.H.; Zhang, R.; Chen, Q.L.; Kan, J.Q. Comprehensive identification of non-volatile bitter-tasting compounds in Zanthoxylum bungeanum Maxim. by untargeted metabolomics combined with sensory-guided fractionation technique. Food Chem. 2021, 347, 129085. [Google Scholar] [CrossRef]
- Zhang, W.L.; Tan, S.; Xi, W.P.; Yang, J.L.; Liao, Q.H.; Lan, J.B.; Lv, Y.K.; Tang, J.M. Comparison of volatile components in fresh and dried Zanthoxylum bungeanum Maxim. Food Sci. Biotechnol. 2019, 28, 1083–1092. [Google Scholar] [CrossRef]
- Zhao, M.; Li, T.; Yang, F.; Cui, X.Y.; Zou, T.T.; Song, H.L.; Liu, Y. Characterization of key aroma-active compounds in Hanyuan Zanthoxylum bungeanum by GC-O-MS and switchable GC × GC-O-MS. Food Chem. 2022, 385, 132659. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Luo, F.Y.; Zhu, T.H.; Han, S.; Li, S.J. Leaf Spot Disease Caused by Pestalotiopsis kenyana on Zanthoxylum schinifolium in Sichuan Province, China. Plant Dis. 2021, 105, 3747. [Google Scholar] [CrossRef] [PubMed]
- Matich, E.K.; Chavez Soria, N.G.; Aga, D.S.; Atilla-Gokcumen, G.E. Applications of metabolomics in assessing ecological effects of emerging contaminants and pollutants on plants. J. Hazard. Mater. 2019, 373, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Gong, Z.G.; Hu, J.; Wu, X.; Xu, Y.J. The recent developments in sample preparation for mass spectrometry-based metabolomics. Crit. Rev. Anal. Chem. 2017, 47, 325–331. [Google Scholar] [CrossRef]
- Chen, F.F.; Ma, R.J.; Chen, X.L. Advances of metabolomics in fungal pathogen-plant interactions. Metabolites 2019, 9, 169. [Google Scholar] [CrossRef]
- Kang, D.; Kim, J.; Choi, J.N.; Liu, K.; Lee, C.H. Chemotaxonomy of Trichoderma spp. using mass spectrometry-based metabolite profiling. J. Microbiol. Biotechnol. 2011, 21, 5–13. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Cubeta, M.A.; Jabaji, S. Chemotaxonomy of fungi in the Rhizoctonia solani species complex performing GC/MS metabolite profiling. Metabolomics 2013, 9, 159–169. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Faubert, D.; Jabaji, S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE 2014, 9, e111930. [Google Scholar] [CrossRef]
- Tenenboim, H.; Brotman, Y. Omic relief for the biotically stressed: Metabolomics of plant biotic interactions. Trends Plant Sci. 2016, 21, 781–791. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.; Piater, L.; Dubery, I.A. Metabolomics in plant priming research: The way forward. Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.G.T.; Allwood, J.W.; Galster, A.M.; Urban, M.; Daudi, A.; Canning, G.; Ward, J.L.; Beale, M.H.; Hammondkosack, K.E. A combined 1 h nuclear magnetic resonance and electrospray ionization-mass spectrometry analysis to understand the basal metabolism of plant-pathogenic fusarium spp. Plant Microbe Interact 2010, 23, 1605–1618. