Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design
2.3. Soil Sampling and Soil Biochemical Analyses
2.4. DNA Extraction, PCR Amplification, and Illumina MiSeq Sequencing
2.5. Microbial Co-Occurrence Network Construction
2.6. Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
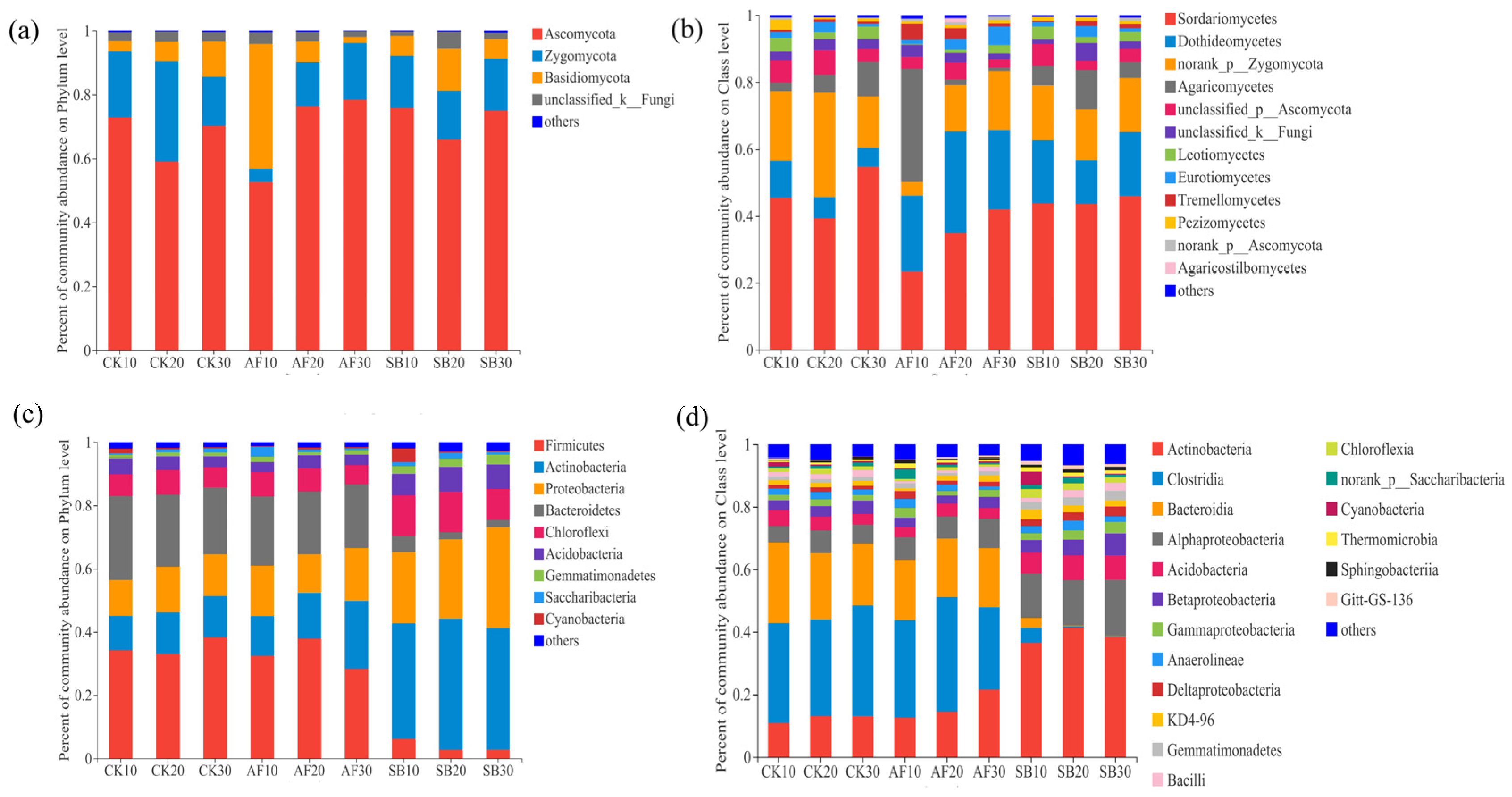
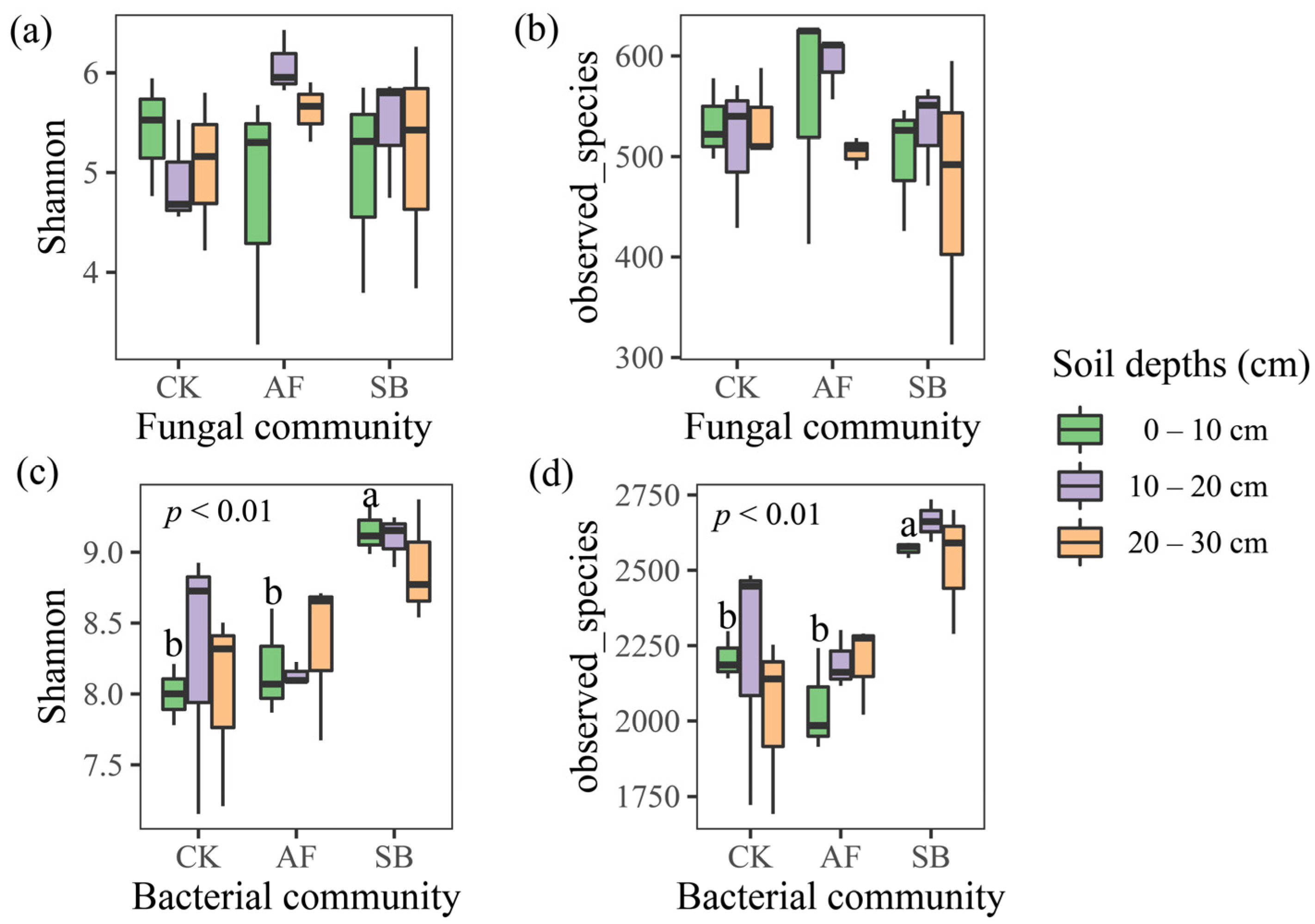
3.2. Bacterial and Fungal Community Structure and Species Diversity
3.3. Relationship between Environmental Variables and Community Structure of Bacteria and Fungi
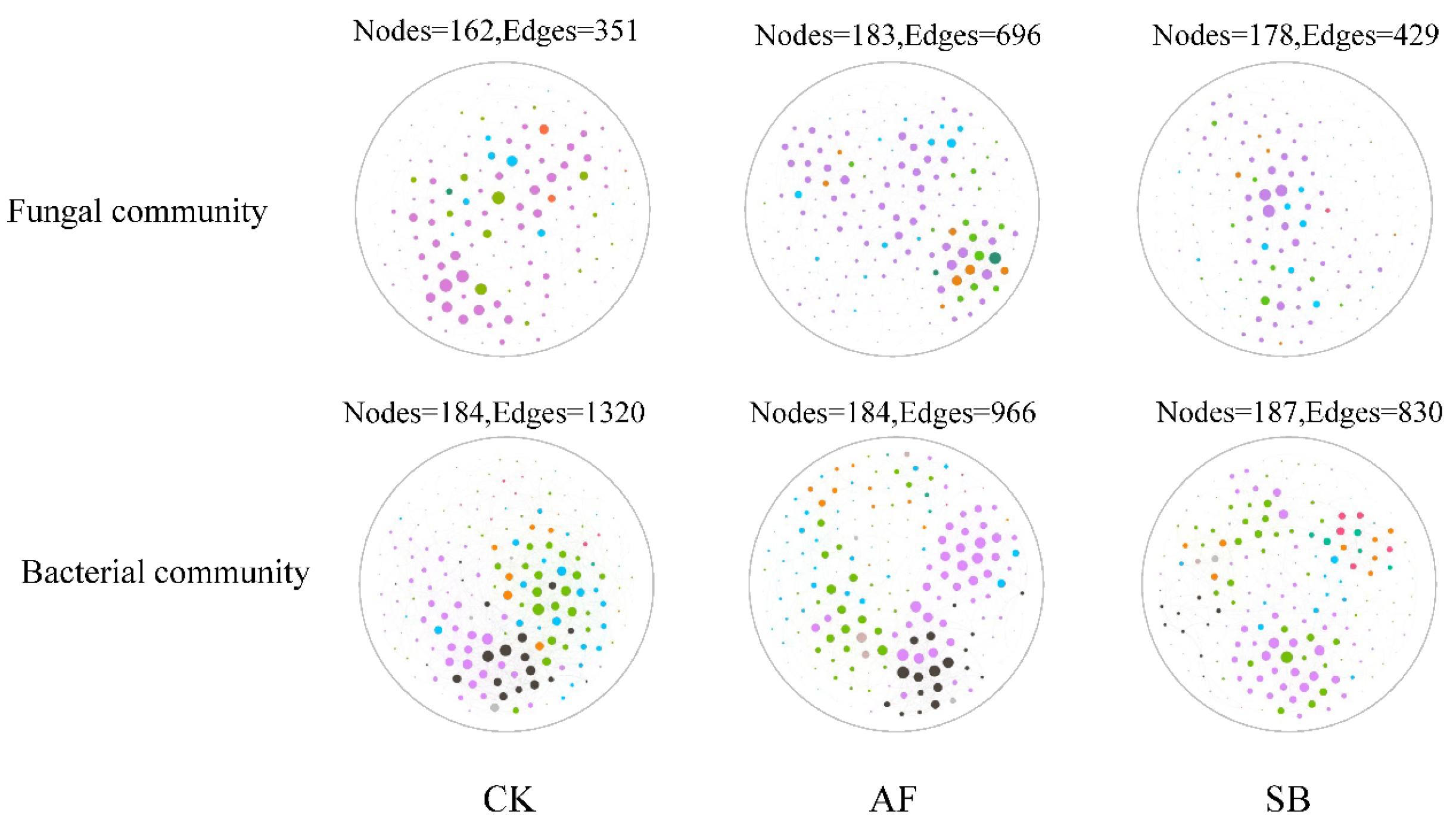
3.4. Soil Microbial Network Complexity
4. Discussion
4.1. Impact of Vegetation Restoration on Soil Properties
4.2. Effects of Vegetation Restoration on the Soil Fungal Community Structures
4.3. Effects of Vegetation Restoration on the Soil Bacterial Community Structures
4.4. Effects of Vegetation Restoration on the Soil Microbial Network Complexity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bardgett, R.D.; Bullock, J.M.; Lavorel, S.; Manning, P.; Schaffner, U.; Ostle, N.; Chomel, M.; Durigan, G.; Fry, E.L.; Johnson, D.; et al. Combatting global grassland degradation. Nat. Rev. Earth Environ. 2021, 2, 720–735. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Zhang, Z.Y.; Tong, L.J.; Khalifa, M.; Wang, Q.; Gang, C.C.; Wang, Z.Q.; Li, J.L.; Sun, Z.G. Assessing the effects of climate variation and human activities on grassland degradation and restoration across the globe. Ecol. Indic. 2019, 106, 105504. [Google Scholar] [CrossRef]
- Steffen, W.; Richardson, K.; Rockstrom, J.; Cornell, S.E.; Fetzer, I.; Bennett, E.M.; Biggs, R.; Carpenter, S.R.; de Vries, W.; de Wit, C.A.; et al. Planetary boundaries: Guiding human development on a changing planet. Science 2015, 347, 1259855. [Google Scholar] [CrossRef] [PubMed]
- Suding, K.; Higgs, E.; Palmer, M.; Callicott, J.B.; Anderson, C.B.; Baker, M.; Gutrich, J.J.; Hondula, K.L.; LaFevor, M.C.; Larson, B.M.H.; et al. Committing to ecological restoration. Science 2015, 348, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.M. Estimating natural landscape changes from 1992 to 2030 in the conterminous US. Landsc. Ecol. 2010, 25, 999–1011. [Google Scholar] [CrossRef]
- Zhou, H.C.; Ma, A.Z.; Liu, G.H.; Zhou, X.R.; Yin, J.; Liang, Y.; Wang, F.; Zhuang, G.Q. Reduced interactivity during microbial community degradation leads to the extinction of Tricholomas matsutake. Land Degrad. Dev. 2021, 32, 5118–5128. [Google Scholar] [CrossRef]
