Morphological, Pathological and Genetic Diversity of the Colletotrichum Species, Pathogenic on Solanaceous Vegetable Crops in Bulgaria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Infected Plant Material and Colletotrichum Isolation
2.2. Morphological and Cultural Characterization
2.3. Pathogenicity Assay of Colletotrichum Isolates
2.4. Preparation of Fungal Biomass for Phylogenetic Analysis
2.5. DNA Extraction and PCR Amplification
2.6. DNA Sequencing and Data Analysis
3. Results
3.1. Morphological and Cultural Characterization of Colletotrichum Isolates
3.2. Pathogenicity of Colletotrichum Isolates
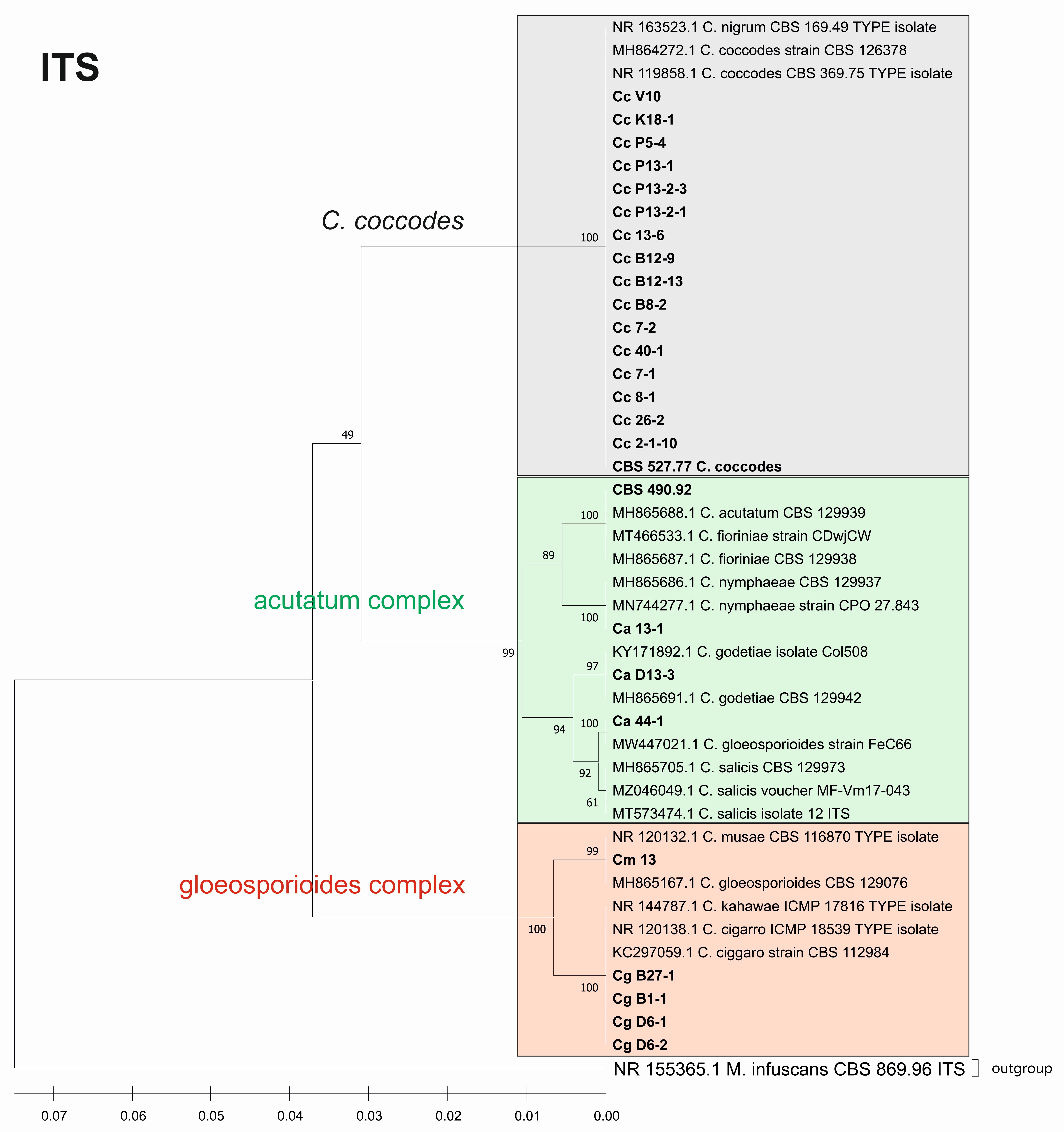
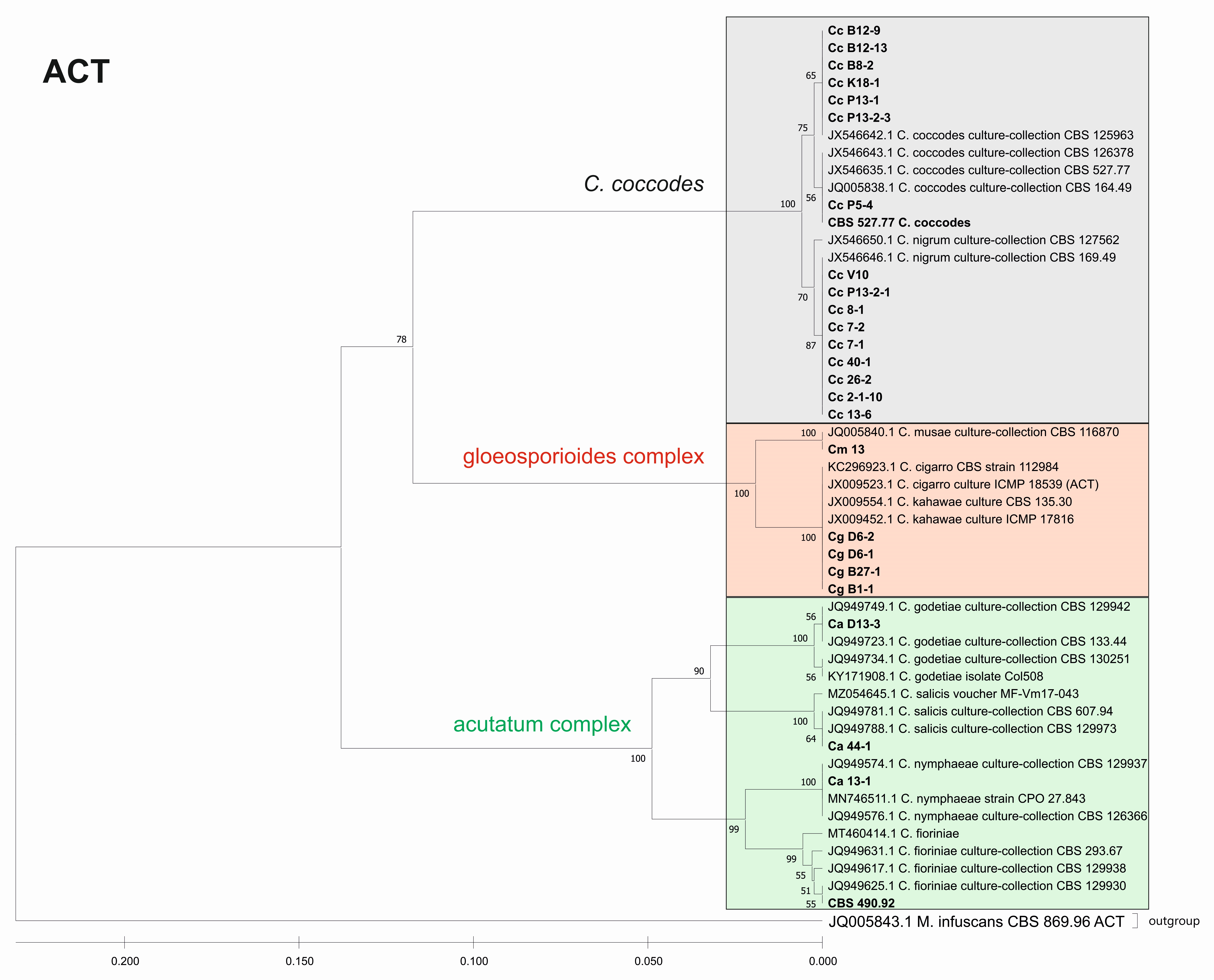
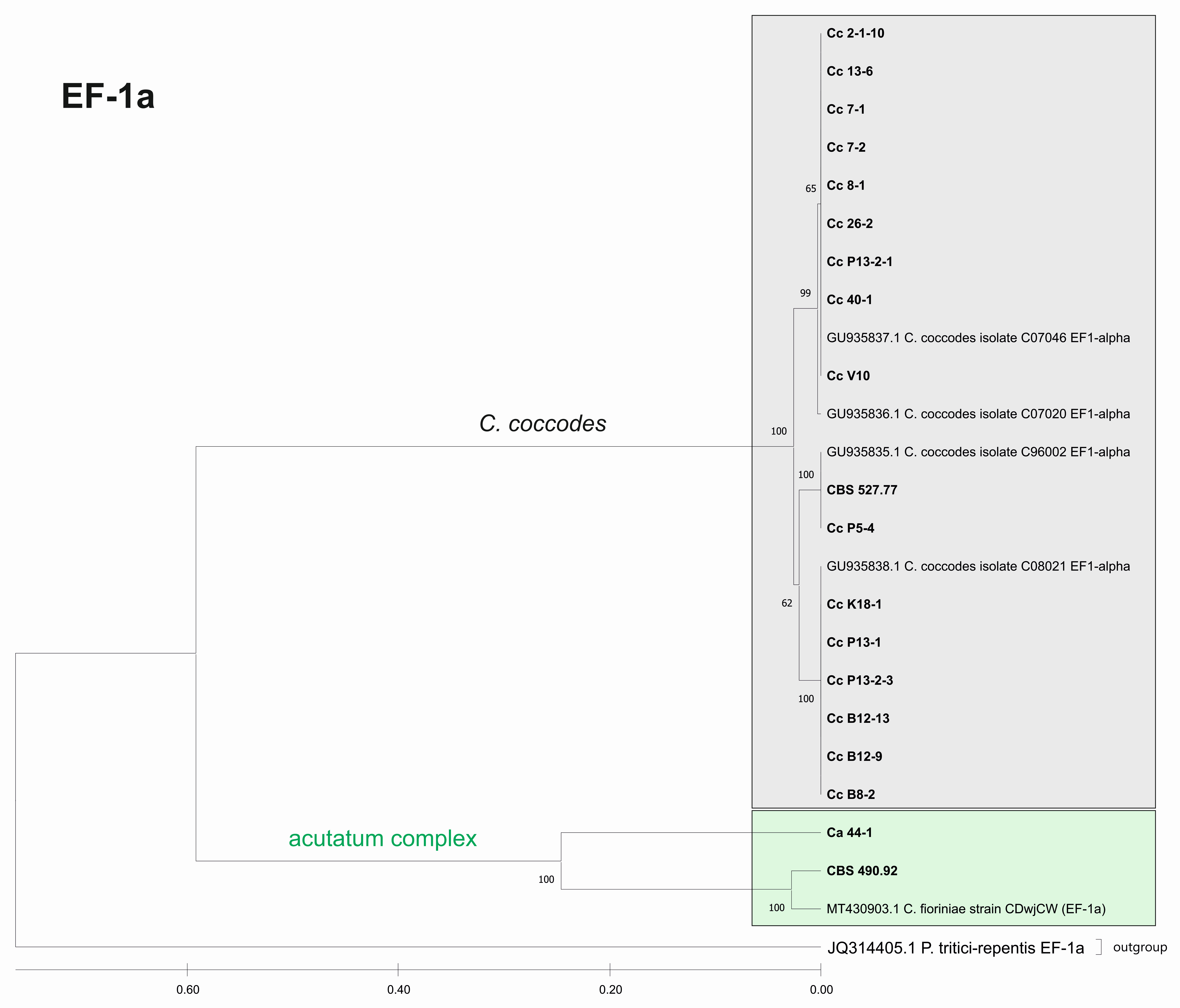
3.3. Genetic Diversity and Phylogenetic Analysis
3.3.1. Efficiency of PCR Amplification and DNA Sequencing
3.3.2. Genetic Diversity and Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

Appendix B
| Isolate | ITS | ACT | EF-1a | TUB2 |
| Ca13-1 | Acc. No OP587502 | Acc. N xx-xx | Acc. N xx-xx | Acc. N xx-xx |
| CaD13-3 | Acc. No OP587503 | Sub2630242 | Sub2630459 | Sub2630466 |
| Ca44-1 | Acc. No OP587504 | |||
| CBS 490.92 | Acc. No OP587505 | |||
| CgB1-1 | Acc. No OP587506 | |||
| CgB27-1 | Acc. No OP587507 | |||
| CgD6-1 | Acc. No OP587508 | |||
| CgD6-2 | Acc. No OP587509 | |||
| Cm13 | Acc. No OP587510 | |||
| Cc2-1-10 | Acc. No OP587511 | |||
| CcB8-1 | Acc. No OP587512 | |||
| CcB8-2 | Acc. No OP587513 | |||
| Cc7-1 | Acc. No OP587514 | |||
| Cc7-2 | Acc. No OP587515 | |||
| Cc26-2 | Acc. No OP587516 | |||
| CcB12-9 | Acc. No OP587517 | |||
| CcB12-13 | Acc. No OP587518 | |||
| CcK18-1 | Acc. No OP587519 | |||
| Cc13-6 | Acc. No OP587520 | |||
| CcV10 | Acc. No OP587521 | |||
| Cc40-1 | Acc. No OP587522 | |||
| P5-4 | Acc. No OP587523 | |||
| P13-1 | Acc. No OP587524 | |||
| P13-2-1 | Acc. No OP587525 | |||
| P13-2-3 | Acc. No OP587526 |
References
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—Current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Shenoy, B.D. Colletotrichum systematics: Past, present and prospects. Mycosphere 2016, 7, 1093–1102. [Google Scholar] [CrossRef]
- Cano, J.; Guarro, J.; Gené, J. Molecular and morphological identification of Colletotrichum species of clinical interest. J. Clin. Microbiol. 2004, 42, 2450–2454. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, J.A.; Gouli, S.; Parker, B.L.; Skinner, M.; Giordano, R. Entomopathogenic activity of a variety of the fungus, Colletotrichum acutatum, recovered from the elongate hemlock scale, Fiorinia externa. J. Insect Sci. 2009, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Han, J.H.; Shim, H.; Shin, J.H.; Kim, K.S. Antagonistic activities of Bacillus spp. Strains isolated from tidal flat sediment towards anthracnose pathogens Colletotrichum acutatum and C. gloeosporioides in South Korea. Plant Pathol. J. 2015, 31, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, R.S.; Hyde, K.D.; Jeewon, R.; Li, X.H.; Liu, M.; Yan, J.Y. Mycosphere Essay 6: Why is it important to correctly name Colletotrichum species. Mycosphere 2016, 7, 1076–1092. [Google Scholar] [CrossRef]
