Abstract
L-Sorbose induces hyperbranching of hyphae, which results in colonial growth in Neurospora crassa. The sor-4 gene, which encodes a glucose sensor that acts in carbon catabolite repression (CCR), has been identified as a sorbose resistance gene. In this study, we found that the deletion mutant of col-26, which encodes an AmyR-like transcription factor that acts in CCR, displayed sorbose resistance. In contrast, the deletion mutants of other CCR genes, such as a hexokinase (hxk-2), an AMP-activated S/T protein kinase (prk-10), and a transcription factor (cre-1), showed no sorbose resistance. Double mutant analysis revealed that the deletion of hxk-2, prk-10, and cre-1 did not affect the sorbose resistance of the col-26 mutant. Genes for a glucoamylase (gla-1), an invertase (inv), and glucose transporters (glt-1 and hgt-1) were highly expressed in the cre-1 mutant, even in glucose-rich conditions, but this upregulation was suppressed in the Δcre-1; Δcol-26a double-deletion mutant. Furthermore, we found that a dgr-2(L1)a mutant with a single amino-acid substitution, S11L, in the F-box protein exo-1 displayed sorbose resistance, unlike the deletion mutants of exo-1, suggesting that the function of exo-1 is crucial for the resistance. Our data strongly suggest that CCR directly participates in sorbose resistance, and that col-26 and exo-1 play important roles in regulating the amylase and glucose transporter genes during CCR.
1. Introduction
Filamentous fungi have elongated hyphae at their growing tips with hyphal branches, and undergo radial growth on agar medium. Sorbose, a rare sugar, exhibits toxic effects on several fungi, including Neurospora crassa. When 1% sorbose is added to medium in the presence of 0.2% sucrose, N. crassa propagates hyper-branched hyphae and forms compact colonies [1,2,3,4]. The formation of compact colonies in the presence of sorbose is a useful methodological tool for the isolation of mutants, allowing high-resolution genetic analyses and contributing to the establishment of N. crassa as a model organism for molecular genetics and biochemistry. Various studies have been conducted to identify the way in which sorbose induces such compact colonies; however, the underlying mechanisms remain largely unclear. Sorbose induces changes in the polysaccharide composition of the cell wall, such as a marked decrease in beta-1,3-glucan [3,4], possibly because a beta-1,3-glucan synthetase is inhibited by the sugar [5]. In contrast, a beta-1,3-glucan synthetase inhibitor, micafungin, and a GPI-anchor biosynthesis inhibitor, aminopyrifen, induced abnormal morphology on a sorbose medium in N. crassa mutant strains in the cell wall integrity MAP kinase genes mak-1 and mak-2, and chitin synthetase genes chs-5 and chs-7 [6,7]. These observations suggest that sorbose may disturb the synthesis of the fungal cell wall.
Several sorbose-resistant mutants have been isolated and characterized in fungi. Aspergillus nidulans sorA mutants show cross-resistance to a glucose analog, 2-deoxyglucose (2-DG), and are defective in sugar uptake [8]. The sorA gene has been shown to encode the high-affinity glucose transporter protein MstC [9,10]. In contrast, sorB mutation in A. nidulans did not confer 2-DG resistance, but instead led to a considerable reduction in phosphoglucomutase activity [8]. In N. crassa, six sorbose-resistant mutants (sor-1, sor-2, sor-3, sor-4, sor-5, and sor-6) have been isolated and characterized [11,12,13]. Among them, only the function of the sor-4 gene has been identified: it encodes an ortholog of Snf3 that acts as a low-affinity glucose sensor in Saccharomyces cerevisiae. In contrast, two other N. crassa mutations—dgr-3, which confers 2-DG resistance, and rco-3, which encodes a regulator for conidiation—are localized to sor-4. Therefore, the mutation is referred to as sor-4/DGR-3/RCO-3 [14,15]. In S. cerevisiae, the function of Snf3 is coupled with that of another glucose sensor, Rgt2, which transduces the glucose signal to Rgt1, the transcription regulator of several glucose transporters (Hxt) for adjusting HXT gene expression [16,17]. Furthermore, Snf3 and Rgt2 regulate the expression of carbon catabolite repression (CCR) genes [18]. Mig1, a Cys2–His2 type zinc-finger transcription factor, acts as a central regulator for CCR gene expression. In glucose-rich conditions, Mig1 is dephosphorylated by the Glc7 phosphatase complex and locates in the nucleus to repress Mig1-dependent genes [19]. A hexose kinase, Hxk2, interacts with Mig1 and regulates the phosphorylation status of Mig1 [20]. When cells are in glucose-depleted conditions, Mig1 is phosphorylated by an AMP-activated protein kinase Snf1, which relocates from the nucleus to the cytoplasm [21]; then, Mig1-dependent genes are derepressed and then expressed to adapt glucose starvation.
Although most fungi use glucose as a primary carbon source, some possess various deconstructing enzymes that hydrolyze polysaccharide for alternative carbon sources such as starch, cellulose, and hemicellulose [18,22,23]. Genes encoding these enzymes are generally regulated by the glucose concentration in their environment and are induced to utilize alternative carbon compounds when cells are exposed to glucose-starved conditions. It is well known that Mig1 ortholog transcription factors also act as a central regulator in filamentous fungi [18]. In Aspergillus sp., CreA, the ortholog of Mig1, controls gene expression associated with the utilization of alternative carbon compounds such as starch, arabinose, and xylose [24,25,26]. CreA from A. nidulans or cre-1 from Trichoderma reesei bind to the promoters of target genes to repress their expression via the consensus motif 5′-SYGGRG-3′ [27,28]. In N. crassa, the Mig1 ortholog cre-1 acts as a central regulator, and the loss of its function leads to the constitutive expression of cellulase and amylase genes, even under glucose nondepleting conditions [29,30]. The expression of polysaccharide-hydrolyzing enzymes such as amylase, cellulase, and hemi-cellulase in Aspergillus species is known to be regulated not only by CreA, but also by another transcriptional regulator, AmyR, a fungal-specific Zn(II)Cys6-type transcription factor [26,31]. The deletion mutant of amyR in A. oryzae exhibited growth defects on starch medium and decreased expression levels of alpha-glucosidases [31]. In T. reesei, the mutation of bglR, an ortholog of amyR, caused growth defects on maltose and starch media and the downregulation of a beta-glucosidase gene [32]. col-26, the ortholog of AmyR in N. crassa, is considered to play a crucial rule in carbon metabolism in the fungus [33,34]. The deletion mutant of col-26 presented a growth defect on various carbon sources, including glucose, fructose, and maltose. col-26 has been shown to be involved in the regulation of the expression of amylolytic and cellulolytic enzyme genes. Recently, Li et al. [35] reported that the transcription factor col-26 and the glucose sensor sor-4/DGR-3/RCO-3 regulated a low-affinity glucose transporter, glt-1, along with two high-affinity glucose transporters, hgt-1 and hgt-2 [35]. In N. crassa, mutants for a probable transcription factor exoamylase-1 (exo-1) were isolated as hypersecretion strains of alpha-amylase, glucoamylase, invertase, pectinase, and trehalase. Recently, Gabriel et al. [36] revealed that exo-1 encodes an F-box protein, and that exo-1 is identical to the 2-DG resistance gene dgr-2. Filamentous fungi tend to have many F-box proteins (39 F-box protein in N. crassa), but their function largely remains unclear. The frp-1 gene, an ortholog of exo-1, was indispensable for the pathogenicity of a plant pathogenic fungus Fusarium oxysporum. This result suggests that a SCF (Skp-Cullin-F-box) ubiquitin ligase complex is involved in the emergence of pathogenicity [37]. In N. crassa, high secretion of amylases and invertase in Δexo-1 was dependent on the transcriptional regulator col-26 but not cre-1 [36].