- Cheng, C.; Gao, M.; Zhang, Y.; Long, M.; Wu, Y.; Li, X. Effects of disturbance to moss biocrusts on soil nutrients, enzyme activities, and microbial communities in degraded karst landscapes in southwest China. Soil Biol. Biochem. 2021, 152, 108065. [Google Scholar] [CrossRef]
- Tucker, C.; Antoninka, A.; Day, N.; Poff, B.; Reed, S. Biological soil crust salvage for dryland restoration: An opportunity for natural resource restoration. Restor. Ecol. 2020, 28, S9–S16. [Google Scholar] [CrossRef]
- Strickland, M.S.; Callaham, M.A.; Gardiner, E.S.; Stanturf, J.A.; Leff, J.W.; Fierer, N.; Bradford, M.A. Response of soil microbial community composition and function to a bottomland forest restoration intensity gradient. Appl. Soil Ecol. 2017, 119, 317–326. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Zhao, C.; Shi, R.; Xue, R.; Li, X. Revegetation by sowing reduces soil bacterial and fungal diversity. Ecol. Evol. 2020, 10, 431–440. [Google Scholar] [CrossRef]
- Stanturf, J.A.; Palik, B.J.; Dumroese, R.K. Contemporary forest restoration: A review emphasizing function. For. Ecol. Manag. 2014, 331, 292–323. [Google Scholar] [CrossRef]
- Xu, R.; Zhao, H.; Liu, G.; You, Y.; Ma, L.; Liu, N.; Zhang, Y. Effects of nitrogen and maize plant density on forage yield and nitrogen uptake in an alfalfa-silage maize relay intercropping system in the North China Plain. Field Crops Res. 2021, 263, 108068. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Zhang, F. Abundant and rare bacteria possess different diversity and function in crop monoculture and rotation systems across regional farmland. Soil Biol. Biochem. 2022, 171, 108742. [Google Scholar] [CrossRef]
- Dong, W.-H.; Zhang, S.; Rao, X.; Liu, C.-A. Newly-reclaimed alfalfa forage land improved soil properties comparison to farmland in wheat-maize cropping systems at the margins of oases. Ecol. Eng. 2016, 94, 57–64. [Google Scholar] [CrossRef]
- Xu, R.; Zhao, H.; Liu, G.; Li, Y.; Li, S.; Zhang, Y.; Liu, N.; Ma, L. Alfalfa and silage maize intercropping provides comparable productivity and profitability with lower environmental impacts than wheat-maize system in the North China plain. Agric. Syst. 2022, 195, 103305. [Google Scholar] [CrossRef]
- Hafner, S.; Kuzyakov, Y. Carbon input and partitioning in subsoil by chicory and alfalfa. Plant Soil 2016, 406, 29–42. [Google Scholar] [CrossRef]
- Clement, C.; Sleiderink, J.; Svane, S.F.; Smith, A.G.; Diamantopoulos, E.; Desbroll, D.B.; Thorup-Kristensen, K. Comparing the deep root growth and water uptake of intermediate wheatgrass (Kernza (R)) to alfalfa. Plant Soil 2022, 472, 369–390. [Google Scholar] [CrossRef]