- Freeman, S.; Katan, T.; Shabi, E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Dis. 1998, 82, 596–605. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.; Waller, J.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Shivas, R.G. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009, 39, 183–204. [Google Scholar]
- Gomes, S.; Azevedo-Nogueira, F.; Martins-Lopes, P. Editorial Comments to the Special Issue: “Colletotrichum spp. on Fruit Crops—State of the Art, Perspectives and Drawbacks”. Pathogens 2021, 10, 478. [Google Scholar] [CrossRef]
- Mills, P.R.; Sreenivasaprasad, S.; Brown, A.E. Detection and differentiation of Colletotrichum gloeosporioides isolates using PCR. FEMS Microbiol. Lett. 1992, 98, 137–143. [Google Scholar] [CrossRef]
- Sreenivasaprasad, S.; Sharada, K.; Brown, A.E.; Mills, P.R. PCR-based detection of Colletotrichum acutatum on strawberry. Plant Pathol. 1996, 45, 650–655. [Google Scholar] [CrossRef]
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022, 101, 1–86. [Google Scholar] [CrossRef]
- Hyde, K.D.; Abd-Elsalam, K.; Cai, L. Morphology: Still essential in a molecular world. Mycotaxon 2010, 114, 439–451. [Google Scholar] [CrossRef]
- Da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.F.; de Queiroz, M.V. Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; de Farias, A.R.G.; Bhunjun, C.S.; Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Herath, I.S.; Thambugala, K.M.; Manawasinghe, I.S.; et al. What is a species in fungal plant pathogens? Fungal Divers. 2021, 109, 239–266. [Google Scholar] [CrossRef]
- Khodadadi, F.; González, J.B.; Martin, P.L.; Giroux, E.; Bilodeau, G.J.; Peter, K.A.; Aćimović, S.G. Identification and characterization of Colletotrichum species causing apple bitter rot in New York and description of C. noveboracense sp. Nov. Sci. Rep. 2020, 10, 11043. [Google Scholar]
- Dowling, M.; Peres, N.; Villani, S.; Schnabel, G. Managing Colletotrichum on fruit crops: A “complex” challenge. Plant Dis. 2020, 104, 2301–2316. [Google Scholar] [CrossRef]
- Hyde, K.D.; Cai, L.; McKenzie, E.H.C.; Yang, Y.L.; Zhang, J.Z.; Prihastuti, H. Colletotrichum: A catalogue of confusion. Fungal Divers. 2009, 39, 1–17. [Google Scholar]
- Crouch, J.; O’Connell, R.; Gan, P.; Buiate, E.; Torres, M.F.; Beirn, L.; Shirasu, K.; Vaillancourt, L. The genomics of Colletotrichum. In Genomics of Plant-Associated Fungi: Monocot Pathogens; Dean, R.A., Lichens-Park, A., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 69–102. [Google Scholar]
- Crouch, J.A.; Clarke, B.B.; Hillman, B.I. What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia 2009, 101, 648–656. [Google Scholar] [CrossRef]
- Hyde, K.D.; Cai, L.; Cannon, P.F.; Crouch, J.A.; Crous, P.W.; Damm, U.; Goodwin, P.H.; Chen, H.; Johnston, P.R.; Jones, E.B.G.; et al. Colletotrichum—Names in current use. Fungal Divers. 2009, 39, 147–182. [Google Scholar]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [PubMed]