2. Materials and Methods
2.1. Fungal Strain and Growth Medium
The N. crassa strains used in this study are listed in Table 1. The gene deletion strains generated by the Neurospora Genome Project were obtained from the Fungal Genetics Stock Center (FGSC) [38]. As cre-1 het strain (FGSC#18633) from KO library was heterokaryon, homo Δcre-1a strains were obtained from crossing with wild-type strains. The Δcol-26a strain was crossed with Δhxk-2A and Δprk-10A to produce Δcol-26;Δhxk-2A and Δcol-26;Δprk-10A, respectively. The Δcol-26; Δcre-1a was obtained by crossing Δcol-26A with Δcre-1a. In the same way, the Δsor-4a strain was used to isolate the Δsor-4;Δhxk-2a and Δsor-4; Δprk-10a strains. The strains were grown on Vogel’s minimal agar medium containing 1.2% sucrose medium (Vm medium) for filamentous growth, and medium containing 1.0% sorbose and 0.2% sucrose (sor medium) for colonial growth at 25 °C [39]. Sorbose resistance was evaluated by filamentous growth on sor medium 4 days after conidia inoculation.

Table 1.
Strain list used in this study.
2.2. Sequencing Analysis of dgr Mutants and Confirming of Gene Replacement of Double Deletion Mutants
Genomic DNAs of the wild-type strain, dgr mutants, and double deletion mutants were isolated as described previously [7]. col-26 and hxk-2 genes in dgr-1 or dgr-4 mutants were amplified by PCR using primers, col-26_seq_F1 and col-26_seq_R4 for col-26 gene, and hxk-2_seq_F1 and hxk-2_seq_R4 for hxk-2 gene (Table S1). PCR products were purified using the QiAquick PCR Purification kit (Qiagen, Tokyo, Japan) and sequenced using sequencing primers listed in Table S1. For confirming of double mutants, Δcol-26;Δhxk-2A, Δcol-26; Δprk-10A, Δcol-26;Δcre-1a, Δsor-4;Δhxk-2a, and Δsor-4;Δprk-10a were used; deletion of each gene was confirmed by PCR analysis using primers sets shown in Table S2 (Figure S3).
2.3. Sensitivity to 2-Deoxyglucose and Mitochondrial Respiration Inhibitors
A glucose analog, 2-deoxyglucose (2-DG), and two mitochondrial complex III inhibitors, azoxystrobin and antimycin A, were purchased from FUJIFILM Wako Pure Chemical Corporation (Tokyo, Japan). To measure the sensitivity to 2-DG, mycelial disks that were precultured on Vogel’s minimal agar medium containing 1.0% fructose were transferred onto 1% fructose Vogel medium containing 2-DG (0.1% to 0.4%), and fungal growth was assessed after 18 h of cultivation at 25 °C. Azoxystrobin and antimycin A act as specific inhibitors of the mitochondrial respiratory chain by binding to the Qo and Qi sites of the cytochrome bc1 complex, respectively. To determine the sensitivity to azoxystrobin and antimycin A, a series of conidia suspensions (1 × 107 to 1 × 103 cells/mL) was spotted on sor medium containing azoxystrobin (0.4 mg/L) and antimycin A (0.4 mg/L), and colony formation was photographed after 2 or 5 days of cultivation at 25 °C.
2.4. Gene Expression Analysis by qPCR
Total RNA was isolated as previously described by Noguchi et al. [40]. The conidia (1.0 × 106 cells/mL) were inoculated and precultured in Vogel’s liquid medium containing 1.2% glucose for 22 h at 25 °C on a reciprocating shaker (135 rpm) and then, the growing hyphae were transferred to fresh Vogel’s medium containing glucose or maltose and cultured for 2 h at 25 °C. The resultant mycelia were harvested by filtration using an aspirator, and frozen in liquid nitrogen until RNA isolation. Total RNA was isolated from each sample using a FastRNA Pro Red kit (MP Biomedicals, Tokyo, Japan). Each RNA sample (1 µg of total RNA) was subjected to cDNA synthesis and quantitative real-time reverse transcription PCR (RT-qPCR) using the LightCycler system (Roche Diagnostics, Tokyo, Japan), as described by Yamashita et al. [41]. The mRNA expression of the target genes was quantified using Universal ProbeLibrary (Roche Diagnostics, Tokyo, Japan). Primers and probes used in this study are summarized in Table S3. The relative gene expression value was calculated by comparing the threshold cycle (Cp), with actin used as the reference gene in the RT-qPCR analysis. A minimum of three biological replicates were performed for each experiment.
2.5. Glucose and Sorbose Consumption of Sorbose-Resistant Strains
To measure glucose or sorbose consumption during liquid culture, the conidia (106 cells/mL) were inoculated in Vogel’s minimal liquid medium with 1% glucose or 1% sorbose + 0.2% glucose, and incubated at 25 °C. The culture filtrates were harvested after 18, 24, and 48 h. Glucose concentration in the medium was measured using the F-kit for glucose (J.K. International, Tokyo, Japan) in accordance with the manufacturer’s protocol. The sorbose concentration was measured using the Si–Mo method as described by Katano et al. [42]. To measure the sorbose concentration, 200 μL of culture filtrate was mixed with 800 μL of reaction buffer (6 mL of 1 M Na2MoO4 and 2 mL of 0.25 M Na2SiO3 were mixed and adjusted to pH 4.5 using 5 M HCl) and incubated 70 °C for 30 min. The absorbance of the resultant samples was measured at 750 nm. Sorbose concentration in the cultures were calculated from the standard curves. Three biological replicates were performed.