- Jokela, W.; Posner, J.; Hedtcke, J.; Balser, T.; Read, H. Midwest Cropping System Effects on Soil Properties and on a Soil Quality Index. Agron. J. 2011, 103, 1552–1562. [Google Scholar] [CrossRef]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.J. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Xu, H.; Liu, N.; Zhang, Y. Short-Term Snow Removal Alters Fungal but Not Bacterial Beta Diversity and Structure during the Spring Snowmelt Period in a Meadow Steppe of China. J. Fungi 2022, 8, 234. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Deng, Y.; Shen, L.N.; Wen, C.Q.; Yan, Q.Y.; Ning, D.L.; Qin, Y.J.; Xue, K.; Wu, L.Y.; He, Z.L.; et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 2016, 7, 12083. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.K.; Bernhardt, E.S.; Bier, R.L.; Bradford, M.A.; Boot, C.M.; Cotner, J.B.; del Giorgio, P.A.; Evans, S.E.; Graham, E.B.; Jones, S.E.; et al. Understanding how microbiomes influence the systems they inhabit. Nat. Microbiol. 2018, 3, 977–982. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.S.; Lauber, C.; Fierer, N.; Bradford, M.A. Testing the functional significance of microbial community composition. Ecology 2009, 90, 441–451. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Firestone, M.K. Seasonal dynamics of microbial community composition and function in oak canopy and open grassland soils. Microb. Ecol. 2006, 52, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, Z.J.; Sun, Q.Y.; Zhan, J.; Yang, Y.; Wang, D. Ecological restoration alters microbial communities in mine tailings profiles. Sci. Rep. 2016, 6, 25193. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.F.; Mills, J.G.; Gellie, N.J.C.; Bissett, A.; Lowe, A.J.; Breed, M.F. High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biol. Conserv. 2018, 217, 113–120. [Google Scholar] [CrossRef]
- Maul, J.; Drinkwater, L. Short-term plant species impact on microbial community structure in soils with long-term agricultural history. Plant Soil 2010, 330, 369–382. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Y.; Peng, L.; Liu, G.; Wan, X.; Hua, Y.; Zhu, D.; Hamilton, D.P. Diversity of anammox bacteria and abundance of functional genes for nitrogen cycling in the rhizosphere of submerged macrophytes in a freshwater lake in summer. J. Soils Sediments 2019, 19, 3648–3656. [Google Scholar] [CrossRef]