- Stielow, J.B.; Levesque, C.A.; Seifert, K.A.; Meyer, W.; Irinyi, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 2015, 35, 242–263. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.D.; Ades, P.K.; Crous, P.W.; Taylor, P.W.J. Colletotrichum species associated with chili anthracnose in Australia. Plant Pathol. 2017, 66, 254–267. [Google Scholar] [CrossRef]
- Silva, D.N.; Talhinhas, P.; Várzea, V.; Cai, L.; Paulo, O.S.; Batista, D. Application of the Apn1/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: An example from coffee (Coffea spp.) hosts. Mycologia 2012, 104, 396–409. [Google Scholar] [CrossRef]
- Hofer, K.M.; Braithwaite, M.; Braithwaite, L.J.; Sorensen, S.; Siebert, B.; Pather, V.; Goudie, L.; Williamson, L.; Alexander, B.J.R.; Toome-Heller, M. First report of Colletotrichum fructicola, C. perseae, and C. siamense causing anthracnose disease of avocado (Persea americana) in New Zealand. Plant Dis. 2021, 105, 1564. [Google Scholar] [CrossRef]
- Vieira, W.A.S.; Bezerra, P.A.; Silva, A.C.; Veloso, J.S.; Câmara, M.P.S.; Doyle, V.P. Optimal markers for the identification of Colletotrichum species. Mol. Phylogenet. Evol. 2020, 143, 181–202. [Google Scholar] [CrossRef]
- MacKenzie, S.J.; Peres, N.A.; Barquero, M.P.; Arauz, L.F.; Timmer, L.W. Host range and genetic relatedness of Colletotrichum acutatum isolates from fruit crops and leatherleaf fern in Florida. Phytopathology 2009, 99, 620–631. [Google Scholar] [CrossRef]
- Katoch, A.; Sharma, P.; Sharma, P.N. Identification of Colletotrichum spp. Associated with fruit rot of Capsicum annuum in North Western Himalayan region of India using fungal DNA barcode markers. J. Plant Biochem. Biotechnol. 2017, 26, 216–223. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, X.; Khaskheli, M.I.; Sun, X.; Chang, X.; Gong, G. Identification of Colletotrichum species associated with blueberry anthracnose in Sichuan, China. Pathogens 2020, 9, 718. [Google Scholar] [CrossRef]
- Giacomin, R.M.; Ruas, C.F.; Baba, V.Y.; De Godoy, S.M.; Sudré, C.P.; Bento, C.D.S.; Da Cunha, M.; Da Costa Geronimo, I.G.; Rodrigues, R.; Gonçalves, L.S. Phenotypic, molecular and pathogenic characterization of Colletotrichum scovillei infecting Capsicum species in Rio de Janeiro, Brazil. PeerJ 2021, 9, e10782. [Google Scholar] [CrossRef]
- Xue, L.; Zhang, L.; Yang, X.X.; Huang, X.; Wu, W.; Zhou, X.; White, J.F.; Liu, Y.; Li, C. Characterization, phylogenetic analyses and pathogenicity of Colletotrichum species on Morus alba in Sichuan Province, China. Plant Dis. 2019, 103, 2624–2633. [Google Scholar] [CrossRef]
- Hassan, O.; Kim, J.S.; Romain, B.B.N.D.; Chang, T. An account of Colletotrichum species associated with anthracnose of Atractylodes ovata in South Korea based on morphology and molecular data. PLoS ONE 2022, 17, e0263084. [Google Scholar] [CrossRef]
- Doyle, V.P.; Oudemans, P.V.; Rehner, S.A.; Litt, A. Habitat and host indicate lineage identity in Colletotrichum gloeosporioides sl from wild and agricultural landscapes in North America. PLoS ONE 2013, 8, e62394. [Google Scholar] [CrossRef]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Zhang, W.; Zhao, W.S.; Yan, J.Y. Notes on currently accepted species of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [Google Scholar] [CrossRef]