2.6. SDS-PAGE Analysis of Extracellular Proteins
Conidia (106 cells/mL) were cultured in Vogel’s minimal medium containing 2% glucose for 5 days on a reciprocating shaker (135 rpm) at 25 °C. The extracellular protein in culture filtrates was concentrated using a Centrifugal Ultrafiltration Filter Unit 3000 (Amicnon Tokyo, Japan), SDS-PAGE was performed with a 8–16% Mini-PROTEIN TGX Gels (BIO-RAD, Tokyo, Japan), and the gel was stained with Coomassie Brilliant Blue.
3. Results
3.1. Loss of Function of Transcription Factor col-26/dgr-1 Confers Sorbose Resistance in N. Crassa
During fungicide sensitivity screening of the deletion mutants in the transcription factor library of N. crassa, we found that the col-26 deletion mutant was hypersensitive to azoxystrobin and antimycin A, which are both mitochondrial respiratory chain complex III inhibitors (Figure S1). In N. crassa, only the sor-4 gene (alternate names of rco-3 and dgr-3), which encodes a glucose sensor protein, has been identified among sorbose resistance genes. Further characterization of the col-26 mutants revealed that the Δcol-26a strain displayed sorbose resistance, similar to sor-4 mutants (Figure 1A). Suspensions of conidia (approximately 106 cells/mL) were spotted on sor medium containing 1% sorbose and 0.2% sucrose; the Δsor-4a and Δcol-26a strains, but not the wild-type strain, followed a normal growth pattern. We also found the sorbose resistance of a 2-deoxyglucose (2-DG)-resistant strain dgr-1(BE52)A (FGSC#4326). The dgr-1 mutation has been identified as a frame shift at codon 335 of the col-26 protein (Figure S2A). These observations are consistent with the conclusion that the loss of function of the transcription factor col-26 confers sorbose resistance in N. crassa.
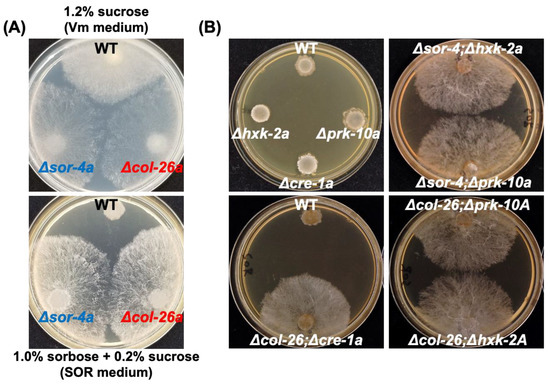
Figure 1.
Sorbose resistance of deletion mutants of sor-4 and col-26 gene, and their double mutant, with the factors involved in carbon catabolite repression. (A) Sorbose resistance of sor-4 and col-26 mutants. Conidia suspensions of wild type, Δsor-4a, and Δcol-26a strains were inoculated and cultured on Vogel’s minimal medium containing 1.2% sucrose (Vm medium, top panel) for 2 days, and on Vogel’s minimal medium containing 1% sorbose and 0.2% sucrose (sor medium, bottom panel) for 4 days. Both Δsor-4a and Δcol-26a displayed filamentous growth on sor medium. (B) Sorbose resistance in mutants of carbon catabolite repression and their double deletion mutants with col-26 or sor-4 mutant. The deletion mutants of hxk-2 (hexokinase), prk-10 (AMPK protein kinase), and cre-1 (transcriptional repressor) genes did not possess sorbose resistance on sor medium, but their double mutants with sor-4 or col-26 deletion were sorbose-resistant.
To compare the growth rates of Δsor-4a and Δcol-26a strains on Vm medium and sor medium, mycelial disks precultured on each medium were transferred and cultured on fresh medium of each type. Both sorbose-resistant strains grew at similar rates to that of the wild-type strain (2.04 mm to 2.16 mm/h) on Vm medium (Figure 2A). On sor medium, the growth rate of the sor-4(DS(r))A strain (1.73 ± 0.13 mm/h) was very similar to that of the sor-4(DS(r))A strain (1.69 ± 0.18 mm/h) (Figure 2B). There was no statically significant difference in growth rate between the Δcol-26a strain and the sor-4(DS(r))A strain. These two precultured sorbose-resistant strains began linear growth on fresh medium without delay, whereas the wild-type strain did not show linear growth on this medium (Figure 2B). When conidia of these strains were inoculated on Vm medium, filamentous growth of the Δsor-4a and Δcol-26a strains, as well as the wild-type strain, were clearly detectable after 18 h. However, on sor medium, all strains formed compact colonies 48 h after inoculation, and only the Δsor-4a and Δcol-26a strains began to grow at the edge of the colonies. These observations suggest that there was a transition stage between colonial growth and linear hyphal growth in the Δsor-4a and Δcol-26a strains on sor medium. In addition, conidia of the Δsor-4a and Δcol-26a strains, as well as the wild-type strain, did not germinate on Vm medium containing sorbose as the sole carbon source; therefore, the addition of a normal carbon source, such as sucrose or glucose, is essential for filamentous growth on medium containing 1% sorbose.
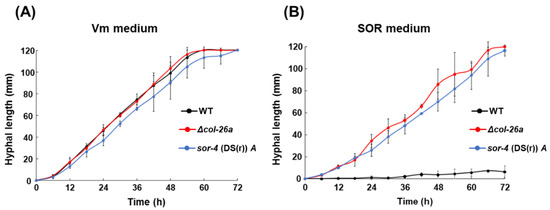
Figure 2.
Comparison of growth rates of the col-26 and sor-4 mutants on Vm medium and sor medium. (A) Growing hyphae of the wild type, sor-4(DS(r))A, and Δcol-26a strains on Vm medium were transferred onto fresh Vm medium. (B) Growing hyphae of the sor-4(DS(r))A and Δcol-26a strains on sor medium were transferred onto fresh sor medium. For the wild-type strain, conidia were spread on the sor medium and incubated for 2 days, and then agar disks containing growing cells were transferred onto fresh sor medium. Hyphal elongation (mm) from the edges of inoculation disks were measured for 3 days. Errors are expressed as standard deviation.