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.G.T.; Lord, M.; Rybak, K.; Trengove, R.D.; Oliver, R.P.; Solomon, P.S. Trehalose biosynthesis is involved in sporulation of Stagonospora nodorum. Fungal Genet. Biol. 2009, 46, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Wahl, R.; Horst, R.J.; Voll, L.M.; Usadel, B.; Poree, F.; Stitt, M.; Pons-Kühnemann, J.; Sonnewald, U.; Kahmann, R.; et al. Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2010, 56, 181–195. [Google Scholar] [CrossRef]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant–Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef]
- Rubert, J.; Righetti, L.; Stranska-Zachariasova, M.; Dzuman, Z.; Chrpova, J.; Dall’Asta, C.; Hajslova, J. Untargeted metabolomics based on ultra-high-performance liquid chromatography–high-resolution mass spectrometry merged with chemometrics: A new predictable tool for an early detection of mycotoxins. Food Chem. 2016, 224, 423–431. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Chukeatirote, E.; Bahkali, A.H.; Hyde, K.D. Pestalotiopsis-morphology, phylogeny, biochemistry and diversity. Fungal Divers. 2011, 50, 167–168. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Cai, L.; Chukeatirote, E.; Wu, W.P.; Sun, X.; Crous, P.W.; Bhat, D.J.; McKenzie, E.H.C.; Bahkali, A.H. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 2012, 56, 95–129. [Google Scholar] [CrossRef]
- Zheng, X.R.; Zhang MJChen, F.M. Occurrence of Pestalotiopsis lushanensis causing leaf blight on Buddhist pine in China. Eur. J. Plant Pathol. 2022, 162, 655–665. [Google Scholar] [CrossRef]
- Morales-Mora, L.A.; Martínez-Salgado, S.J.; Valencia de Ita, M.A.; Andrade-Hoyos, P.; Silva-Rojas, H.V.; Romero-Arenas, O. First Report of Leaf Spot and Anthracnosis Caused by Pestalotiopsis sp. on Strawberry in Puebla, Mexico. Plant Dis. 2019, 103, 2668. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Chen, F.; Chen, J.; Ye, J. First Report of Shoot Blight on Cryptomeria japonica Caused by Pestalotiopsis neglecta in China. Plant Dis. 2019, 103, 2140. [Google Scholar] [CrossRef]
- Qiu, L.; Liu, J.W.; Kuang, W.J.; Zhang, K.; Ma, J. First Report of Pestalotiopsis chamaeropis Causing Leaf Spot on Eurya nitida in China. Plant Dis. 2021, 106, 329. [Google Scholar] [CrossRef] [PubMed]
- Borrero, C.; Castaño, R.; Avilés, M. First Report of Pestalotiopsis clavispora (Neopestalotiopsis clavispora) Causing Canker and Twig Dieback on Blueberry Bushes in Spain. Plant Dis. 2018, 102, 1178. [Google Scholar] [CrossRef]
- Yuan, S.Q.; Wang, I.C.; Lei, L.; Hong, J.Y.; Yi, T.Y.; Hong, Y.Y. First Report of Pestalotiopsis microspora Causing Leaf Spot on Moyeam in China. Plant Dis. 2021, 106, 1996. [Google Scholar] [CrossRef]
- Nalin Rathnayake, G.R.; Savitri Kumar, N.; Jayasinghe, L.; Araya, H.; Fujimoto, Y. Secondary Metabolites Produced by an Endophytic Fungus Pestalotiopsis microspore. Nat. Prod. Bioprospect. 2019, 9, 411–417. [Google Scholar] [CrossRef]
- Xing, J.G.; Deng, H.Y.; Luo, D.Q. Two new compounds from an endophytic fungus Pestalotiopsis heterocornis. J. Asian Nat. Prod. Res. 2011, 13, 1069–1073. [Google Scholar] [CrossRef]