- Termorshuizen, A.J. Fungal and fungus-like pathogens of potato. In Potato Biology and Biotechnology, 1st ed.; Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., Taylor, M., MacKerron, D., Ross, H., Eds.; Elsevier Science BV: Amsterdam, The Netherlands, 2007; pp. 643–665. [Google Scholar]
- Massana-Codina, J.; Schnee, S.; Allard, P.M.; Rutz, A.; Boccard, J.; Michellod, E.; Cléroux, M.; Schürch, S.; Gindro, K.; Wolfender, J.L. Insights on the structural and metabolic resistance of potato (Solanum tuberosum) cultivars to tuber black dot (Colletotrichum coccodes). Front. Plant Sci. 2020, 11, 1287. [Google Scholar] [CrossRef]
- Heilmann, L.J.; Nitzan, N.; Johnson, D.A.; Pasche, J.S.; Doetkott, C.; Gudmestad, N.C. Genetic variability in the potato pathogen Colletotrichum coccodes as determined by amplified fragment length polymorphism and vegetative compatibility group analyses. Phytopathology 2006, 96, 1097–1107. [Google Scholar] [CrossRef][Green Version]
- Liu, F.; Hyde, K.D.; Cai, L. Neotypification of Colletotrichum coccodes, the causal agent of potato black dot disease and tomato anthracnose. Mycology 2011, 2, 248–254. [Google Scholar]
- Liu, F.; Cai, L.; Crous, P.W.; Damm, U. Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C. nigrum. Mycologia 2013, 105, 844–860. [Google Scholar] [CrossRef] [PubMed]
- Rodeva, R.; Stoyanova, Z.; Pandeva, R. Occurrence of Colletotrichum coccodes on pepper fruits in Bulgaria. Plant Prot. 2009, 20, 65–69. [Google Scholar]
- Stoyanova, Z.; Rodeva, R.; Manova, V.; Stoilov, L.; Mijatovic, M. Unusual Colletotrichum sp. associated with pepper fruit anthracnose in Bulgaria and Serbia—Preliminary results. In Proceedings of the International Symposium: Current Trends in Plant Protection, Institute for Plant Protection and Environment, Belgrade, Serbia, 25–28 September 2012; pp. 299–306. [Google Scholar]
- Rodeva, R.; Stoyanova, Z.; Manova, V.; Georgieva, R.; Stoilov, L. Isolation and characterization of Colletotrichum coccodes from potato in Bulgaria. Acta Hortic. 2016, 1142, 127–134. [Google Scholar] [CrossRef]
- Stoyanova, Z. Fungal Pathogens of the Genus Colletotrichum, as Causal Agents of Anthracnose and Root Rot in Vegetable Crops of Solanaceae Family in Bulgaria. Ph.D. Thesis, Institute of Plant Physiology and Genetics, Bulgarian Academy of Sciences, Sofia, Bulgaria, 2021. [Google Scholar]
- Bobev, S.G.; Zveibil, A.; Freeman, S. First report of Colletotrichum acutatum on strawberry in Bulgaria. Plant Dis. 2002, 86, 1178. [Google Scholar] [CrossRef] [PubMed]
- Jelev, Z.J.; Bobev, S.G.; Minz, D.; Maymon, M.; Freeman, S. First report of anthracnose fruit rot caused by Colletotrichum acutatum on pepper and tomato in Bulgaria. Plant Dis. 2008, 92, 172. [Google Scholar] [CrossRef]
- Rodeva, R.; Stoyanova, Z.; Pandeva, R.; Petrov, N. Field reaction to anthracnose caused by Colletotrichum spp. On pepper fruits. Acta Hortic. 2009, 830, 557–562. [Google Scholar] [CrossRef]