- Kamble, P.N.; Baath, E. Carbon and Nitrogen Amendments Lead to Differential Growth of Bacterial and Fungal Communities in a High-pH Soil. Pedosphere 2018, 28, 255–260. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Yu, Y.; Zheng, L.; Zhou, Y.J.; Sang, W.G.; Zhao, J.N.; Liu, L.; Li, C.; Xiao, C.W. Changes in soil microbial community structure and function following degradation in a temperate grassland. J. Plant Ecol. 2021, 14, 384–397. [Google Scholar] [CrossRef]
- Wang, C.Q.; Xue, L.; Dong, Y.H.; Jiao, R.Z. Effects of stand density on soil microbial community composition and enzyme activities in subtropical Cunninghamia lanceolate (Lamb.) Hook plantations. For. Ecol. Manag. 2021, 479, 118559. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Mendez, M.; Garcia, D.; Maestre, F.T.; Escudero, A. More ecology is needed to restore Mediterranean ecosystems: A reply to Valladares and Gianoli. Restor. Ecol. 2008, 16, 210–216. [Google Scholar] [CrossRef]
- Ge, A.H.; Liang, Z.H.; Xiao, J.L.; Zhang, Y.; Zeng, Q.; Xiong, C.; Han, L.L.; Wang, J.T.; Zhang, L.M. Microbial assembly and association network in watermelon rhizosphere after soil fumigation for Fusarium wilt control. Agric. Ecosyst. Environ. 2021, 312, 107336. [Google Scholar] [CrossRef]
- Dong, K.; Yu, Z.; Kerfahi, D.; Lee, S.S.; Li, N.; Yang, T.; Adams, J.M. Soil microbial co-occurrence networks become less connected with soil development in a high Arctic glacier foreland succession. Sci. Total Environ. 2022, 813, 152565. [Google Scholar] [CrossRef]
- Chen, W.; Wang, J.; Chen, X.; Meng, Z.; Xu, R.; Duoji, D.; Zhang, J.; He, J.; Wang, Z.; Chen, J.; et al. Soil microbial network complexity predicts ecosystem function along elevation gradients on the Tibetan Plateau. Soil Biol. Biochem. 2022, 172, 108766. [Google Scholar] [CrossRef]
- Pržulj, N.; Malod-Dognin, N. Network analytics in the age of big data. Science 2016, 353, 123–124. [Google Scholar] [CrossRef]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Xue, L.; Ren, H.D.; Brodribb, T.J.; Wang, J.; Yao, X.H.; Li, S. Long term effects of management practice intensification on soil microbial community structure and co-occurrence network in a non-timber plantation. For. Ecol. Manag. 2020, 459, 117805. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.W.; Zhang, Y.; Xiao, N.J.; Ning, D.L.; Shi, Z.; Zhou, X.S.; Wu, L.Y.; Yang, Y.F.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Bowman, R.A. A Rapid Method to Determine Total Phosphorus in Soils. Soil Sci. Soc. Am. J. 1988, 52, 1301–1304. [Google Scholar] [CrossRef]