3.2. Sorbose Resistance, 2-DG Resistance, and QoI Sensitivity of CCR Mutants
The sorbose-resistance factors col-26 and sor-4 are known to be involved in CCR and their mutant strains are less sensitive to 2-DG, an analog of glucose that cannot be isomerized to fructose; therefore, it is not further metabolized and is often used to select for CCR factors in filamentous fungi [13,43]. The involvement of CCR factors in sorbose resistance prompted us to examine whether the deletion of other CCR factors was involved in sorbose resistance and/or 2-DG resistance. We examined three factors, namely a hexokinase, HXK-2, a major CCR-transcription factor, cre-1, and an AMP-activated S/T protein kinase, PRK-10. None of these N. crassa deletion mutants displayed sorbose resistance (Figure 1B); however, the Δhxk-2a mutant was as resistant to 2-DG as the Δsor-4a and Δcol-26a strains (Figure 3A). In contrast, the Δprk-10a mutant was hypersensitive to 2-DG. The Δcre-1 mutant was previously reported to have 2-DG resistance when Avicel was used as a carbon source [33], but our results showed that the Δcre-1a mutant was somewhat sensitive to 2-DG on agar medium containing fructose as a carbon source. We sequenced the hxk-2 gene of the dgr-4(KHY7)a mutant (FGSC#8287) and found the insertion of a 139 bp fragment within the third exon of hxk-2 (Figure S2B). This insertion resulted in a frame shift at codon 289 and immature termination at codon 292 in hxk-2. This is the first report to show the dgr-4 gene is identical to the hxk-2 gene.
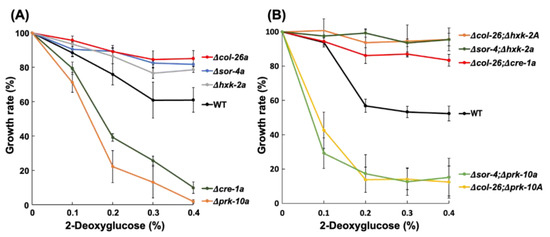
Figure 3.
2-Deoxyglucose resistance of the CCR mutants. (A) Single deletion mutants of hxk-2, prk-10, cre-1, col-26 and sor-4. (B) Their double mutants of col-26; hxk-2, col-26; prk-10, col-26; cre-1, sor-4; hxk-2, and sor-4; prk-10. Mycelium disks precultured on Vogel’s minimal medium containing 1% fructose were inoculated on fructose medium containing 2-deoxyglucose (0.1%, 0.2%, 0.3%, and 0.4%) for 18 h at room temperature. The growth rate was calculated following formula: Growth rate (%) = {hyphal length of treatment (mm)/hyphal length of control (mm)} × 100. Three biological replicates were performed. Errors are expressed as the standard error.
As described above, Δcol-26a showed hyper-sensitivity to antimycin A and azoxystrobin, which inhibit mitochondrial complex III by binding the Qi-site and the Qo-site of cytochrome b, respectively. Both the Δsor-4a and Δcol-26a mutants exhibited very similar sensitivity to these respiration inhibitors (Figure 4A). Not only sorbose-resistant mutants, but also Δhxk-2a and Δprk-10a mutants were hypersensitive to antimycin A and azoxystrobin (Figure 4B). In contrast, QoI- and QoI sensitivity in the Δcre-1a mutant was almost same to that of the wild-type strain. It is well known that the alternative oxidase AOD-1 reduces sensitivity to complex III inhibitors. Therefore, we analyzed aod-1 expression in the Δcol-26a mutants. The aod-1 gene was upregulated by azoxystrobin to the same level as in the wild-type strain, suggesting that QoI sensitivities of CCR mutants are independent with alternative oxidase activity.
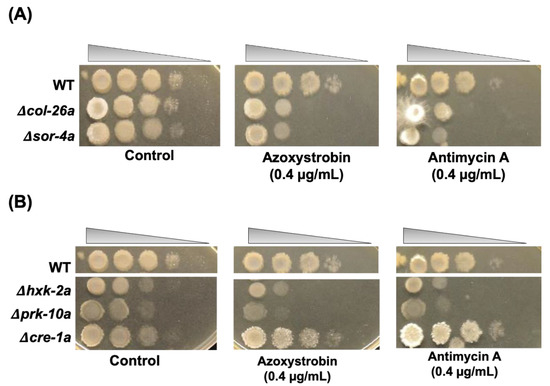
Figure 4.
Sensitivity to the mitochondrial complex III inhibitors azoxystrobin and antimycin A. A series of conidia suspensions (107–103 cells/mL) was spotted on sor medium containing azoxystrobin (0.4 mg/L) and antimycin A (0.4 mg/L) at room temperature. Control plates and fungicide treatment plates were photographed at 2 days and 5 days after inoculation, respectively. (A) col-26 and sor-4 strains. (B) hxk-2, prk-10 and cre-1 strains.
3.3. Comparison of Sorbose and 2-DG Resistance and Gene Expressions of Double Mutants Isolated from Sorbose-Resistant Mutants and CCR Mutants
To investigate the genetic relationship between glucose repression and sorbose resistance, we crossed the col-26 or sor-4 mutant with CCR mutants to obtain corresponding double mutants (Figure S3). All isolated double mutants, Δcol-26; Δcre-1a, Δcol-26; Δhxk-2A, Δcol-26; Δprk-10A, Δsor-4; Δhxk-2a, and Δsor-4; Δprk-10a displayed sorbose resistance similar to the Δcol-26a and Δsor-4a mutants on sor medium (Figure 1B). In contrast, similar to the Δprk-10a mutant, the Δcol-26; Δprk-10A and Δsor-4; Δprk-10a double mutants were hypersensitive to 2-DG, whereas the Δcol-26; Δhxk-2A, Δsor-4; Δhxk-2a, and Δcol-26; Δcre-1a mutants were resistant to 2-DG (Figure 3B).
col-26 and cre-1 have been shown to regulate several carbohydrate-related enzymes involved in CCR. We compared the expression pattern of relevant genes in the Δcre-1a, Δcol-26a, and Δcol-26; Δcre-1a mutants. First, we selected six genes, gla-1 (glucoamylase), inv (invertase), gh31-3 (alpha-glucosidase), gh13-2 (alpha-amylase), glt-1 (low-affinity glucose transporter), and hgt-1 (high-affinity glucose transporter), and compared their expression in medium with glucose or maltose as the carbon source. These genes, except for gh13-2 and glt-1, were highly induced in the wild-type strain grown on maltose medium (Figure 5A). As previously reported [29], the deletion of the negative regulator cre-1 resulted in high constitutive expression of CCR-related genes (Figure 5B). The inductions of gla-1, inv, gh31-3, gh13-2, and hgt-1 in the Δcre-1a mutant was quite evident in glucose-rich conditions, but minimal in glucose-depleted conditions. In contrast, the expression profiles of all six genes were almost the same in the Δcol-26a and Δsor-4a mutants in both conditions. The expression of gla-1, inv, gh31-3, and hgt-1 was slightly upregulated in both the Δcol-26a and Δsor-4a mutants in glucose-rich condition, whereas the expression of gh13-2 and glt-1 was significantly downregulated in glucose-rich and glucose-depleted conditions. It should be noted that the expression pattern of the Δcol-26;Δcre-1a double mutant was almost the same as that of the Δcol-26a and Δsor-4a mutants (Figure 5B). These data indicated that derepression by the loss of cre-1 function might be overcome by the deletion of the col-26 gene.