- Talhinhas, P.; Muthumeenakshi, S.; Neves-Martins, J.; Oliveira, H.; Sreenivasaprasad, S. Agrobacterium-mediated transformation and insertional mutagenesis in Colletotrichum acutatum for investigating varied pathogenicity lifestyles. Mol. Biotechnol. 2008, 39, 57–67. [Google Scholar] [CrossRef][Green Version]
- Sreenivasaprasad, S.; Brown, A.E.; Mills, R.P. DNA sequence variation and interrelationships among Colletotrichum species causing strawberry anthracnose. Physiol. Mol. Plant Pathol. 1992, 41, 265–281. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two Divergent Intragenomic rDNA ITS2 Types within a Monophyletic Lineage of the Fungus Fusarium are Nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; WF Freeman Co.: New York, NY, USA, 1973; p. 573. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Stoyanova, Z.B.; Rodeva, R.M.; Karov, I.; Kovacevik, B.; Manova, V.I.; Georgieva, R.G. Morphological and molecular characterization of Colletotrichum coccodes isolated from pepper cultivated in Bulgaria and Macedonia. J. Nat. Sci. 2013, 124, 249–262. [Google Scholar]
- Johnston, P.R.; Jones, D. Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia 1997, 89, 420–430. [Google Scholar] [CrossRef]
- Guerber, J.C.; Correll, J.C. Characterization of Glomerella acutata, the teleomorph of Colletotrichum acutatum. Mycologia 2001, 93, 216–229. [Google Scholar] [CrossRef]
- Talhinhas, P.; Baroncelli, R. Colletotrichum species and complexes: Geographic distribution, host range and conservation status. Fungal Divers. 2021, 110, 109–198. [Google Scholar] [CrossRef]
- Nasehi, A.; Kadir, J.; Rashid, T.S.; Awla, H.K.; Golkhandan, E.; Mahmodi, F. Occurrence of anthracnose fruit rot caused by Colletotrichum nymphaeae on pepper (Capsicum annuum) in Malaysia. Plant Dis. 2016, 100, 1244. [Google Scholar] [CrossRef]
- Lardner, R.; Johnston, P.R.; Plummer, K.M.; Pearson, M.N. Morphological and molecular analysis of Colletotrichum acutatum sensu lato. Mycol. Res. 1999, 103, 275–285. [Google Scholar] [CrossRef]
- Jelev, Z.J.; Bobev, S.G.; Minz, D.; Maymon, M.; Freeman, S. Characterization of Colletotrichum species causing strawberry anthracnose in Bulgaria. J. Phytopathol. 2008, 156, 668–677. [Google Scholar] [CrossRef]
- Förster, H.; Adaskaveg, J.E. Identification of subpopulations of Colletotrichum acutatum and epidemiology of almond anthracnose in California. Phytopathology 1999, 89, 1056–1065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sreenivasaprasad, S.; Talhinhas, P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Mol. Plant Pathol. 2005, 6, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.Z.; Zhang, C.; Liu, F.; Wang, W.Z.; Liu, L.; Cai, L.; Liu, X.L. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 2017, 38, 20–37. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Bhunjun, C.S.; Hyde, K.D.; Gentekaki, E.; Itthayakorn, P. Colletotrichum: Lifestyles, biology, morpho-species, species complexes and accepted species. Mycosphere 2021, 12, 519–669. [Google Scholar] [CrossRef]
- Waller, J.M.; Bridge, P.D.; Black, R.; Hakiza, G. Characterization of the coffee berry disease pathogen, C. kahawae sp. Nov. Mycol. Res. 1993, 97, 989–994. [Google Scholar] [CrossRef]