- Xiao-Na, L.I.; Zhang, W.W.; Zhao, C.Q.; Song, J.K.; Shi, R.S.; Xue, R.B.; Wang, C. Plant Diversity and Soil Physicochemical Properties in the Wasteland of Yanqing District. Acta Agrestia Sin. 2019, 27, 695–701. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- McGuire, K.L.; Payne, S.G.; Palmer, M.I.; Gillikin, C.M.; Keefe, D.; Kim, S.J.; Gedallovich, S.M.; Discenza, J.; Rangamannar, R.; Koshner, J.A.; et al. Digging the New York City Skyline: Soil Fungal Communities in Green Roofs and City Parks. PLoS ONE 2013, 8, e58020. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Montoya, J.M.; Pimm, S.L.; Solé, R.V. Ecological networks and their fragility. Nature 2006, 442, 259–264. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Zhu, R.; Chen, N.; Ding, L.; Chen, C. Vegetation richness, species identity and soil nutrients drive the shifts in soil bacterial communities during restoration process. Environ. Microbiol. Rep. 2021, 13, 411–424. [Google Scholar] [CrossRef]
- Bienes, R.; Marques, M.J.; Sastre, B.; García-Díaz, A.; Ruiz-Colmenero, M. Eleven years after shrub revegetation in semiarid eroded soils. Influence in soil properties. Geoderma 2016, 273, 106–114. [Google Scholar] [CrossRef]
- Zhang, K.; Li, X.; Cheng, X.; Zhang, Z.; Zhang, Q. Changes in soil properties rather than functional gene abundance control carbon and nitrogen mineralization rates during long-term natural revegetation. Plant Soil 2019, 443, 293–306. [Google Scholar] [CrossRef]
- Huang, C.; Zeng, Y.; Wang, L.; Wang, S. Responses of soil nutrients to vegetation restoration in China. Reg. Environ. Change 2020, 20, 82. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, D.; Jin, Y.; Wang, M.; Song, Y.; Li, G. Effects of fencing on vegetation and soil restoration in a degraded alkaline grassland in northeast China. J. Arid Land 2014, 6, 478–487. [Google Scholar] [CrossRef]
- Liang, Y.; Pan, F.; He, X.; Chen, X.; Su, Y. Effect of vegetation types on soil arbuscular mycorrhizal fungi and nitrogen-fixing bacterial communities in a karst region. Environ. Sci. Pollut. Res. 2016, 23, 18482–18491. [Google Scholar] [CrossRef]
- Hu, P.; Zhao, Y.; Xiao, D.; Xu, Z.; Zhang, W.; Xiao, J.; Wang, K. Dynamics of soil nitrogen availability following vegetation restoration along a climatic gradient of a subtropical karst region in China. J. Soils Sediments 2021, 21, 2167–2178. [Google Scholar] [CrossRef]
- Zhu, Y.; Guo, B.; Liu, C.; Lin, Y.; Fu, Q.; Li, N.; Li, H. Soil fertility, enzyme activity, and microbial community structure diversity among different soil textures under different land use types in coastal saline soil. J. Soils Sediments 2021, 21, 2240–2252. [Google Scholar] [CrossRef]