- Suryanarayan, T.S.; Kumaresan, V. Endophytic fungi of some halophytes from an estuarine mangrove forest. Mycol. Res. 2000, 104, 1465–1467. [Google Scholar] [CrossRef]
- Liu, C.M.; Wang, Y.L.; Wang, X.; Li, G.H. Active Metabolites from the Fungus Pestalotiopsis sp. YMF1.0474. Chem. Nat. Compd. 2021, 57, 257–259. [Google Scholar] [CrossRef]
- Song, R.Y.; Liu, Y.; Liu, R.H.; Wang, X.B.; Li, T.X.; Kong, L.Y.; Yang, M.H. Benzophenone derivatives from the plant endophytic fungus, Pestalotiopsis sp. Phytochem. Lett. 2017, 22, 189–193. [Google Scholar] [CrossRef]
- Liao, G.F.; Wu, P.; Liu, Z.F.; Xue, J.H.; Li, H.X.; Wei, X.Y. 2H-pyranone and isocoumarin derivatives from the endophytic fungus Pestalotiopsis microspora SC3082 derived from Scaevola taccada (Gaertn.) Roxb. Nat. Prod. Res. 2020, 35, 3644–3651. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.H.; Ye, H.Z.; Luo, M.J. Isolation and purification of Pf-toxin from Pestalotia funereal. Acta Phytopathol. Sin. 2003, 33, 541–545. [Google Scholar]
- Gao, F.; Chu, J.M.; Li, J.H.; Wang, M.L. Research Progress on Pathogenic Mechanism of Plant Pathogenic Fungi. Jiangsu J. Agric. 2014, 30, 1174–1179. [Google Scholar]
- Zhao, L.; Zhang, T.Y. Degrading enzymes produced by plant pathogens and their functions. Microbiol. Bull. 2002, 29, 89–93. [Google Scholar]
- Cano-Canchola, C.; Acevedo, L.; Ponce-Noyola, P.; Flores-Martinez, A.; Flores-Carreon, A.; Leal-Morales, C.A. Induction of Lytic Enzymes by the Interaction of Ustilago maydis with Zea mays Tissues. Fungal Genet. Biol. 2000, 29, 145–151. [Google Scholar] [CrossRef]
- Siewers, V.; Viaud, M.; Jimenez-Teja, D.; Collado, I.G.; Gronover, C.S.; Pradier, J.M.; Tudzynski, B.; Tudzynski, P. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol. Plant-Microbe Interact. 2005, 18, 602–612. [Google Scholar] [CrossRef]
- Zhang, Y.S. Plant Pathophysiology; Jiangsu Science and Technology Press: Nanjing, China, 1993. [Google Scholar]
- Qi, G.F.; Yang, B.; Ye, J.R. Advances in research on phytopathogenic mycotoxins. J. Nanjing For. Univ. 2000, 24, 66–70. [Google Scholar]
- Chen, L.Q.; Yang, J.H.; Wang, H.Y.; Yang, X.L.; Zhang, C.K.; Zhao, Z.H.; Wang, J.H. NX toxins: New threat posed by Fusarium graminearum species complex. Trends Food Sci. Technol. 2022, 119, 179–191. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC–MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.L.; Zeng, C.W.; Liu, S.Q. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.Z.; Qiu, H.Z.; Hu, X.Y.; Luo, A.H. Identification of root exudates from continuous cropping potato and its effect on Fusarium oxysporum. Desert China 2021, 41, 7–15. [Google Scholar]
- Li, W.J.; Yang, B.; Xu, J.Y.; Peng, L.L.; Sun, S.; Huang, Z.B.; Jiang, X.H.; He, Y.Q.; Wang, Z.F. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice. Plant J. Cell Mol. Biol. 2021, 108, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, A.V. Palmitic acid and its role in the structure and functions of plant cell membranes. Russ. J. Plant Physiol. 2015, 62, 706–713. [Google Scholar] [CrossRef]