- Liu, F.; Damm, U.; Cai, L.; Crous, P.W. Species of the Colletototrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Divers. 2013, 61, 89–105. [Google Scholar] [CrossRef]
- Afanador-Kafuri, L.; González, A.; Gañán, L.; Mejía, J.F.; Cardona, N.; Álvarez, E. Characterization of Colletotrichum species causing anthracnose in Andean blackberry in Colombia. Plant Dis. 2014, 98, 1503–1513. [Google Scholar] [CrossRef]
- Ismail, A.M.; Cirvilleri, G.; Yaseen, T.; Epifani, F.; Perrone, G.; Polizzi, G. Characterisation of Colletotrichum species causing anthracnose disease of mango in Italy. J. Plant Pathol. 2015, 97, 167–171. [Google Scholar]
- Perrone, G.; Magistà, D.; Ismail, A.M. First report of Colletotrichum kahawae subsp. ciggaro on mandarin in Italy. J. Plant Pathol. 2016, 98, 682. [Google Scholar]
- Zhang, H.; Wei, Y.; Shi, H. First report of anthracnose caused by Colletotrichum kahawae subsp. ciggaro on Areca in China. Plant Dis. 2020, 104, 1871. [Google Scholar]
- Cabral, A.; Azinheira, H.G.; Talhinhas, P.; Batista, D.; Ramos, A.P.; Silva, M.C.; Oliveira, H.; Várzea, V. Pathological, morphological, cytogenomic, biochemical and molecular data support the distinction between Colletotrichum cigarro comb. et stat. nov. and Colletotrichum kahawae. Plants 2020, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Rivera, Y.; Stommel, J.; Dumm, J.; Ismaiel, A.; Wyenandt, C.A.; Crouch, J.A. First report of Colletotrichum nigrum causing anthracnose disease on tomato fruit in New Jersey. Plant Dis. 2016, 100, 2162. [Google Scholar] [CrossRef]
- Chesters, C.G.C.; Hornby, D. Studies on Colletotrichum coccodes: I. The taxonomic significance of variation in isolates from tomato roots. Trans. Br. Mycol. Soc. 1965, 48, 573–581. [Google Scholar] [CrossRef]
- Chesters, C.G.C.; Hornby, D. Studies on Colletotrichum coccodes: II. Alternative host tests and tomato fruit inoculations using a typical tomato root isolate. Trans. Br. Mycol. Soc. 1965, 48, 583–594. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
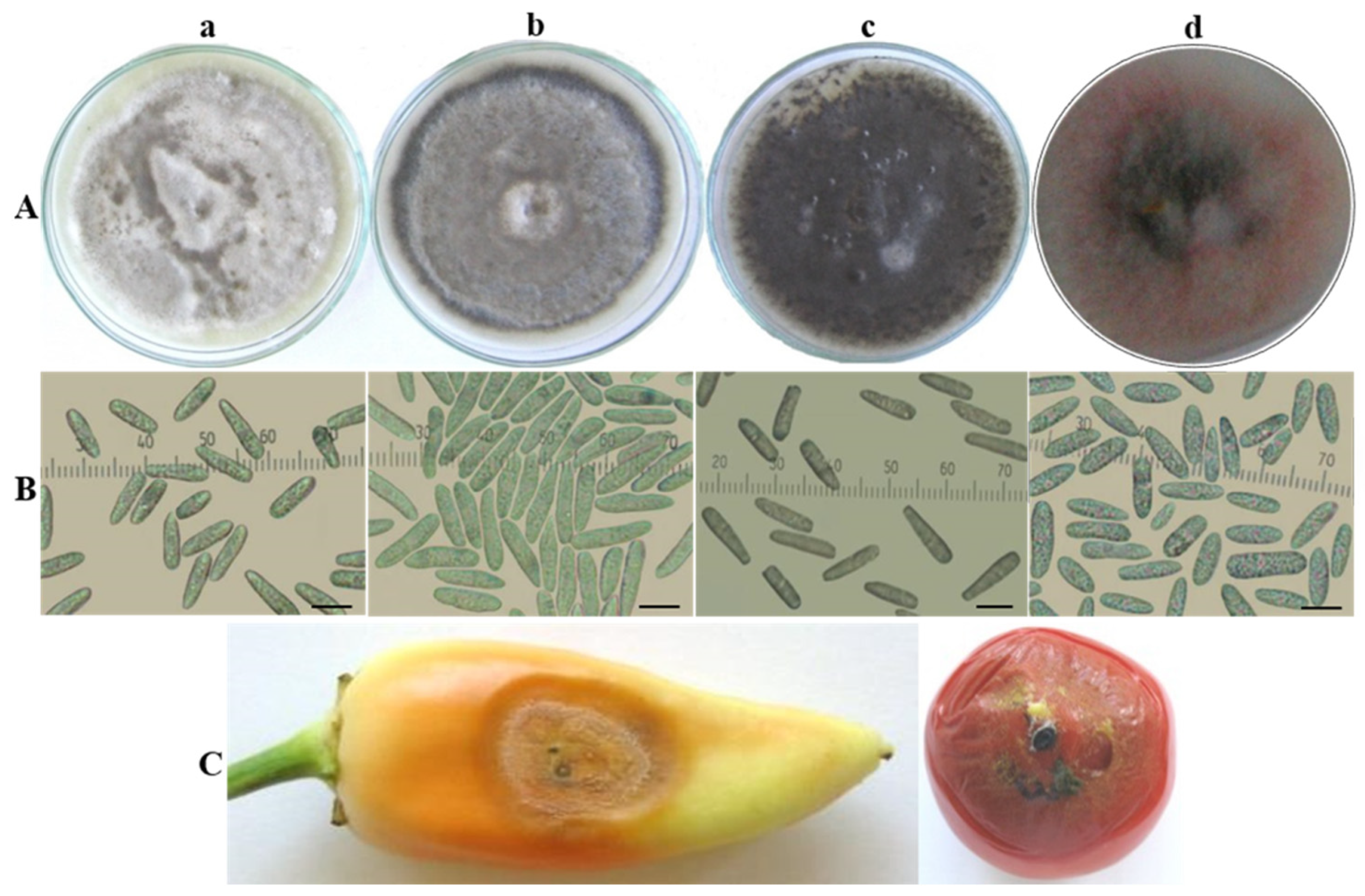
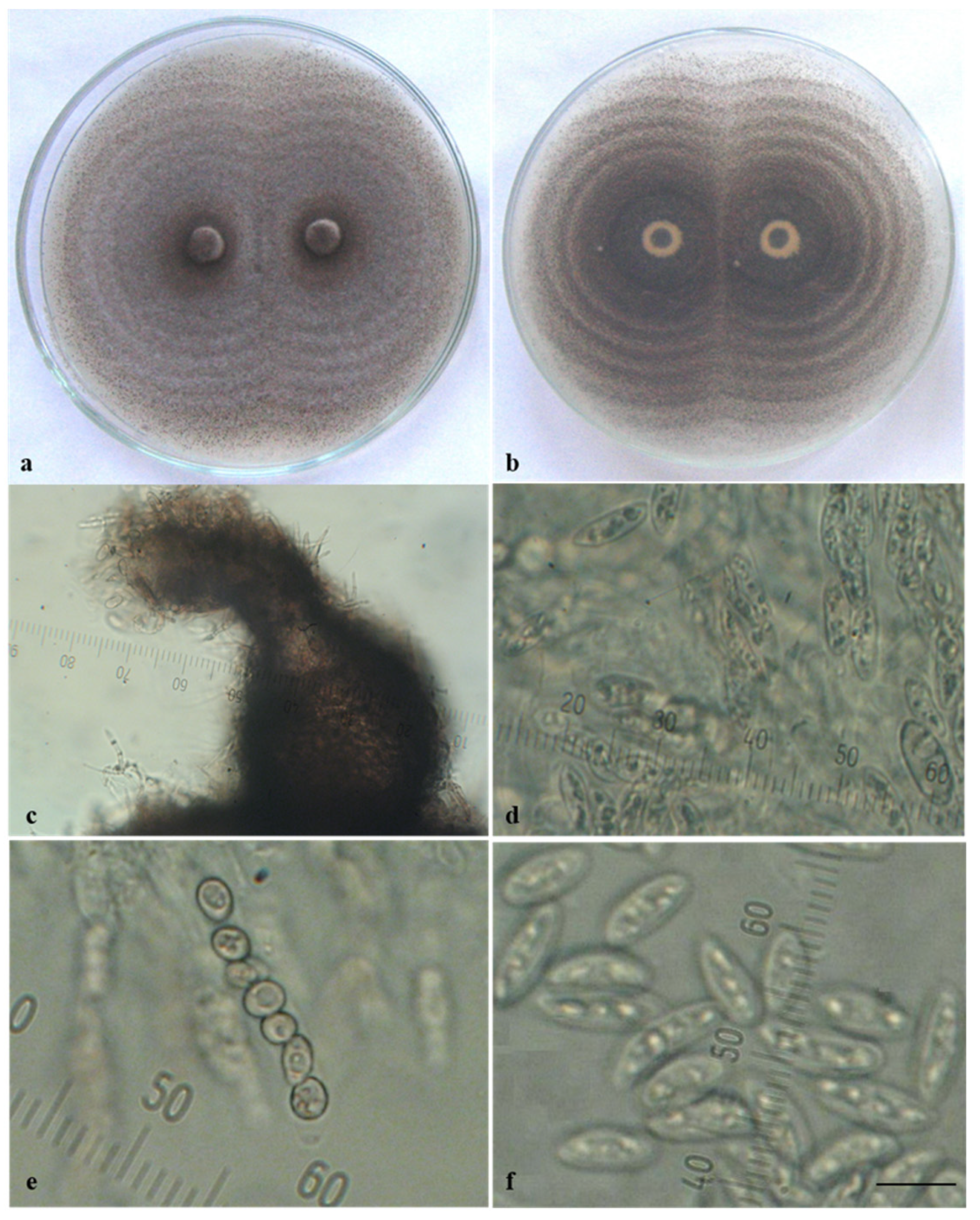
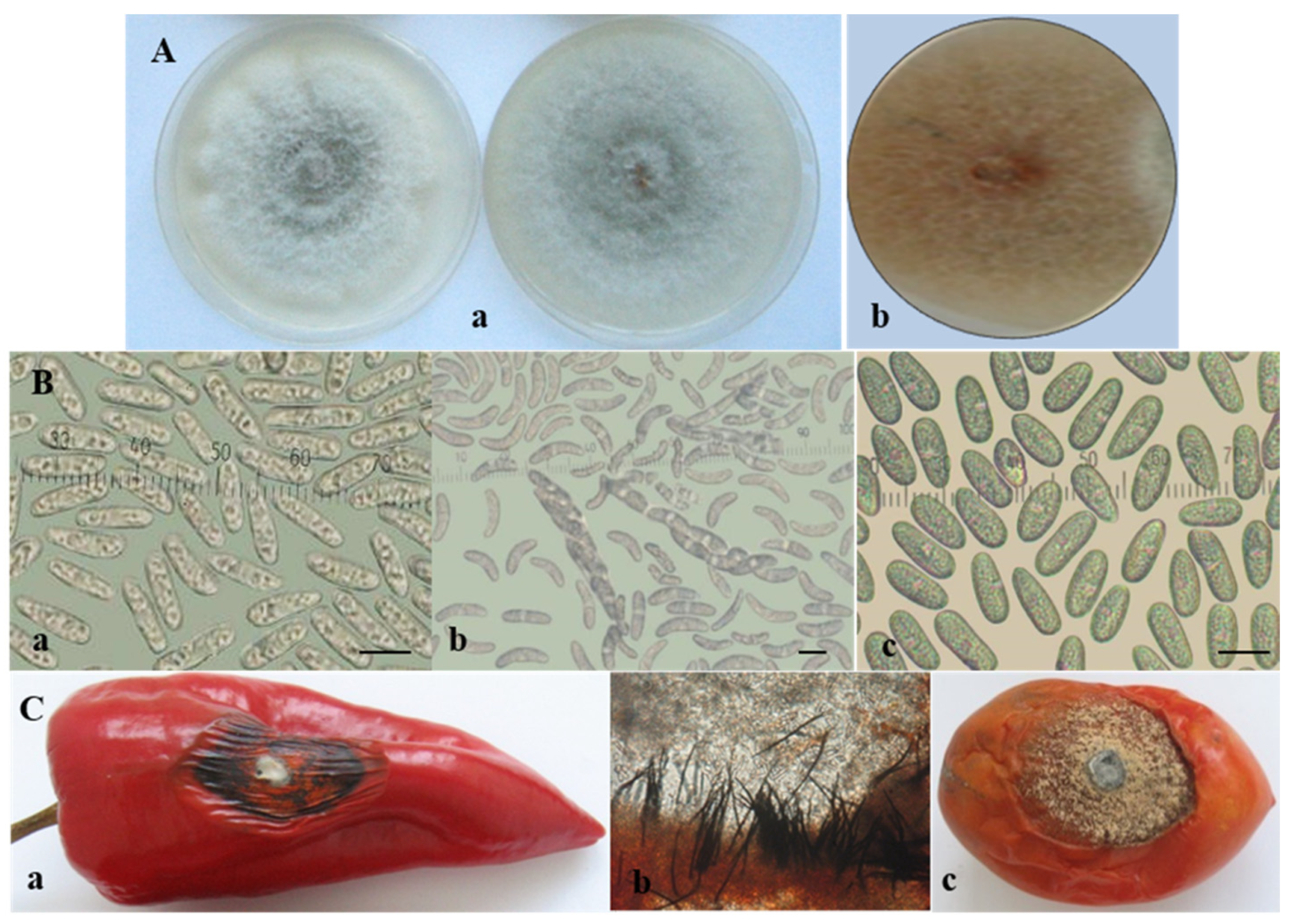

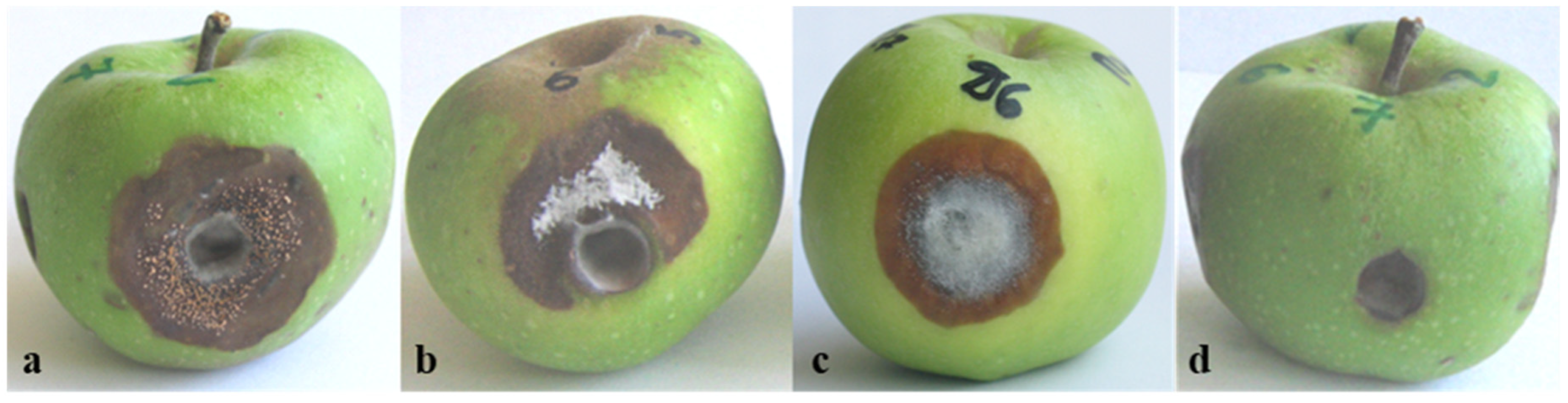

| Isolate Designation | Host/Substrate | Locality | Year of Isolation |
|---|---|---|---|
| Ca13-1 | Capsicum annuum, fruit | Sofia, Bulgaria | 2007 |
| CaD13-3 | Solanum lycopersicum, fruit | Sofia, Bulgaria | 2013 |
| Ca44-1 | Solanum lycopersicum, fruit | Lozen, Bulgaria | 2008 |
| CBS490.92 | Solanum lycopersicum | CBS | |
| CgB1-1 | Capsicum annuum, fruit | Sofia, Bulgaria | 2012 |
| CgB27-1 | Capsicum annuum, fruit | Sofia, Bulgaria | 2012 |
| CgD6-1 | Solanum lycopersicum, fruit | Sofia, Bulgaria | 2012 |
| CgD6-2 | Solanum lycopersicum, fruit | Sofia, Bulgaria | 2012 |
| Cm13 | Musa sp., fruit | imported from Ecuador | 2013 |
| Cc2-1-10 | Solanum lycopersicum, fruit | Sofia, Bulgaria | 2010 |
| CcB8-1 | Capsicum annuum, fruit | Sofia, Bulgaria | 2010 |
| CcB8-2 | Capsicum annuum, root | Sofia, Bulgaria | 2011 |
