Deletion of the col-26 Transcription Factor Gene and a Point Mutation in the exo-1 F-Box Protein Gene Confer Sorbose Resistance in Neurospora crassa
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strain and Growth Medium
2.2. Sequencing Analysis of dgr Mutants and Confirming of Gene Replacement of Double Deletion Mutants
2.3. Sensitivity to 2-Deoxyglucose and Mitochondrial Respiration Inhibitors
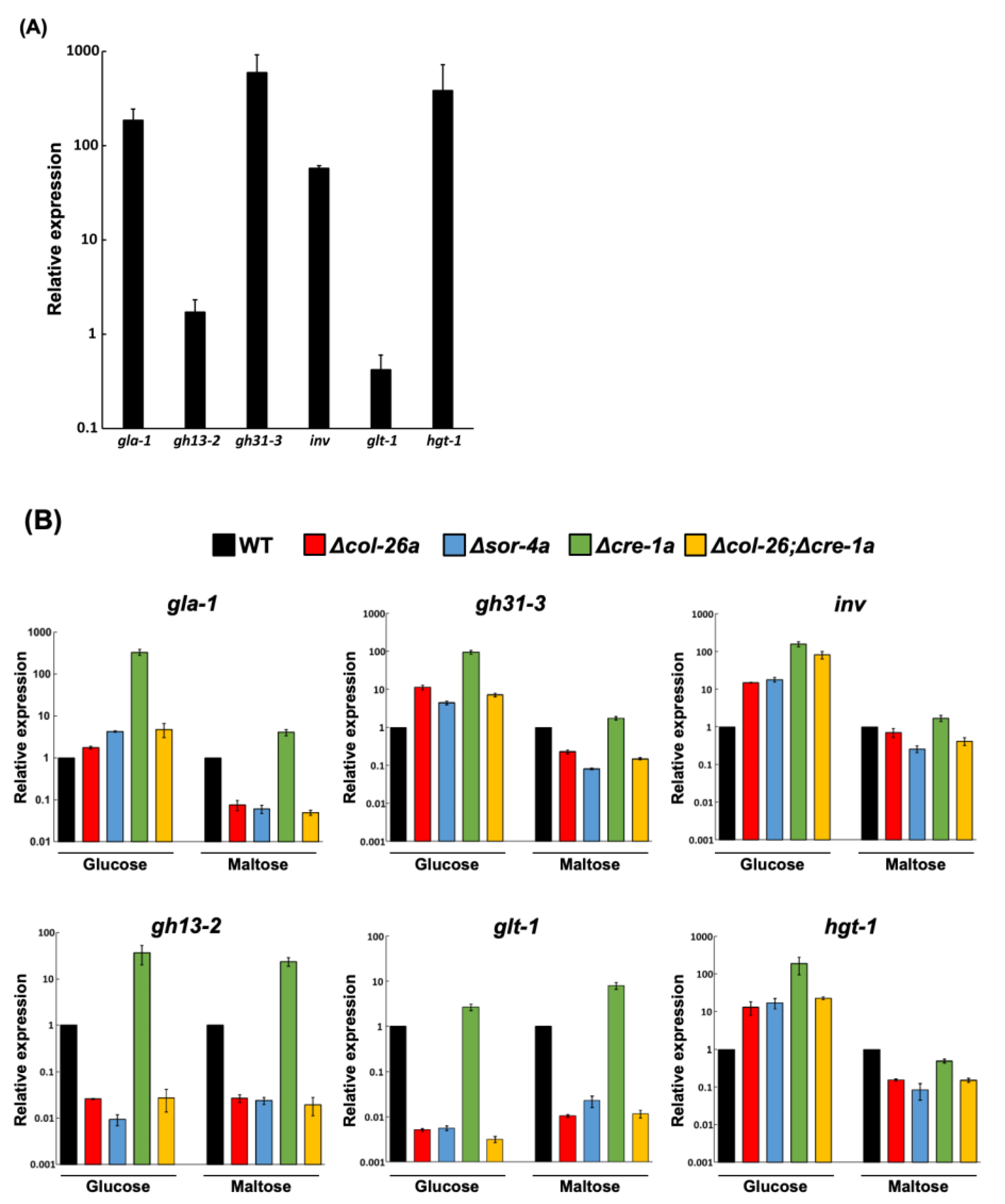
2.4. Gene Expression Analysis by qPCR
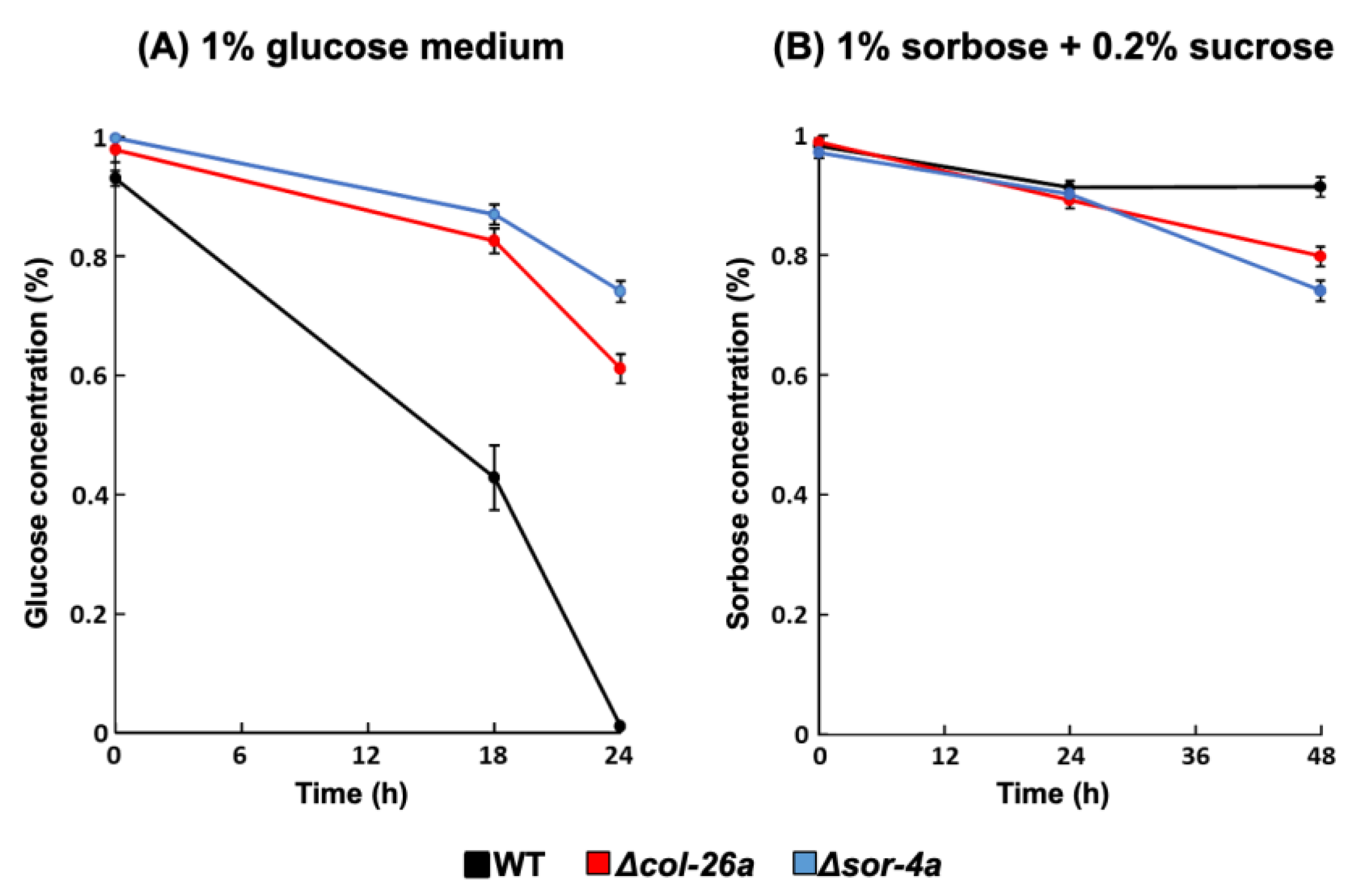
2.5. Glucose and Sorbose Consumption of Sorbose-Resistant Strains
2.6. SDS-PAGE Analysis of Extracellular Proteins
3. Results
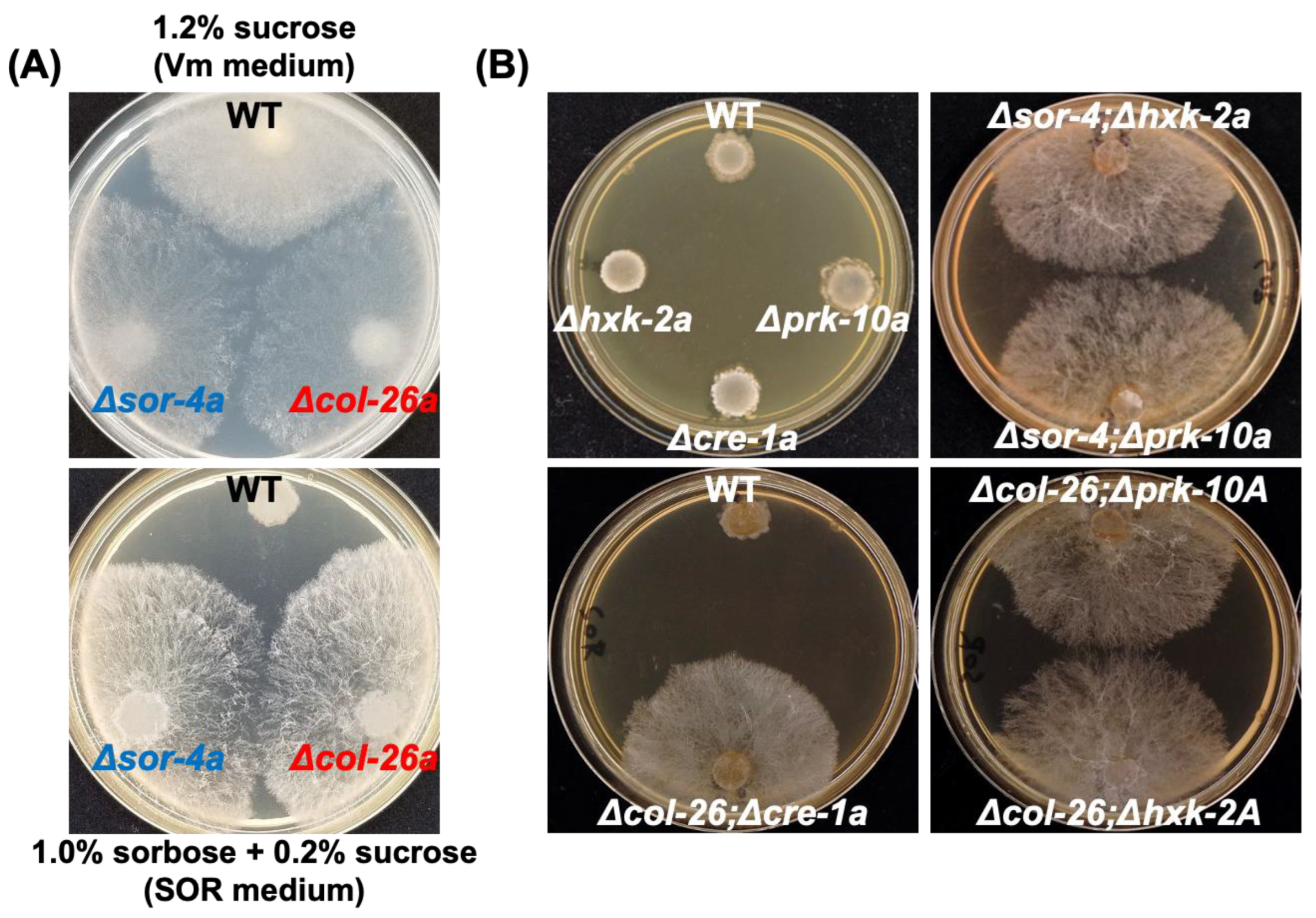
3.1. Loss of Function of Transcription Factor col-26/dgr-1 Confers Sorbose Resistance in N. Crassa
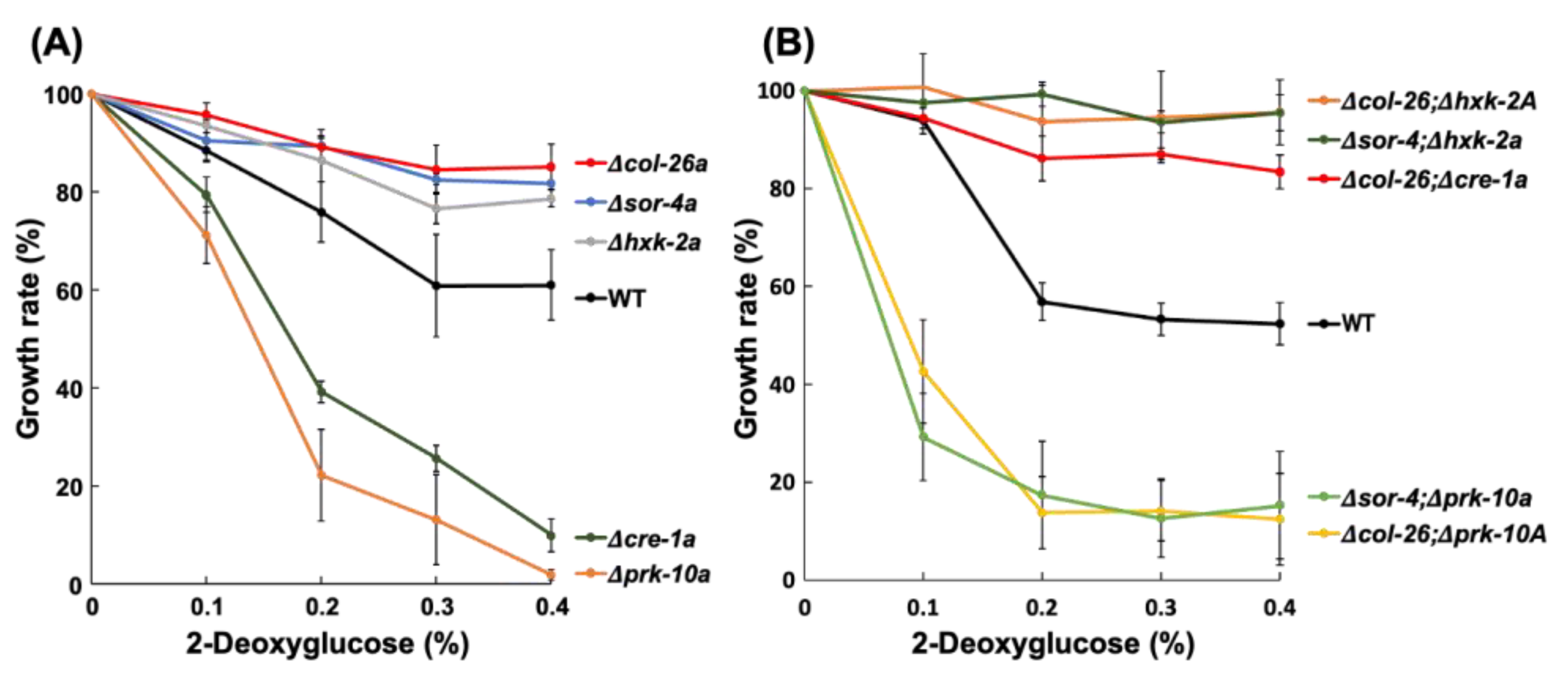
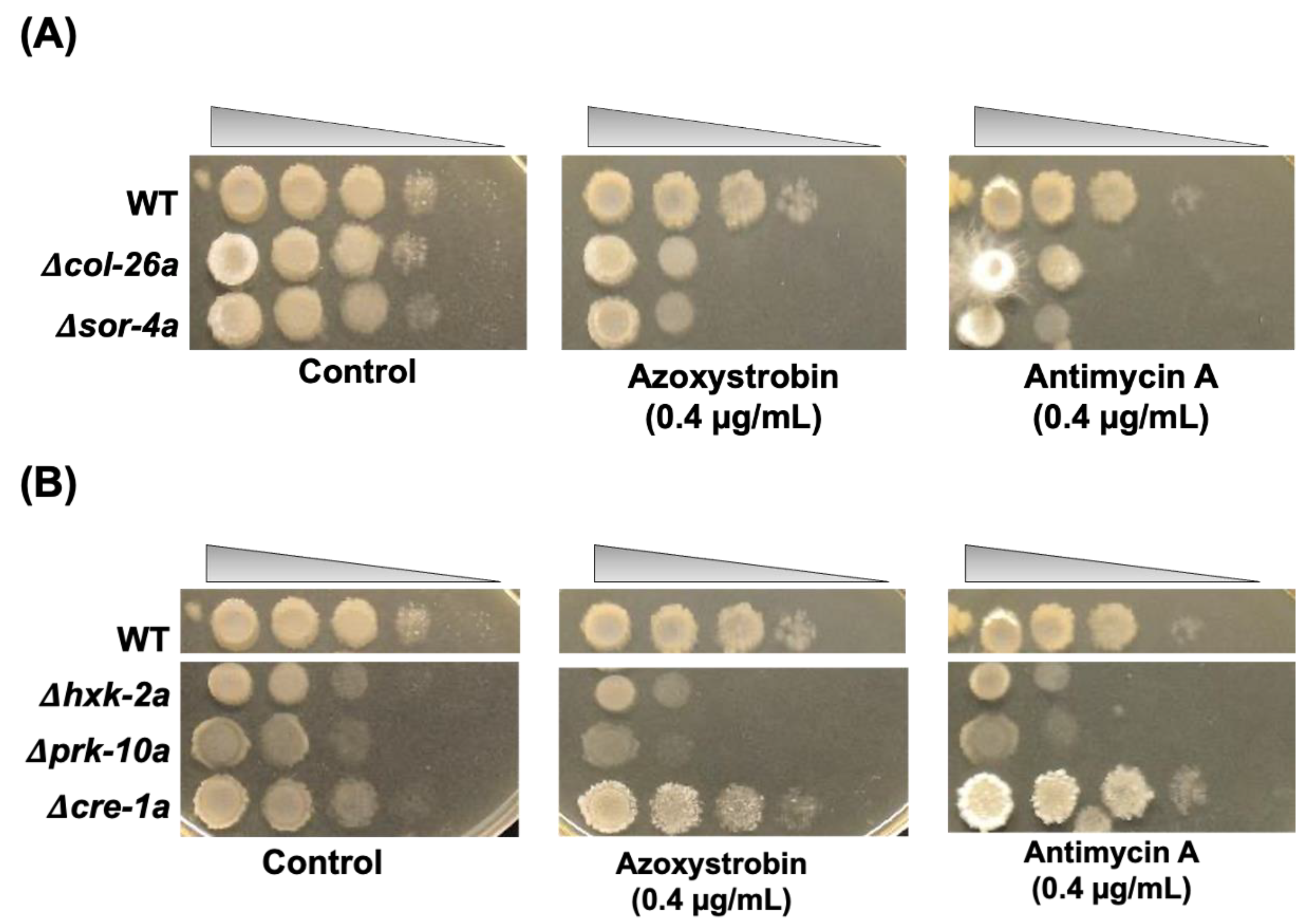
3.2. Sorbose Resistance, 2-DG Resistance, and QoI Sensitivity of CCR Mutants
3.3. Comparison of Sorbose and 2-DG Resistance and Gene Expressions of Double Mutants Isolated from Sorbose-Resistant Mutants and CCR Mutants
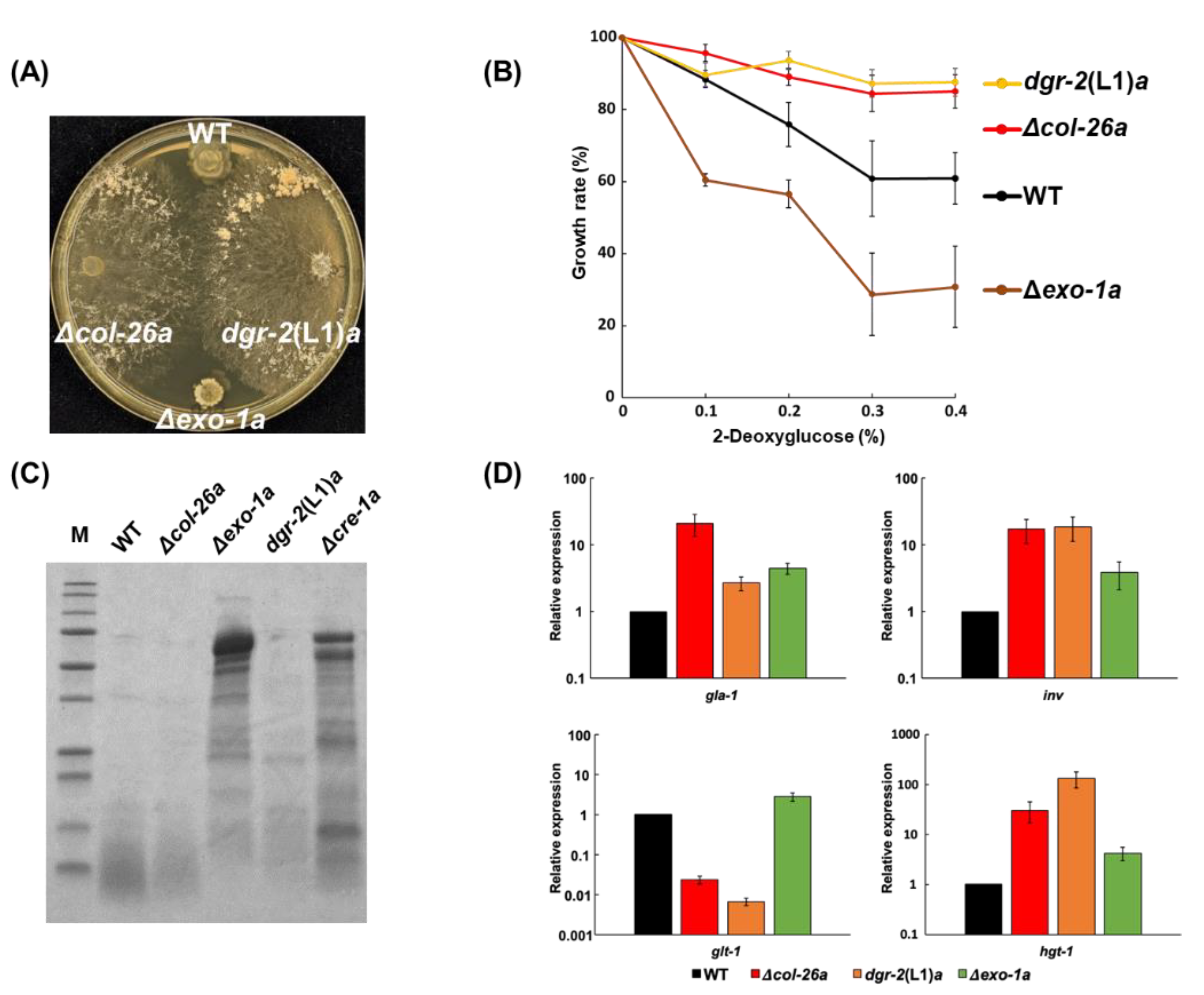
3.4. A Single S11L Mutation, but Not the Loss-of-function Mutation of F-box Protein exo-1, Confers Sorbose, and 2-DG Resistances in N. Crassa
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tatum, E.L.; Barratt, R.W.; Cutter, V.M. Chemical Induction of Colonial Paramorphs in Neurospora and Syncephalastrum. Science 1949, 109, 509–511. [Google Scholar] [CrossRef] [PubMed]