- Fu, B.; Qi, Y.; Chang, Q. Impacts of revegetation management modes on soil properties and vegetation ecological restoration in degraded sandy grassland in farming-pastoral ecotone. Int. J. Agric. Biol. Eng. 2015, 8, 26–34. [Google Scholar] [CrossRef]
- Qi, Y.; Yang, F.; Shukla, M.K.; Pu, J.; Chang, Q.; Chu, W. Desert Soil Properties after Thirty Years of Vegetation Restoration in Northern Shaanxi Province of China. Arid Land Res. Manag. 2015, 29, 454–472. [Google Scholar] [CrossRef]
- Lin, J.; Roswanjaya, Y.P.; Kohlen, W.; Stougaard, J.; Reid, D. Nitrate restricts nodule organogenesis through inhibition of cytokinin biosynthesis in Lotus japonicus. Nat. Commun. 2021, 12, 6544. [Google Scholar] [CrossRef]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Shao, X.; Liu, N. Soil nitrogen and climate drive the positive effect of biological soil crusts on soil organic carbon sequestration in drylands: A Meta-analysis. Sci. Total Environ. 2021, 803, 150030. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, X.; Fei, H.; Wan, C.; Han, F. Changes in diversity, composition and assembly processes of soil microbial communities during Robinia pseudoacacia L. restoration on the Loess Plateau, China. J. Arid Land 2022, 14, 561–575. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.; Liu, L.; Dou, Y.; An, S. Comparison of soil microbial community between planted woodland and natural grass vegetation on the Loess Plateau. For. Ecol. Manag. 2020, 460, 117817. [Google Scholar] [CrossRef]
- Zhang, X.X.; Wang, L.J.; Zhou, W.X.; Hu, W.; Hu, J.W.; Hu, M. Changes in litter traits induced by vegetation restoration accelerate litter decomposition in Robinia pseudoacacia plantations. Land Degrad. Dev. 2022, 33, 179–192. [Google Scholar] [CrossRef]
- Xu, M.-p.; Wang, J.-y.; Zhu, Y.-f.; Han, X.-h.; Ren, C.-j.; Yang, G.-h. Plant Biomass and Soil Nutrients Mainly Explain the Variation of Soil Microbial Communities During Secondary Succession on the Loess Plateau. Microb. Ecol. 2022, 83, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, T.; Wang, Y.; Dou, Y.; Cheng, H.; Liu, L.; An, S. Linkage between soil ectoenzyme stoichiometry ratios and microbial diversity following the conversion of cropland into grassland. Agric. Ecosyst. Environ. 2021, 314, 107418. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, H.; Zhao, P.; Wei, X.; Ding, G.; Gao, G.; Shi, M. Vegetation Restoration Alters Fungal Community Composition and Functional Groups in a Desert Ecosystem. Front. Environ. Sci. 2021, 9, 589068. [Google Scholar] [CrossRef]