| Cc7-1 | Capsicum annuum, fruit | Sveti Nikole, The Republic of North Macedonia | 2011 |
| Cc7-2 | Capsicum annuum, fruit | Sveti Nikole, The Republic of North Macedonia | 2011 |
| Cc26-2 | Capsicum annuum, fruit | Strumitsa, The Republic of North Macedonia | 2011 |
| CcB12-9 | Capsicum annuum, root | Sofia, Bulgaria | 2012 |
| CcB12-13 | Capsicum annuum, root | Sofia, Bulgaria | 2012 |
| CcK18-1 | Capsicum annuum, root | Sofia, Bulgaria | 2012 |
| Cc13-6 | Capsicum annuum, fruit | Sofia, Bulgaria | 2013 |
| CcV10 | Capsicum annuum, fruit | Sofia, Bulgaria | 2014 |
| Cc40-1 | Solanum melongenae, fruit | Sofia, Bulgaria | 2009 |
| P5-4 | Solanum tuberosum, tuber | Kyustendil, Bulgaria | 2013 |
| P13-1 | Solanum tuberosum, root | Lozen, Bulgaria | 2013 |
| P13-2-1 | Solanum tuberosum, stolon | Sofia, Bulgaria | 2013 |
| P13-2-3 | Solanum tuberosum, stem basis | Sofia, Bulgaria | 2013 |
| CBS 527.77 | Solanum lycopersicum, root | CBS (Bulgaria) | 1977 |
| Barcode Region | Primer Name | Primer Sequences 5′-3′ | T ann | Reference |
|---|---|---|---|---|
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | 56 °C | [53] |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| ACT | ACT-512F | ATGTGCAAGGCCGGTTTCGC | 59 °C | [54] |
| ACT-783R | TACGAGTCCTTCTGGCCCAT | |||
| EF-1a | EF1-728F | CATCGAGAAGTTCGAGAAGG | 58 °C | [54] |
| EF1-986R | TACTTGAAGGAACCCTTACC | |||
| Tub2 | T1 | AACATGCGTGAGATTGTAAGT | 60 °C | [55] |
| T2 | TAGTGACCCTTGGCCCAGTTG |
| Isolate Designation | Length | Width | Shape |
|---|---|---|---|
| Ca13-1 | (8.1−) 10.8 ± 0.1 (−13.4) | (3.3−) 4.2 ± 0.1 (−5.5) | Cylindrical to cylindric–clavate, pointed at one end and rounded at the other |
| CaD13-3 | (9.2−) 12.1 ± 0.1 (−15.8) | (3.6−) 4.7 ± 0.1 (−5.9) | Cylindrical to fusiform with both ends acute or one end round and one end slightly acute |
| Ca44-1 | (12.0−) 15.5 ± 0.2 (−21.5) | (3.5−) 4.5 ± 0.1 (−5.8) | Cylindrical to clavate with one end round and one end acute to truncate |
| CBS 490.92 | (11.1−) 14.5 ± 0.9 (19.2) | (3.3−) 4.4 ± 0.1 (−5.9) | Fusiform to cylindrical with both ends acute |
| CgB1-1 | (12.8−) 15.5 ± 1.1 (−17.6) | (3.7−) 4.7 ± 0.4 (−6.2) | Cylindrical with two to seven oil globules, rounded at both ends or pointed at one end and rounded at the other |
| CgB27-1 | (12.5−) 15.1 ± 0.9 (−16.7) | (3.5−) 4.6 ± 0.3 (−6.3) | |
| CgD6-1 | (13.0−) 15.8 ± 0.7 (−17.8) | (3.9−) 4.9 ± 0.4 (−6.5) | |
| CgD6-2 | (12.6−) 14.9 ± 0.8 (−16.6) | (3.4−) 4.5 ± 0.5 (−6.1) | |
| Cm13 | (11.4−) 13.3 ± 0.2 (−16.1) | (5.4−) 6.1 ± 0.1 (−7.1) | Cylindrical, with rounded ends |
| Cc2-1-10 | (11.3−) 16.3 ± 0.2 (−20.6) | (2.9−) 4.6 ± 0.1 (−6.8) | Cylindrical, rounded at both ends |
| CcB8-1 | (19.1−) 21.3 ± 1.7 (−24.6) | (3.1−) 4.1 ± 0.4 (−4.7) | Cylindrical, rounded at both ends |
| CcB8-2 | (11.8−) 20.1 ± 0.1 (−21.6) | (4.0−) 5.1 ± 0,1 (−6.4) | Fusiform to cylindrical |
| Cc7-1 | (9.4−) 15.8 ± 1.0 (−25.7) | (3.4−) 4.2 ± 0.1 (−5.0) | Cylindrical, rounded at both ends with two to seven globules |
| Cc7-2 | (8.0−) 13.7 ± 0.5 (−21.2) | (3.4−) 4.7 ± 0.1 (−5.9) | Cylindrical, rounded at both ends |