- De Terra, N.; Tatum, E.L. Colonial growth of Neurospora. Sorbose and enzymes alter the composition of the cell wall and induce morphological changes. Science 1961, 134, 1066–1068. [Google Scholar] [CrossRef]
- Mahadevan, P.R.; Tatum, E.L. Relationship of the Major Constituents of the Neurospora crassa Cell Wall to Wild-type and Colonial Morphology. J. Bacteriol. 1965, 90, 1073–1081. [Google Scholar] [CrossRef]
- Crocken, B.; Tatum, E.L. The effect of sorbose on metabolism and morphology of Neurospora. Biochim. Biophys. Acta 1968, 156, 1–8. [Google Scholar] [CrossRef]
- Mishra, N.C.; Tatum, E.L. Effect of L-sorbose on Polysaccharide Synthetases of Neurospora crassa (glycogen- -1,3-glucan-morphology-cell wall-digitonin-particulate enzymes). Proc. Natl. Acad. Sci. USA 1972, 69, 313–317. [Google Scholar] [CrossRef]
- Kamei, M.; Yamashita, K.; Takahashi, M.; Fukumori, F.; Ichiishi, A.; Fujimura, M. Involvement of MAK-1 and MAK-2 MAP kinases in cell wall integrity in Neurospora crassa. Biosci. Biotechnol. Biochem. 2016, 80, 1843–1852. [Google Scholar] [CrossRef]
- Hatamoto, M.; Aizawa, R.; Kobayashi, Y.; Fujimura, M. A novel fungicide aminopyrifen inhibits GWT-1 protein in glycosylphosphatidylinositol-anchor biosynthesis in Neurospora crassa. Pestic. Biochem. Physiol. 2019, 156, 1–8. [Google Scholar] [CrossRef]
- Elorza, M.V.; Arst, H.N. Sorbose resistant mutants of Aspergillus nidulans. Mol. Gen. Genet. 1971, 111, 185–193. [Google Scholar] [CrossRef]
- MacCabe, A.P.; Miró, P.; Ventura, L.; Ramon, D. Glucose uptake in germinating Aspergillus nidulans conidia: Involvement of the creA and sorA genes. Microbiology 2003, 149, 2129–2136. [Google Scholar] [CrossRef][Green Version]