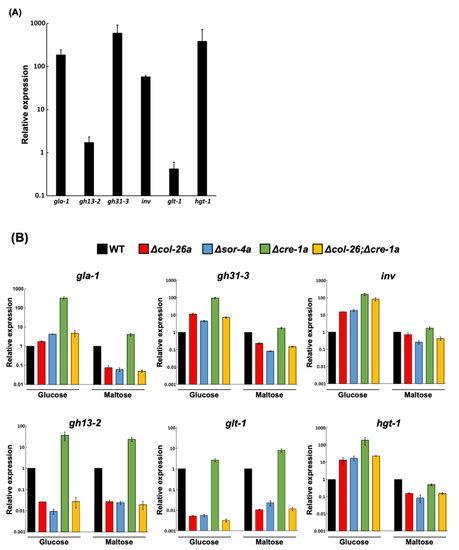
Figure 5.
Gene expression analysis of the col-26, sor-4, cre-1 and cre-1; col-26 mutants. (A) Gene upregulation in the wild-type strain under glucose derepression conditions shifted from 1.2% glucose medium to 1.2% maltose medium (see Materials and Methods). Six genes, gla-1 (glucoamylase), gh13-2 (alpha-amylase), gh31-3 (alpha-glucosidase), inv (invertase), glt-1 (low-affinity glucose transporter), and hgt-1 (high-affinity glucose transporter) were selected as target genes for qPCR analysis. With the exception of gh13-2 and glt-1, all genes were highly upregulated in maltose medium. (B) Comparison of gene expression patterns in the Δcol-26a, Δsor-4a, Δcre-1a and Δcol-26; Δcre-1a mutants. Expression levels in each mutant strain were calculated relative to the wild-type strain grown under glucose-rich conditions (Glucose) and glucose derepression conditions (Maltose). Errors are expressed as the standard error. At least three biological replicates of each experiment were performed.
Several glucose transporters have been reported to be downregulated by the Δcol-26a and Δsor-4a mutants [35]; therefore, we measured the concentrations of extracellular glucose and sorbose during cultivation (Figure 6). The wild-type strain consumed the most glucose after incubation for 24 h when the initial glucose concentration was 1%, meanwhile, more than 60% of glucose remained in the culture medium in the cases of the Δcol-26a and Δsor-4a mutants, suggesting their low uptake of glucose (Figure 6A). In contrast, after 24 h incubation, approximately 90% of sorbose remained unincorporated in the case of the Δcol-26a and Δsor-4a mutants, as well as the wild-type strain (Figure 6B). These data suggest that these sorbose-resistance mutants do not incorporate sorbose more than the wild type and do not assimilate it.
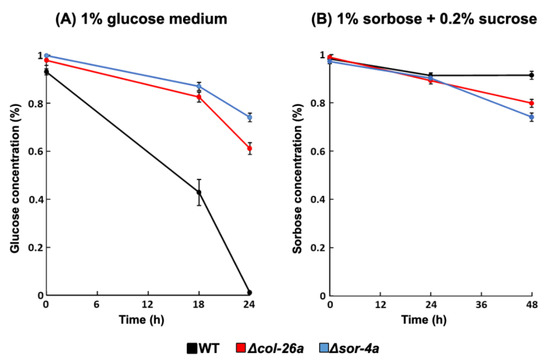
Figure 6.
Consumption of glucose or sorbose in sorbose-resistant sor-4 and col-26 mutants. Glucose (A) and sorbose (B) concentration in the culture medium in the wild type, Δcol-26a, and Δsor-4a strains. The concentration of glucose and sorbose in culture filtrate was measured using the F-kit for glucose and the Si–Mo method (see Materials and Methods), respectively. Errors are expressed as the standard error. Three biological replicates were performed.
3.4. A Single S11L Mutation, but Not the Loss-of-function Mutation of F-box Protein exo-1, Confers Sorbose, and 2-DG Resistances in N. Crassa
A 2-DG-resistant strain in dgr-2, dgr-2(L1)a, has sorbose resistance (Figure 7A). Recently, Gabriel et al. [36] revealed that the exo-1 gene encoded a F-box protein, and identified a S11L missense mutation within the exo-1 gene in dgr-2(L1)a strain (Figure S4) [36]. In N. crassa, the exo-1 mutant produced the maximum extracellular glucoamylase activity in starch-supplemented medium [44]; however, the exo-1 deletion mutant did not show any sorbose resistance (Figure 7A), indicating that the single amino-acid substitution in exo-1/DGR-2, S11L, confers sorbose resistance as well as 2-DG resistance. As shown in Figure 7B, the dgr-2(L1)a strain was resistant to 2-DG but Δexo-1a was somewhat sensitive to the sugar. Furthermore, we confirmed that the Δexo-1a strain displayed hypersecretion of proteins, as described previously [36] (Figure 7C), even though the dgr-2(L1)a strain did not. In glucose-rich conditions, all four genes—namely gla-1, inv, glt-1, and hgt-1—were slightly upregulated in the Δexo-1a strain, whereas the expression pattern of the exo-1S11L mutant resembles to that of the Δcol-26a strain (Figure 7D). These results suggest that the phenotypes of Δexo-1a differ from those of the exo-1S11L strain.
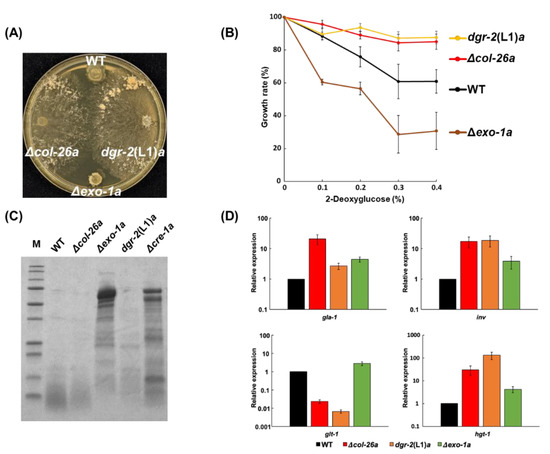
Figure 7.