- Garbe, E.; Vylkova, S. Correction to: Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi. Curr. Clin. Microbiol. Rep. 2019, 6, 120. [Google Scholar] [CrossRef]
- Lu, S.W. Genome-Wide Analysis of Small Secreted Cysteine-Rich Proteins Identifies Candidate Effector Proteins Potentially Involved in Fusarium graminearum Wheat Interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef]
- Zeng, R.; Gao, S.G.; Xu, L.H.; Liu, X.; Dai, F.M. Prediction of pathogenesis-related secreted proteins from Stemphylium lycopersici. BMC Microbiol. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Dong, X.; Ling, N.; Wang, M.; Shen, Q.R.; Guo, S.W. Fusaric acid is a crucial factor in the disturbance of leaf water imbalance in Fusarium-infected banana plants. Plant Physiol. Biochem. 2012, 60, 171–179. [Google Scholar] [CrossRef]
- Li, J.L.; Luo, J.; Shan, H.Y.; Cheng, J.L.; Chen, D.X.; An, D.R. Causal agent of tobacco mould in Shaanxi Province and preventive effects of Trametes versicolor polysaccharides on the disease. Acta Fungi 2013, 32, 168–178. [Google Scholar]
- Hu, Z.H.; Chang, X.N.; Dai, F.; Wu, J.C.; Liu, P.F. Metabolomics analysis of Botrytis cinerea based on GC-MS. Anal. Test. J. 2017, 36, 633–639. [Google Scholar]
- Cutler, H.G.; Jacyno, J.M.; Harwood, J.S.; Dulik, D.; Goodrich, P.D.; Roberts, R.G. Botcinolide: A biologically active natural product from Botrytis cinereal. Biosci. Biotech. Biochem. 1993, 57, 1980–1982. [Google Scholar] [CrossRef]
- Movahedi, S.; Heale, J.B. The role of aspartic proteinase and endo-pectinlyaseenzymes in the primary stages of infection and pathogenesis of various host tissues by different isolates of Botrytis cinerea Ex Pers. Physiol. Mol. Plant Pathol. 1990, 36, 303–324. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, L.; Wang, Y.; Chen, Y.Y.; Hu, K.M.; Yang, W.; Zuo, S.M.; Xu, J.D.; Kang, Z.S.; Xiao, X.Q.; et al. A High-Quality Genome of Rhizoctonia solani, a Devastating Fungal Pathogen with a Wide Host Range. Mol. Plant-Microbe Interact. MPMI 2022, 35, MPMI06220126A. [Google Scholar] [CrossRef] [PubMed]
- Dauda, W.P.; Singh Rana, V.; Solanke, A.U.; Krishnan, G.; Bashya, B.M.; Aggarwal, R.; Shanmugam, V. Metabolomic analysis of sheath blight disease of rice (Oryza sativa L.) induced by Rhizoctonia solani phytotoxin. J. Appl. Microbiol. 2022, 133, 3215–3227. [Google Scholar] [CrossRef]
- Wang, S.W.; Liang, H.; Wei, Y.; Zhang, P.H.; Dang YJLi GHZhang Shi, H. Alternative Splicing of MoPTEN Is Important for Growth and Pathogenesis in Magnaporthe oryzae
. Front. Microbiol. 2021, 12, 715773. [Google Scholar]
- Dang, Y.J.; Wei, Y.; Batool, W.; Sun, X.C.; Li, X.Q.; Zhang, S.H. Contribution of the Mitochondrial Carbonic Anhydrase (MoCA1) to Conidiogenesis and Pathogenesis in Magnaporthe oryzae. Front. Microbiol. 2022, 13, 845570. [Google Scholar] [CrossRef]