- Forment, J.V.; Flipphi, M.; Ventura, L.; Gonzalez, R.; Ramón, D.; Maccabe, A.P. High-Affinity Glucose Transport in Aspergillus nidulans is Mediated by the Products of Two Related but Differentially Expressed Genes. PLoS ONE 2014, 9, e94662. [Google Scholar] [CrossRef]
- Klingmuller, W. Crossing analysis of sorbose resistant mutants of Neurospora crassa. Mol. Gen. Genet. 1967, 100, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Quigley, D.R. Sorbose-resistant mutants of Neurospora crassa do not have altered beta (1-3)glucan synthase activity. Curr. Microbiol. 1987, 15, 185–192. [Google Scholar] [CrossRef]
- Allen, K.E.; McNally, M.T.; Lowendorf, H.S.; Slayman, C.W.; Free, S.J. Deoxyglucose-resistant mutants of Neurospora crassa: Isolation, mapping, and biochemical characterization. J. Bacteriol. 1989, 171, 53–58. [Google Scholar] [CrossRef]
- Madi, L.; McBride, S.A.; Bailey, L.A.; Ebbole, D.J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 1997, 146, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.; Radford, A.; Sachs, M. The Neurospora Compendium Chromosomal Loci; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Ozcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 2566–2573. [Google Scholar] [CrossRef]
- Polish, J.A.; Kim, J.H.; Johnston, M. How the Rgt1 Transcription Factor of Saccharomyces cerevisiae Is Regulated by Glucose. Genetics 2005, 169, 583–594. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef]