| Cc26-2 | (10.8) 15.0 ± 0.6 (−19.2) | (3.6−) 4.7 ± 0.2 (−5.8) | Cylindrical, rounded at both ends |
| CcB12-9 | (12.7−) 17.0 ± 0.7 (−18.6) | (4.5−) 5.2 ± 0.1 (−5.9) | Fusiform to cylindrical |
| CcB12-13 | (15.8−) 20.2 ± 0.1 (−23.6) | (3.2−) 4.2 ± 0.1 (−5.9) | Fusiform, attenuated at the ends |
| CcK18-1 | (15.4−) 19.7 ± 0.2 (−23.4) | (3.3−) 4.8 ± 0.1 (−6.8) | Fusiform, attenuated at the ends |
| Cc13-6 | (10.8−) 18.7 ± 0,4 (−27.5) | (3.8−) 5.8 ± 0,1 (−8.6) | Cylindrical, rounded at both ends |
| CcV10 | (9.8−) 17.7 ± 0.4 (−26.3) | (3.2−) 4.9 ± 0.1 (−6.5) | Cylindrical, rounded at both ends |
| Cc40-1 | (11.1−) 16.9 ± 0.5 (−20.2) | (3.1−) 4.8 ± 0.1 (−6.7) | Cylindrical, rounded at both ends |
| P5-4 | (16.0−) 20.7 ± 0.1 (−25.4) | (3.3−) 4.5 ± 0.1 (−5.6) | Cylindrical, rounded at both ends |
| P13-1 | (17.7−) 21.0 ± 0.1 (−25.2) | (3.5−) 5.0 ± 0.1 (−6.5) | Fusiform to cylindrical |
| P13-2-1 | (7.8−) 15.0 ± 0.4 (−21.0) | (3.4−) 5.1 ± 0.1 (−7.0) | Cusiform, attenuated at the ends |
| P13-2-3 | (18.7−) 21.3 ± 0.1 (−25.4) | (3.7−) 5.0 ± 0.1 (−6.2) | Fusiform to cylindrical |
| CBS 527.77 | (12.9−) 18.6 ± 0.3 (−23.2) | (2.8−) 4.3 ± 0.1 (−5.6) | Fusiform to cylindrical |
| Barcode Region | N | Alignment Length (bp) | % AE | % SE |
|---|---|---|---|---|
| ITS | 26 | 556 | 100 | 100 |
| ACT | 26 | 246 | 100 | 100 |
| EF-1a | 19 | 242 | 75.0 | 100 |
| Tub2 | 26 | 750 | 100 | 100 |
| DNA Barcode Region | N Taxa | Ns | C | V | Pi | S | Average Pairwise Distance/SE |
|---|---|---|---|---|---|---|---|
| ITS | 46 | 556 | 434 | 115 | 68 | 47 | 0.051/0.006 |
| ACT | 52 | 246 | 127 | 119 | 85 | 34 | 0.191/0.020 |
| EF-1a | 25 | 242 | 25 | 128 | 85 | 43 | 0.397/0.053 |
| Tub2 | 57 | 750 | 421 | 328 | 270 | 58 | 0.170/0.011 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manova, V.; Stoyanova, Z.; Rodeva, R.; Boycheva, I.; Korpelainen, H.; Vesterinen, E.; Wirta, H.; Bonchev, G. Morphological, Pathological and Genetic Diversity of the Colletotrichum Species, Pathogenic on Solanaceous Vegetable Crops in Bulgaria. J. Fungi 2022, 8, 1123. https://doi.org/10.3390/jof8111123
Manova V, Stoyanova Z, Rodeva R, Boycheva I, Korpelainen H, Vesterinen E, Wirta H, Bonchev G. Morphological, Pathological and Genetic Diversity of the Colletotrichum Species, Pathogenic on Solanaceous Vegetable Crops in Bulgaria. Journal of Fungi. 2022; 8(11):1123. https://doi.org/10.3390/jof8111123
Chicago/Turabian StyleManova, Vasilissa, Zornitsa Stoyanova, Rossitza Rodeva, Irina Boycheva, Helena Korpelainen, Eero Vesterinen, Helena Wirta, and Georgi Bonchev. 2022. "Morphological, Pathological and Genetic Diversity of the Colletotrichum Species, Pathogenic on Solanaceous Vegetable Crops in Bulgaria" Journal of Fungi 8, no. 11: 1123. https://doi.org/10.3390/jof8111123
APA StyleManova, V., Stoyanova, Z., Rodeva, R., Boycheva, I., Korpelainen, H., Vesterinen, E., Wirta, H., & Bonchev, G. (2022). Morphological, Pathological and Genetic Diversity of the Colletotrichum Species, Pathogenic on Solanaceous Vegetable Crops in Bulgaria. Journal of Fungi, 8(11), 1123. https://doi.org/10.3390/jof8111123