- Liu, H.T.; Ji, Y.; Liu, Y.; Tian, S.H.; Gao, Q.H.; Zou, X.H.; Yang, J.; Dong, C.; Tan, J.H.; Ni, D.A.; et al. The sugar transporter system of strawberry: Genome-wide identification and expression correlation with fruit soluble sugar-related traits in a Fragaria×ananassa germplasm collection. Hortic. Res. 2020, 7, 132. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Zhao, H.; Liu, Y.H. Expression of Invertase and Sugar Transporter Genes in Both Tomato and Botrytis cinerea during the Development of Grey Mold. J. Anhui Agric. Sci. 2021, 49, 101–107+124. [Google Scholar]
- Luo, L.F.; Jiang, B.B.; Deng, L.M.; Mei, X.Y.; Huang, H.C.; Liu, Y.X.; Yang, M.; Zhu, S.S. Effects of some components in root exudates of Panax noto-ginseng (Bark)F. H. Chen on the growth of root rot pathogens. J. South. Agric. 2020, 51, 2952–2961. [Google Scholar]
- Li, P.; Li, Z.; Li, D.; Li, J. Report of Leaf Spot on Pueraria lobata Caused by Bipolaris sorokiniana in China. Plant Dis. 2019, 103, 1040. [Google Scholar] [CrossRef]
- Phan, C.S.; Li, H.; Kessler, S.; Solomon, P.S.; Piggott, A.M.; Chooi, Y.H. Bipolenins K-N: New sesquiterpenoids from the fungal plant pathogen Bipolaris sorokiniana. Beilstein J. Org. Chem. 2019, 15, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Soulie, M.C.; Koka, S.M.; Floch, K.; Vancostenoble, B.; Barbe, D.; Daviere, A.; Soubigou, T.L.; Brunaud, V.; Poussereau, N.; Loisel, E.; et al. Plant nitrogen supply affects the Botrytis cinerea infection process and modulates known and novel virulence factors. Mol. Plant Pathol. 2020, 21, 1436–1450. [Google Scholar] [CrossRef] [PubMed]
- Adachi, T.; Inagaki, K. Phytotoxin producued by Rhizoctonia oryzae Ryker et Gooch. Agric. Biol. Chem. 1998, 52, 2625. [Google Scholar] [CrossRef][Green Version]




| Sample Group | Group Name | Number of Samples in the Group | Sample Status |
|---|---|---|---|
| 1 | PKP3 | 6 | solid |
| 2 | PKZ3 | 6 | solid |
| 3 | PKP6 | 6 | solid |
| 4 | PKZ6 | 6 | solid |
| 5 | PKP15 | 6 | solid |
| 6 | PKZ15 | 6 | solid |
| 7 | QC | 5 | liquid |
| Sample Comparison Group | Ion Mode | A | R2Y (cum) | Q2Y (cum) | R2 Intercept | Q2 Intercept |
|---|---|---|---|---|---|---|
| PKP3 vs. PKZ3 | + | 2 | 0.94 | 0.75 | 0.81 | −0.92 |
| PKP3 vs. PKZ3 | − | 2 | 0.91 | 0.67 | 0.76 | −0.98 |
| PKP6 vs. PKZ6 | + | 2 | 0.99 | 0.82 | 0.88 | −0.88 |
| PKP6 vs. PKZ6 | − | 2 | 0.99 | 0.79 | 0.89 | −0.8 |
| PKP15 vs. PKZ15 | + | 2 | 0.97 | 0.86 | 0.75 | −0.89 |
| PKP15 vs. PKZ15 | − | 2 | 0.95 | 0.82 | 0.74 | −1.04 |
| PKP3 vs. PKP6 | + | 2 | 0.98 | 0.89 | 0.81 | −0.97 |
| PKP3 vs. PKP6 | − | 2 | 0.96 | 0.83 | 0.78 | −0.99 |
| PKP3 vs. PKP15 | + | 2 | 0.99 | 0.98 | 0.73 | −1.32 |
| PKP3 vs. PKP15 | − | 2 | 0.98 | 0.94 | 0.71 | −1.17 |
| PKP6 vs. PKP15 | + | 2 | 0.99 | 0.96 | 0.8 | −1.08 |
| PKP6 vs. PKP15 | − | 2 | 0.98 | 0.9 | 0.78 | −1.2 |
| PKZ3 vs. PKZ6 | + | 2 | 0.99 | 0.73 | 0.87 | −0.8 |
| PKZ3 vs. PKZ6 | − | 2 | 0.96 | 0.65 | 0.85 | −0.79 |
| PKZ3 vs. PKZ15 | + | 2 | 1 | 0.96 | 0.84 | −0.74 |
| PKZ3 vs. PKZ15 | − | 2 | 0.99 | 0.93 | 0.81 | −0.85 |
| PKZ6 vs. PKZ15 | + | 2 | 1 | 0.9 | 0.89 | −0.8 |