- Tu, J.; Carlson, M. The GLC7 type 1 Protein Phosphatase is Required for Glucose Repression in Saccharomyces cerevisiae. Mol. Cell Biol. 1994, 14, 6789–6796. [Google Scholar] [CrossRef]
- Ahuatzi, D.; Riera, A.; Pela Ez, R.; Herrero, P.; Moreno, F. Hxk2 Regulates the Phosphorylation State of Mig1 and Therefore Its Nucleocytoplasmic Distribution. J. Biol. Chem. 2007, 282, 4485–4493. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Gligoris, T.; Tzamarias, D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 2004, 5, 368–372. [Google Scholar] [CrossRef]
- Tian, C.; Beeson, W.T.; Iavarone, A.T.; Sun, J.; Marletta, M.A.; Cate, J.H.; Glass, N.L. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. USA 2009, 106, 22157–22162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Dowzer, C.E.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell Biol. 1991, 11, 5701–5709. [Google Scholar] [PubMed]
- Prathumpai, W.; McIntyre, M.; Nielsen, J. The effect of CreA in glucose and xylose catabolism in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2004, 63, 748–753. [Google Scholar] [CrossRef]
- Nakamura, T.; Maeda, Y.; Tanoue, N.; Makita, T.; Kato, M.; Kobayashi, T. Expression Profile of Amylolytic Genes in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2006, 70, 2363–2370. [Google Scholar] [CrossRef]
- Cubero, B.; Scazzocchio, C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994, 13, 407–415. [Google Scholar] [CrossRef]
- Strauss, J.; Mach, R.L.; Zeilinger, S.; Hartler, G.; Stoffler, G.; Wolschek, M.; Kubicek, C.P. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 1995, 376, 103–107. [Google Scholar] [CrossRef]