Comparison of the phenotypes of the exo-1S11L (dgr-2(L1)a) and Δexo-1a mutants. Sorbose resistance (A) and 2-DG resistance (B) of wild-type, Δcol-26a, exo-1S11L (dgr-2(L1)a), and Δexo-1a strains. (C) SDS-PAGE analysis of proteins secreted by each strain. The extracellular protein in culture filtrates was concentrated and applied for SDS-PAGE analysis. (D) Gene expression of gla-1, inv, glt-1, and hgt-1 under glucose-rich conditions (see Figure 5) in each strain. Errors are expressed as the standard error.
4. Discussion
L-Sorbose induces hyperbranching of hyphae and results in colonial growth on agar media in Neurospora crassa. Among the six genes identified as conferring sorbose resistance (sor-1 to sor-6), only sor-4, which encodes a glucose sensor protein, has been thoroughly investigated [14,15]. In this work, we revealed two more genes, col-26, which encodes a transcription factor, and exo-1, which encodes a F-box protein, that are likely involved in sorbose resistance (Figure 1A and Figure 7A).
It is well known that sor-4 and col-26 are factors with important roles in CCR in N. crassa. This prompted us to investigate whether any gene mutants in CCR displayed sorbose resistance. cre-1, a homolog of S. cerevisiae Mig1 and A. nidulans CreA transcription factors, controls glucose repression along with a glucose sensor sor-4 and a hexokinase HXK-2 [18]. Similar to homologs in other fungi, N. crassa cre-1 acts as a negative regulator of regulons for the utilization of carbohydrates other than glucose; therefore, the deletion of the cre-1 gene results in the increased secretion of cellulases, amylases, and beta-galactosidases even in glucose-rich condition [29]. In contrast, the AMP-activated S/T protein kinase PRK-10, an ortholog of Snf1 of S. cerevisiae, probably phosphorylates cre-1 and results in the release of cre-1-mediated repression. As described in S. cerevisiae [43], the deletion of hxk-2 and prk-10 in N. crassa resulted in cells that were resistant and hypersensitive to 2-DG, respectively (Figure 3). However, neither the cre-1, hxk-2, nor prk-10 mutants showed sorbose resistance (Figure 1B). These results suggest that sorbose resistance may correlate with catabolite repression, but independently of cre-1, hxk-2, and prk-10.
As sor-4 and col-26 strains are resistant to 2-DG, we further characterized 2-DG resistance (dgr) mutants to identify the missing factors that might connect the sor-4 glucose sensor and the col-26 transcription factor [13]. Among the four dgr mutants dgr-1 to dgr-4, dgr-3 is allelic to sor-4 [15], the dgr-1 and dgr-2 strains were sorbose-resistant, but the dgr-4 strain was sensitive to the chemical. From the mapping data of dgr-4 (Figure S2B) [13], this gene was shown to be closely linked to al-2 (0.6%). Physical mapping of data from the genome sequence database indicated that hxk-2 localizes very close to al-2. The direct sequencing of hxk-2 from the dgr-4(KHY7)a strain revealed that the hxk-2 gene was disrupted by an insertional mutation (Figure S2B). We also sequenced sor-4 and col-26 of the dgr-1(BE52)A strain and found that the dgr-1 gene is allelic to the col-26 gene (Figure S2A). Mapping data indicated the localization of dgr-1 and col-26 on edge of the left arm of chromosome V was consistent with the interpretation that dgr-1 and col-26 are the same gene. In contrast, there was no mutation in sor-4 and col-26 in the dgr-2 mutants, suggesting that it is a new gene that confers sorbose resistance. Recently, Gabriel et al. [36] revealed that dgr-2 is allelic to exo-1, which encodes an F-box protein, by means of dgr-2 and exo-1 mutant genomes [36]. We confirmed that the exo-1S11L missense mutation in sorbose-resistance progenies by genome sequencing of dgr-2(L1)a and dgr-2(L5)A; however, deletion mutants of dgr-2/exo-1 did not display sorbose-resistance (Figure 7A). We also confirmed that an exo-1 deletion mutant strain, Δexo-1a, secreted exocellular enzymes in large amounts, but the dgr-2(L1)a strain with the exo-1S11L mutation did not (Figure 7C). Moreover, the dgr-2(L1)a strain displayed 2-DG resistance, but the exo-1 deletion mutant did not show clear 2-DG resistance (Figure 7B). Our results indicate that the phenotypes of the exo-1 point mutation strain dgr-2(L1)a and the deletion mutant strain Δexo-1a are quite different in these aspects. A serine residue at amino-acid position 11 of the F-box protein is conserved in other fungi, such as Magnaporthe grisea, Fusarium graminearum, Botrytis cinerea, and Trichoderma reesei (Figure S4). Although the function of the N-terminal conserved region (consisting of approximately 20 amino acids) is unclear, the serine at 11 may be phosphorylated and affect the function of F-box domain (amino acids 112–144 in exo-1). Although its function is still unknown, the F-box protein exo-1 may form SCF complexes and induce ubiquitination of protein(s) targeted for degradation by the 26S proteasome: col-26 is a possible candidate target. Phenotypes of exo-1S11L strain resemble col-26 deletion mutants but not exo-1 deletion mutants regarding sorbose resistance and gene expression (Figure 7). Gabriel et al. [36] reported that the loss of function of exo-1 induces glucoside enzymes in cre-1 independently [36]. One possible explanation is that the exo-1 deletion suppresses col-26 degradation and leads to the overexpression of glucosidases. In contrast, the S11L mutation results in the constitutive activation of exo-1, hyper-ubiquitination, and the degradation of col-26.
As previously reported [29,30], the loss of cre-1 function leads to the overexpression of gla-1, gh13-2, gh31-3, inv, glt-1, and hgt-1, even in glucose-rich conditions (Figure 5A). cre-1-like transcription factors are the main negative regulators in glucose repression in many fungi [18]. The expression of glt-1, which encodes a low-affinity glucose transporter, in three sorbose-resistance mutants, namely Δcol-26a, Δsor-4a, and Δcol-26;Δcre-1a was significantly reduced, even in glucose-rich conditions, suggesting that the glucose sensor sor-4 and the transcription factor col-26 are essential for the expression of glt-1, as described elsewhere [35]. In addition, the expression of gh13-2, which encodes an alpha-amylase, in these three mutants was comparable to that of glt-1, suggesting that these two genes would be controlled under the same system. We noticed that the gene expression profiles for carbohydrate-related genes in col-26 and sor-4 were similar. Moreover, a double mutant Δcol-26;Δcre-1a had a very similar gene expression profile to Δcol-26a and Δsor-4a. It should be noted that the upregulation found in the cre-1 strain was mostly negated in the Δcol-26;Δcre-1a strain. The col-26 transcription factor could positively control its target genes, which should be suppressed by unphosphorylated cre-1, and the loss cannot be compensated for because of the absence of the negative regulator cre-1. Indeed, the Δcol-26;Δcre-1a strain displayed a more similar phenotype to the col-26 single mutant than the cre-1 mutant. Meanwhile, the expression of gla-1, inv, gh31-3, and hgt-1 in these three sorbose-resistant mutants in glucose-rich conditions was slightly upregulated. The presence of any cre-1 and col-26 independent regulatory system (s) cannot be eliminated.