- Bastida, F.; Hernandez, T.; Albaladejo, J.; Garcia, C. Phylogenetic and functional changes in the microbial community of long-term restored soils under semiarid climate. Soil Biol. Biochem. 2013, 65, 12–21. [Google Scholar] [CrossRef]
- Lu, Z.X.; Wang, P.; Ou, H.B.; Wei, S.X.; Wu, L.C.; Jiang, Y.; Wang, R.J.; Liu, X.S.; Wang, Z.H.; Chen, L.J.; et al. Effects of different vegetation restoration on soil nutrients, enzyme activities, and microbial communities in degraded karst landscapes in southwest China. For. Ecol. Manag. 2022, 508, 120002. [Google Scholar] [CrossRef]
- Araujo, A.S.F.; Borges, C.D.; Tsai, S.M.; Cesarz, S.; Eisenhauer, N. Soil bacterial diversity in degraded and restored lands of Northeast Brazil. Antonie Van Leeuwenhoek 2014, 106, 891–899. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Hao, M.; Cui, Y.; Zhu, S.; Zhang, Y. Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China. Sustainability 2019, 11, 2295. [Google Scholar] [CrossRef]
- Xue, L.; Ren, H.D.; Li, S.; Leng, X.H.; Yao, X.H. Soil Bacterial Community Structure and Co-occurrence Pattern during Vegetation Restoration in Karst Rocky Desertification Area. Front. Microbiol. 2017, 8, 2377. [Google Scholar] [CrossRef]
- Song, S.Z.; Xiong, K.N.; Chi, Y.K.; He, C.; Fang, J.Z.; He, S.Y. Effect of Cultivated Pastures on Soil Bacterial Communities in the Karst Rocky Desertification Area. Front. Microbiol. 2022, 13, 922989. [Google Scholar] [CrossRef]
- Hu, L.; Li, Q.; Yan, J.H.; Liu, C.; Zhong, J.X. Vegetation restoration facilitates belowground microbial network complexity and recalcitrant soil organic carbon storage in southwest China karst region. Sci. Total Environ. 2022, 820, 153137. [Google Scholar] [CrossRef]
- Liang, C.; Schimel, J.P.; Jastrow, J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017, 2, 17105. [Google Scholar] [CrossRef]


| Treatment | Soil Depth | pH | Organic Matter (mg/kg) | Total Nitrogen (mg/kg) | Total Phosphorus (mg/kg) | Total Potassium (mg/kg) | Available Phosphorus (mg/kg) | Available Potassium (mg/kg) | Available Nitrogen (mg/kg) | Ammonium Nitrogen (mg/kg) | Nitrate Nitrogen (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–10 cm | 7.81 ± 0.01 | 6.55 ± 1.75b | 472.67 ± 75.97b | 486 ± 18.03cd | 239 ± 7.22 | 2.7 ± 0.16c | 72.52 ± 3.54c | 36.05 ± 6.00b | 16.82 ± 1.69b | 5.08 ± 0.16c | |
| CK | 10–20 cm | 7.81 ± 0.01 | 7.34 ± 1.54b | 510.67 ± 89.55b | 438 ± 18.19d | 246 ± 9.24 | 1.54 ± 0.47c | 61.6 ± 6.84c | 37.1 ± 7.31b | 15.72 ± 2.67b | 5.5 ± 0.35bc |