- Sun, J.; Glass, N.L. Identification of the cre-1 Cellulolytic Regulon in Neurospora crassa. PLoS ONE 2011, 6, e25654. [Google Scholar] [CrossRef]
- Ziv, C.; Gorovits, R.; Yarden, O. Carbon source affects PKA-dependent polarity of Neurospora crassa in a cre-1-dependent and independent manner. Fungal Genet. Biol. 2008, 45, 103–116. [Google Scholar] [CrossRef]
- Gomi, K.; Akeno, T.; Minetoki, T.; Ozeki, K.; Kumagai, C.; Okazaki, N.; Iimura, Y. Molecular Cloning and Characterization of a Transcriptional Activator Gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2000, 64, 816–827. [Google Scholar] [CrossRef]
- Nitta, M.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates β-glucosidase expression in Trichoderma reesei. Fungal Genet. Biol. 2012, 49, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sun, J.; Glass, N.L. VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, is Essential for Plant Cell Wall Degradation by Neurospora crassa. PLoS Genet. 2014, 10, e1004500. [Google Scholar] [CrossRef]
- Xiong, Y.; Wu, V.W.; Lubbe, A.; Qin, L.; Deng, S.; Kennedy, M.; Bauer, D.; Singan, V.R.; Barry, K.; Northen, T.R.; et al. A fungal transcription factor essential for starch degradation affects integration of carbon and nitrogen metabolism. PLoS Genet. 2017, 13, e1006737. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Q.; Lin, L.; Li, X.; Zhang, Y.; Tian, C. RCO-3 and col-26 form an external-to-internal module that regulates the dual-affinity glucose transport system in Neurospora crassa. Biotechnol. Biofuels 2021, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; Thieme, N.; Liu, Q.; Li, F.; Meyer, L.T.; Harth, S.; Jecmenica, M.; Ramamurthy, M.; Gorman, J.; Simmons, B.A.; et al. The F-box protein gene exo-1 is a target for reverse engineering hypersecretion in filamentous fungi. Proc. Natl. Acad. Sci. USA 2021, 118, e2025689118. [Google Scholar] [CrossRef]
- Duyvesteijn, R.G.; van Wijk, R.; Boer, Y.; Rep, M.; Cornelissen, B.J.; Haring, M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005, 57, 1051–1063. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Vogel, H.J. Distribution of Lysine Pathways Amomg Fungi: Evolutionary Implications. Am. Nat. 1964, 98, 435–446. [Google Scholar] [CrossRef]
- Noguchi, R.; Banno, S.; Ichikawa, R.; Fukumori, F.; Ichiishi, A.; Kimura, M.; Yamaguchi, I.; Fujimura, M. Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genet. Biol. 2007, 44, 208–218. [Google Scholar] [CrossRef]
- Yamashita, K.; Shiozawa, A.; Watanabe, S.; Fukumori, F.; Kimura, M.; Fujimura, M. ATF-1 transcription factor regulates the expression of ccg-1 and cat-1 genes in response to fludioxonil under OS-2 MAP kinase in Neurospora crassa. Fungal Genet. Biol. 2008, 45, 1562–1569. [Google Scholar] [CrossRef]
- Katano, H.; Takakuwa, M.; Itoh, T.; Hibi, T. Colorimetric determination of fructose for the high-throughput microtiter plate assay of glucose isomerase. Biosci. Biotechnol. Biochem. 2015, 79, 1057–1060. [Google Scholar] [CrossRef] [PubMed]