The mechanism underlying sorbose resistance in N. crassa is still unclear. In our study, we revealed that, in addition to the glucose sensor sor-4, two factors—namely col-26 and exo-1—are involved in sorbose resistance. All evidence obtained regarding sorbose resistance indicates the connection of the expression of carbohydrate-related genes in glucose-depleted conditions and the toxicity of the chemical. Sorbose might disturb signaling pathway(s) concerning carbohydrate-related gene expression by interacting with the sugar-sensing system. col-26 upregulates glucose transporter genes, including glt-1 and hgt-1 [35]; indeed, col-26 mutant uptake less glucose than the wild-type strain. We speculate that sorbose-resistant mutants hardly assimilate the sugar, as its incorporation by the col-26 mutants was only marginally different to the that of the wild-type strain (Figure 6).
CCR is fundamental mechanism using proper energy sources; therefore, it is conserved in prokaryote and eukaryotes. In fungi, CCR is directly connected with various applications, such as the production of Japanese sake and miso by Aspergillus species, bioethanol production by T. reesei, and plant protection from plant pathogenic fungi. Recent studies indicate that catabolite repression in fungi is connected to several signaling molecules, such as cAMP-dependent protein kinase and the stress-response MAP kinase. Therefore, our results in the model fungus N. crassa will contribute to elucidation of the complex mechanism of fungal CCR.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8111169/s1, Figure S1 Screening of transcription factor knockout library for sensitivity to the mitochondria complex III inhibitors, azoxystrobin and antimycin A, Figure S2A Physical and genetical mapping of the col-26 and dgr-1genes, and the mutation of dgr-1(BE52)A strain, S2B Physical and genetical mapping of the hxk-2 and dgr-4 genes, and the mutation of the dgr-4(KHY7)a strain, Figure S3 PCR confirmation of the deletion of cre-1 and col-26 genes in Δcol-26; Δcre-1 double mutants, Figure S4A Amino-acid homology of N-terminal region of F-box protein exo-1 protein in fungi, and Figure S4B Alignment of exo-1 amino-acid sequences in fungi. The dgr-2(L1)a strain has S11L mutation in exo-1. Table S1 List of sequencing primers for col-26 and hxk-2 genes, Table S2 List of primers used for PCR detection of gene deletions, Table S3 List of RT-qPCR primers and universal probes.
Author Contributions
K.H. and M.F. designed the research plan with assistance from A.I. and F.F. K.H., T.I. and S.K. performed experiments. K.H., A.I., F.F. and M.F. analyzed and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This works was supported by JST SPRING, Grant Number JPMJSP2159 and the Inoue Enryo memorial grant, Toyo University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available in the main text or the Supplementary Files.
Acknowledgments
We are grateful to the Fungal Genetics Stock Center for strains.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tatum, E.L.; Barratt, R.W.; Cutter, V.M. Chemical Induction of Colonial Paramorphs in Neurospora and Syncephalastrum. Science 1949, 109, 509–511. [Google Scholar] [CrossRef] [PubMed]
- De Terra, N.; Tatum, E.L. Colonial growth of Neurospora. Sorbose and enzymes alter the composition of the cell wall and induce morphological changes. Science 1961, 134, 1066–1068. [Google Scholar] [CrossRef]
- Mahadevan, P.R.; Tatum, E.L. Relationship of the Major Constituents of the Neurospora crassa Cell Wall to Wild-type and Colonial Morphology. J. Bacteriol. 1965, 90, 1073–1081. [Google Scholar] [CrossRef]
- Crocken, B.; Tatum, E.L. The effect of sorbose on metabolism and morphology of Neurospora. Biochim. Biophys. Acta 1968, 156, 1–8. [Google Scholar] [CrossRef]
- Mishra, N.C.; Tatum, E.L. Effect of L-sorbose on Polysaccharide Synthetases of Neurospora crassa (glycogen- -1,3-glucan-morphology-cell wall-digitonin-particulate enzymes). Proc. Natl. Acad. Sci. USA 1972, 69, 313–317. [Google Scholar] [CrossRef]
- Kamei, M.; Yamashita, K.; Takahashi, M.; Fukumori, F.; Ichiishi, A.; Fujimura, M. Involvement of MAK-1 and MAK-2 MAP kinases in cell wall integrity in Neurospora crassa. Biosci. Biotechnol. Biochem. 2016, 80, 1843–1852. [Google Scholar] [CrossRef]
- Hatamoto, M.; Aizawa, R.; Kobayashi, Y.; Fujimura, M. A novel fungicide aminopyrifen inhibits GWT-1 protein in glycosylphosphatidylinositol-anchor biosynthesis in Neurospora crassa. Pestic. Biochem. Physiol. 2019, 156, 1–8. [Google Scholar] [CrossRef]
- Elorza, M.V.; Arst, H.N. Sorbose resistant mutants of Aspergillus nidulans. Mol. Gen. Genet. 1971, 111, 185–193. [Google Scholar] [CrossRef]
- MacCabe, A.P.; Miró, P.; Ventura, L.; Ramon, D. Glucose uptake in germinating Aspergillus nidulans conidia: Involvement of the creA and sorA genes. Microbiology 2003, 149, 2129–2136. [Google Scholar] [CrossRef][Green Version]
- Forment, J.V.; Flipphi, M.; Ventura, L.; Gonzalez, R.; Ramón, D.; Maccabe, A.P. High-Affinity Glucose Transport in Aspergillus nidulans is Mediated by the Products of Two Related but Differentially Expressed Genes. PLoS ONE 2014, 9, e94662. [Google Scholar] [CrossRef]
- Klingmuller, W. Crossing analysis of sorbose resistant mutants of Neurospora crassa. Mol. Gen. Genet. 1967, 100, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Quigley, D.R. Sorbose-resistant mutants of Neurospora crassa do not have altered beta (1-3)glucan synthase activity. Curr. Microbiol. 1987, 15, 185–192. [Google Scholar] [CrossRef]
- Allen, K.E.; McNally, M.T.; Lowendorf, H.S.; Slayman, C.W.; Free, S.J. Deoxyglucose-resistant mutants of Neurospora crassa: Isolation, mapping, and biochemical characterization. J. Bacteriol. 1989, 171, 53–58. [Google Scholar] [CrossRef]