| 20–30 cm | 7.81 ± 0.01 | 6.84 ± 1.93b | 449.33 ± 100.86b | 443.33 ± 8.41d | 230 ± 18.55 | 4.11 ± 1.11c | 80.32 ± 11.7c | 31.62 ± 7.72b | 12.92 ± 1.84b | 5.12 ± 0.21c | |
| 0–10 cm | 7.82 ± 0.02 | 13.82 ± 0.45a | 1027.33 ± 20.27a | 686.67 ± 8.09a | 233 ± 2.08 | 34.75 ± 3.01a | 463.12 ± 6.12a | 81.78 ± 3.44a | 67.95 ± 18.56a | 7.03 ± 0.38ab | |
| AF | 10–20 cm | 7.81 ± 0.01 | 8.13 ± 2.01b | 654.67 ± 108.21b | 551.33 ± 21.80bc | 214 ± 16.13 | 7.22 ± 1.15bc | 379.12 ± 23.77b | 52.38 ± 9.23b | 18.12 ± 4.86b | 6.47 ± 0.34ab |
| 20–30 cm | 7.81 ± 0.01 | 7.41 ± 2.04b | 555 ± 125.4b | 520 ± 55.51cd | 248 ± 8.72 | 13.1 ± 4.54bc | 354.45 ± 40.52b | 44.8 ± 9.15b | 20.55 ± 2.66b | 5.75 ± 0.45bc | |
| 0–10 cm | 7.83 ± 0.02 | 6.83 ± 1.61b | 559 ± 124.71b | 507 ± 52.17cd | 256 ± 25.75 | 13.53 ± 4.75bc | 323.62 ± 27.06b | 37.8 ± 7.91b | 7.73 ± 1.62b | 5.98 ± 0.78ab | |
| SB | 10–20 cm | 7.81 ± 0.01 | 8.23 ± 2.03b | 619.33 ± 110.27b | 621 ± 25.81ab | 258 ± 15.54 | 18.14 ± 8.41b | 373.12 ± 25.6b | 42.12 ± 4.81b | 5.05 ± 0.43b | 7.35 ± 0.65a |
| 20–30 cm | 7.83 ± 0.01 | 6.51 ± 0.88b | 526.33 ± 79.67b | 520.33 ± 14.31cd | 239 ± 3.71 | 6.01 ± 1.18bc | 470.12 ± 23.19a | 40.48 ± 7.48b | 4.22 ± 0.44b | 5.62 ± 0.61bc |
| Treatment | Fungal Community | Bacterial Community | ||||
|---|---|---|---|---|---|---|
| CK | AF | SB | CK | AF | SB | |
| Number of nodes | 162 | 183 | 178 | 184 | 184 | 187 |
| Number of edges | 351 | 696 | 429 | 1320 | 966 | 830 |
| Average neighborhood | 4.33 | 7.61 | 4.82 | 14.35 | 10.5 | 8.87 |
| Linkage distance | 14 | 15 | 15 | 13 | 10 | 11 |
| Clustering coefficient | 0.39 | 0.47 | 0.43 | 0.54 | 0.53 | 0.51 |
| Linkage density | 2.16 | 3.8 | 2.4 | 7.17 | 5.25 | 4.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Chen, C.; Pang, Z.; Zhang, G.; Wu, J.; Kan, H. Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network. J. Fungi 2022, 8, 1122. https://doi.org/10.3390/jof8111122
Xu H, Chen C, Pang Z, Zhang G, Wu J, Kan H. Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network. Journal of Fungi. 2022; 8(11):1122. https://doi.org/10.3390/jof8111122
Chicago/Turabian StyleXu, Hengkang, Chao Chen, Zhuo Pang, Guofang Zhang, Juying Wu, and Haiming Kan. 2022. "Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network" Journal of Fungi 8, no. 11: 1122. https://doi.org/10.3390/jof8111122
APA StyleXu, H., Chen, C., Pang, Z., Zhang, G., Wu, J., & Kan, H. (2022). Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network. Journal of Fungi, 8(11), 1122. https://doi.org/10.3390/jof8111122