- McCartney, R.R.; Chandrashekarappa, D.G.; Zhang, B.B.; Schmidt, M.C. Genetic analysis of resistance and sensitivity to 2-deoxyglucose in Saccharomyces cerevisiae. Genetics 2014, 198, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Gratzner, H.G. Cell Wall Alterations Associated with the Hyperproduction of Extracellular Enzymes in Neurospora crassa. J. Bacteriol. 1972, 111, 443–446. [Google Scholar] [CrossRef] [PubMed]

| Strain | Gene Locus | Genotype (Allele) | FGSC No. | Reference |
|---|---|---|---|---|
| wild type | mat a (ORS-SL6) | 4200 | FGSC a | |
| Δcol-26a | NCU07788 | mat a; col-26::Hyg r | 11030 | FGSC a |
| Δcol-26A | NCU07788 | mat A; col-26::Hyg r | 11031 | FGSC a |
| Δsor-4a | NCU02582 | mat a; sor-4::Hyg r | 17928 | FGSC a |
| sor-4(DS(r))A | NCU02582 | mat A; sor-4(DS(r)) | 1741 | FGSC a |
| Δhxk-2a | NCU00575 | mat a; hxk-2::Hyg r | 15921 | FGSC a |
| Δhxk-2A | NCU00575 | mat A; hxk-2::Hygr | 15920 | FGSC a |
| Δprk-10a | NCU04566 | mat a; prk-10::Hyg r | 12420 | FGSC a |
| Δprk-10A | NCU04566 | mat A; prk-10::Hyg r | 12421 | FGSC a |
| cre-1 het | NCU08807 | mat a; cre-1::Hyg r (heterokaryon) | 18633 | FGSC a |
| Δcre-1a | NCU08807 | mat a; cre-1::Hyg r | this study b | |
| Δexo-1a | NCU09899 | mat a; exo-1::Hyg r | 19860 | FGSC a |
| dgr-1(BE52)A | mat A; dgr-1(BE52) | 4326 | FGSC a | |
| dgr-4(KHY7)a | mat a; dgr-4(KHY7) | 8287 | FGSC a | |
| dgr-2(L1)a | mat a; dgr-2(L1) | 4328 | FGSC a | |
| dgr-2(L5)A | mat A; dgr-2(L5) | 4329 | FGSC a | |
| Δcol-26; Δhxk-2A | mat A; col-26::Hyg r; hxk-2::Hyg r | this study b | ||