- Madi, L.; McBride, S.A.; Bailey, L.A.; Ebbole, D.J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 1997, 146, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.; Radford, A.; Sachs, M. The Neurospora Compendium Chromosomal Loci; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Ozcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 2566–2573. [Google Scholar] [CrossRef]
- Polish, J.A.; Kim, J.H.; Johnston, M. How the Rgt1 Transcription Factor of Saccharomyces cerevisiae Is Regulated by Glucose. Genetics 2005, 169, 583–594. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef]
- Tu, J.; Carlson, M. The GLC7 type 1 Protein Phosphatase is Required for Glucose Repression in Saccharomyces cerevisiae. Mol. Cell Biol. 1994, 14, 6789–6796. [Google Scholar] [CrossRef]
- Ahuatzi, D.; Riera, A.; Pela Ez, R.; Herrero, P.; Moreno, F. Hxk2 Regulates the Phosphorylation State of Mig1 and Therefore Its Nucleocytoplasmic Distribution. J. Biol. Chem. 2007, 282, 4485–4493. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Gligoris, T.; Tzamarias, D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 2004, 5, 368–372. [Google Scholar] [CrossRef]
- Tian, C.; Beeson, W.T.; Iavarone, A.T.; Sun, J.; Marletta, M.A.; Cate, J.H.; Glass, N.L. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. USA 2009, 106, 22157–22162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Dowzer, C.E.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell Biol. 1991, 11, 5701–5709. [Google Scholar] [PubMed]
- Prathumpai, W.; McIntyre, M.; Nielsen, J. The effect of CreA in glucose and xylose catabolism in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2004, 63, 748–753. [Google Scholar] [CrossRef]
- Nakamura, T.; Maeda, Y.; Tanoue, N.; Makita, T.; Kato, M.; Kobayashi, T. Expression Profile of Amylolytic Genes in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2006, 70, 2363–2370. [Google Scholar] [CrossRef]
- Cubero, B.; Scazzocchio, C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994, 13, 407–415. [Google Scholar] [CrossRef]
- Strauss, J.; Mach, R.L.; Zeilinger, S.; Hartler, G.; Stoffler, G.; Wolschek, M.; Kubicek, C.P. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 1995, 376, 103–107. [Google Scholar] [CrossRef]
- Sun, J.; Glass, N.L. Identification of the cre-1 Cellulolytic Regulon in Neurospora crassa. PLoS ONE 2011, 6, e25654. [Google Scholar] [CrossRef]
- Ziv, C.; Gorovits, R.; Yarden, O. Carbon source affects PKA-dependent polarity of Neurospora crassa in a cre-1-dependent and independent manner. Fungal Genet. Biol. 2008, 45, 103–116. [Google Scholar] [CrossRef]
- Gomi, K.; Akeno, T.; Minetoki, T.; Ozeki, K.; Kumagai, C.; Okazaki, N.; Iimura, Y. Molecular Cloning and Characterization of a Transcriptional Activator Gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2000, 64, 816–827. [Google Scholar] [CrossRef]
- Nitta, M.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates β-glucosidase expression in Trichoderma reesei. Fungal Genet. Biol. 2012, 49, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sun, J.; Glass, N.L. VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, is Essential for Plant Cell Wall Degradation by Neurospora crassa. PLoS Genet. 2014, 10, e1004500. [Google Scholar] [CrossRef]
- Xiong, Y.; Wu, V.W.; Lubbe, A.; Qin, L.; Deng, S.; Kennedy, M.; Bauer, D.; Singan, V.R.; Barry, K.; Northen, T.R.; et al. A fungal transcription factor essential for starch degradation affects integration of carbon and nitrogen metabolism. PLoS Genet. 2017, 13, e1006737. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Q.; Lin, L.; Li, X.; Zhang, Y.; Tian, C. RCO-3 and col-26 form an external-to-internal module that regulates the dual-affinity glucose transport system in Neurospora crassa. Biotechnol. Biofuels 2021, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; Thieme, N.; Liu, Q.; Li, F.; Meyer, L.T.; Harth, S.; Jecmenica, M.; Ramamurthy, M.; Gorman, J.; Simmons, B.A.; et al. The F-box protein gene exo-1 is a target for reverse engineering hypersecretion in filamentous fungi. Proc. Natl. Acad. Sci. USA 2021, 118, e2025689118. [Google Scholar] [CrossRef]
- Duyvesteijn, R.G.; van Wijk, R.; Boer, Y.; Rep, M.; Cornelissen, B.J.; Haring, M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005, 57, 1051–1063. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Vogel, H.J. Distribution of Lysine Pathways Amomg Fungi: Evolutionary Implications. Am. Nat. 1964, 98, 435–446. [Google Scholar] [CrossRef]
- Noguchi, R.; Banno, S.; Ichikawa, R.; Fukumori, F.; Ichiishi, A.; Kimura, M.; Yamaguchi, I.; Fujimura, M. Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genet. Biol. 2007, 44, 208–218. [Google Scholar] [CrossRef]
- Yamashita, K.; Shiozawa, A.; Watanabe, S.; Fukumori, F.; Kimura, M.; Fujimura, M. ATF-1 transcription factor regulates the expression of ccg-1 and cat-1 genes in response to fludioxonil under OS-2 MAP kinase in Neurospora crassa. Fungal Genet. Biol. 2008, 45, 1562–1569. [Google Scholar] [CrossRef]
- Katano, H.; Takakuwa, M.; Itoh, T.; Hibi, T. Colorimetric determination of fructose for the high-throughput microtiter plate assay of glucose isomerase. Biosci. Biotechnol. Biochem. 2015, 79, 1057–1060. [Google Scholar] [CrossRef] [PubMed]
- McCartney, R.R.; Chandrashekarappa, D.G.; Zhang, B.B.; Schmidt, M.C. Genetic analysis of resistance and sensitivity to 2-deoxyglucose in Saccharomyces cerevisiae. Genetics 2014, 198, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Gratzner, H.G. Cell Wall Alterations Associated with the Hyperproduction of Extracellular Enzymes in Neurospora crassa. J. Bacteriol. 1972, 111, 443–446. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).