| PKZ6 vs. PKZ15 | − | 2 | 0.99 | 0.91 | 0.84 | 0.81 |
| Reagent | Concentration (μg/mL) | ||||
|---|---|---|---|---|---|
| 20 | 40 | 60 | 80 | 100 | |
| 3,5-Dimethoxybenzoic acid | 17.08 ± 0.67 aE | 22.85 ± 0.81 aD | 27.25 ± 0.46 aC | 33.43 ± 0.81 aB | 38.15 ± 0.91 aA |
| S-(5-Adenosy)-l-homocysteine | 16.25 ± 0.35 aE | 21.83 ± 0.51 aD | 24.63 ± 0.31 bC | 32.70 ± 0.63 aB | 37.13 ± 0.25aA |
| 2-(1H-Indol-3-yl)acetic acid | 3.10 ± 0.14 bD | 3.48 ± 0.09 bC | 4.23 ± 0.11 cB | 4.50 ± 0.15 bB | 5.48 ± 0.23 bA |
| l-Glutamic acid | 5.83 ± 0.15 cD | 6.80 ± 0.15 cC | 8.05 ± 0.13 dB | 8.50 ± 0.09 cB | 10.53 ± 0.33 cA |
| 2-(2-acetyl-3,5-dihydroxyphenyl)acetic acid | 1.70 ± 0.13 dD | 2.13 ± 0.06 dC | 2.35 ± 0.06 eB | 2.93 ± 0.09 dB | 3.48 ± 0.18 dA |
| l-Phenylalanine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| N-Acetyl-l-phenylalanine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| 3-Nitro-l-tyrosine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| N-Acetylhistidine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| 2-Aminobenzenesulfonic acid | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| l-Tyrosine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| dl-Lysine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| 3-Methyl-2-oxobutanoic acid | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| Glycyl-l-leucine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| 3-hydroxy-3-methyl pentane dioic acid | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| dl-Tryptophan | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| dl-Arginine | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| 3,5-Dimethoxybenzoic acid | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA | 0.00 ± 0.00 fA | 0.00 ± 0.00 eA | 0.00 ± 0.00 eA |
| CK1 | 0.00 ± 0.00 | ||||
| CK2 | 35.18 ± 1.89 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Guo, H.; Liu, H.; Yu, J.; Li, S.; Zhu, T.; Owusu, A.M.; Li, S. Differential Metabolomics Reveals Pathogenesis of Pestalotiopsis kenyana Causing Leaf Spot Disease of Zanthoxylum schinifolium. J. Fungi 2022, 8, 1208. https://doi.org/10.3390/jof8111208
Liu C, Guo H, Liu H, Yu J, Li S, Zhu T, Owusu AM, Li S. Differential Metabolomics Reveals Pathogenesis of Pestalotiopsis kenyana Causing Leaf Spot Disease of Zanthoxylum schinifolium. Journal of Fungi. 2022; 8(11):1208. https://doi.org/10.3390/jof8111208
Chicago/Turabian StyleLiu, Chang, Haiyao Guo, Han Liu, Jiawen Yu, Shuying Li, Tianhui Zhu, Adjei Mark Owusu, and Shujiang Li. 2022. "Differential Metabolomics Reveals Pathogenesis of Pestalotiopsis kenyana Causing Leaf Spot Disease of Zanthoxylum schinifolium" Journal of Fungi 8, no. 11: 1208. https://doi.org/10.3390/jof8111208
APA StyleLiu, C., Guo, H., Liu, H., Yu, J., Li, S., Zhu, T., Owusu, A. M., & Li, S. (2022). Differential Metabolomics Reveals Pathogenesis of Pestalotiopsis kenyana Causing Leaf Spot Disease of Zanthoxylum schinifolium. Journal of Fungi, 8(11), 1208. https://doi.org/10.3390/jof8111208