| Δcol-26; Δprk-10A | mat A; col-26::Hyg r; prk-10::Hyg | this study b | ||
| Δcol-26; Δcre-1a | mat a; col-26::Hyg r; cre-1::Hyg r | this study b | ||
| Δsor-4; Δhxk-2a | mat a; sor-4::Hyg r; prk-10::Hyg r | this study b | ||
| Δsor-4; Δprk-10a | mat a; sor-4::Hyg r; hxk-2::Hyg r | this study b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirai, K.; Idemoto, T.; Kato, S.; Ichiishi, A.; Fukumori, F.; Fujimura, M. Deletion of the col-26 Transcription Factor Gene and a Point Mutation in the exo-1 F-Box Protein Gene Confer Sorbose Resistance in Neurospora crassa. J. Fungi 2022, 8, 1169. https://doi.org/10.3390/jof8111169
Hirai K, Idemoto T, Kato S, Ichiishi A, Fukumori F, Fujimura M. Deletion of the col-26 Transcription Factor Gene and a Point Mutation in the exo-1 F-Box Protein Gene Confer Sorbose Resistance in Neurospora crassa. Journal of Fungi. 2022; 8(11):1169. https://doi.org/10.3390/jof8111169
Chicago/Turabian StyleHirai, Kenshi, Takuya Idemoto, Shiho Kato, Akihiko Ichiishi, Fumiyasu Fukumori, and Makoto Fujimura. 2022. "Deletion of the col-26 Transcription Factor Gene and a Point Mutation in the exo-1 F-Box Protein Gene Confer Sorbose Resistance in Neurospora crassa" Journal of Fungi 8, no. 11: 1169. https://doi.org/10.3390/jof8111169
APA StyleHirai, K., Idemoto, T., Kato, S., Ichiishi, A., Fukumori, F., & Fujimura, M. (2022). Deletion of the col-26 Transcription Factor Gene and a Point Mutation in the exo-1 F-Box Protein Gene Confer Sorbose Resistance in Neurospora crassa. Journal of Fungi, 8(11), 1169. https://doi.org/10.3390/jof8111169





