Abstract
Dollar spot disease, caused by the fungal pathogen Clarireedia jacksonii, is a major problem in many turfgrass species, particularly creeping bentgrass (Agrostis stolonifera). It is well-established that strong creeping red fescue (Festuca rubra subsp. rubra) exhibits good dollar spot resistance when infected by the fungal endophyte Epichloë festucae. This endophyte-mediated disease resistance is unique to the fine fescues and has not been observed in other grass species infected with other Epichloë spp. The mechanism underlying the unique endophyte-mediated disease resistance in strong creeping red fescue has not yet been established. We pursued the possibility that it may be due to the presence of an abundant secreted antifungal protein produced by E. festucae. Here, we compare the activity of the antifungal protein expressed in Escherichia coli, Pichia pastoris, and Penicillium chrysogenum. Active protein was recovered from all systems, with the best activity being from Pe. chrysogenum. In greenhouse assays, topical application of the purified antifungal protein to creeping bentgrass and endophyte-free strong creeping red fescue protected the plants from developing severe symptoms caused by C. jacksonii. These results support the hypothesis that Efe-AfpA is a major contributor to the dollar spot resistance observed with E. festucae-infected strong creeping red fescue in the field, and that this protein could be developed as an alternative or complement to fungicides for the management of this disease on turfgrasses.
1. Introduction
Antifungal proteins have been reported from several fungal species [1,2]. The most well-characterized are those from Penicillium chrysogenum, designated PAF and PAFB [3,4,5], and Aspergillus spp., designated AFP and NFAP [6,7,8]. These proteins are of considerable interest for use as possible therapeutic agents against fungal diseases in humans, in food preservation, and in plant disease protection [9,10,11,12].
We are interested in a similar antifungal protein produced by Epichloë festucae, a fungal endophytic symbiont of the grass strong creeping red fescue (Festuca rubra subsp. rubra). Strong creeping red fescue is a commercially important low maintenance turfgrass species [13,14]. Epichloë spp. are endophytes of several cool-season grass species, often conferring insect resistance to their grass hosts due to the production of toxic alkaloids [15]. In Epichloë-infected forage grasses, the presence of some of the alkaloids can be toxic to grazing animals. Efforts to reduce or eliminate animal toxins while maintaining the insect toxins of the fungal endophytes is a topic of considerable research [16]. For turfgrasses, cultivars containing Epichloë endophytes are desired because of the enhanced insect resistance [17]. In addition to insect resistance, a unique feature of the strong creeping red fescue/E. festucae symbiosis is field level endophyte-mediated resistance to fungal pathogens [18,19]. Such endophyte-mediated disease resistance has not been reported in cultivated perennial ryegrasses (Lolium perenne) or tall fescues (Lolium arundinaceum) [19,20,21].
The mechanism underlying the unique endophyte-mediated disease resistance in strong creeping red fescue has not yet been established. We are pursuing the possibility that it may be due to the presence of an abundant secreted antifungal protein (gene model EfM3.063660; NCBI accession AWO72254) produced by the fungal endophyte E. festucae. The presence of the antifungal protein in E. festucae was first identified from a transcriptome study of endophyte-infected strong creeping red fescue [22]. The antifungal protein transcript was the second most abundant fungal transcript recovered in the study. The antifungal protein gene found in E. festucae infecting strong creeping red fescue is not present in most Epichloë spp. genomes for which whole genome sequences are available [20,22]. The transcript abundance and the limited existence of the gene among Epichloë spp. suggested the E. festucae antifungal protein may be a component of the unique endophyte-mediated disease resistance observed in strong creeping red fescue. Other antifungal compounds have been reported from some Epichloë spp. [16,23,24], but for commercially produced grasses, the field level disease resistance seen in E. festucae-infected fine fescues is unique.
The antifungal protein was expressed in the yeast Pichia pastoris, and the recombinant protein was shown to inhibit growth in culture of Clarireedia jacksonii (formerly Sclerotinia homoeocarpa), the causal agent of dollar spot disease [20,25]. C. jacksonii is a destructive fungal pathogen on many cultivated turfgrass species, particularly creeping bentgrass (Agrostis stolonifera), often requiring repeated applications of fungicides to suppress disease development [26,27]. Treatment of C. jacksonii mycelium with the antifungal protein resulted in uptake of the viability stains SYTOX Green and Evans blue, indicating that observed growth inhibition of the pathogen was due to damage to the plasma membrane of the pathogen [20]. These results support the hypothesis that the E. festucae antifungal protein is a component of the disease resistance seen in strong creeping red fescue.
The objective of this study was to determine if direct application of the E. festucae antifungal protein to susceptible turfgrass plants could protect them from injury caused by dollar spot pathogen. To do this, a simple method of protein purification was required. The yeast system was valuable for the initial demonstration that the purified E. festucae antifungal protein was active [20], but would be cumbersome for large-scale production of the protein. Unlike the similar proteins from Penicillium and Aspergillus, where their antifungal proteins could be easily purified from the culture filtrate, the E. festucae antifungal protein was not found in the culture filtrate, although it was highly expressed in the infected grass host [22]. Quantitative PCR analysis revealed the antifungal protein gene was expressed at a level greater than 700-fold higher in infected leaf sheath tissue than in culture [28]. Therefore, a simpler method of producing large quantities of the protein was sought. Here, we compare the activities and yields of the E. festucae antifungal protein expressed in Escherichia coli, Pi. pastoris and Pe. chrysogenum, and report the ability of the purified protein to reduce disease symptoms on plants of endophyte-free strong creeping red fescue and creeping bentgrass. We also report differences in the activity of Efe-AfpA with that of the similar protein PAF against C. jacksonii as well as wild type and glucosylceramide mutant strains of Neurospora crassa. Throughout this manuscript, we use the suggested nomenclature for Epichloë spp. genes and proteins [29]. The E. festucae antifungal protein gene is designated Efe-afpA and the protein is designated Efe-AfpA.
2. Materials and Methods
2.1. Primer Sequences
All oligonucleotide primer sequences used in this study are presented in Table S1.
2.2. Cloning of Modified E. festucae Antifungal Protein Coding Sequences in Escherichia coli
The mature Efe-afpA coding sequence [20] was modified and cloned into the pETite™ N-His SUMO vector according to the instructions of the Expresso T7 SUMO Cloning and Expression System (Lucigen Corp., Middleton, WI, USA). According to the system manual, large aliphatic residues following the SUMO tag, such as the N-terminal isoleucine of the mature Efe-AfpA, may result in slower cleavage rates by the SUMO protease. Both the mature Pe. chrysogenum antifungal protein (PAF) and the A. giganteus antifungal protein (AFP) start with an alanine. Therefore, the N-terminal isoleucine of Efe-AfpA cloned in the Pi. pastoris expression plasmid pPICZα A [20] was changed to alanine by using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs Inc., Ipswich, MA, USA) according to the manufacturer’s instructions.
The N-terminal Ala-modified Efe-afpA coding sequence was then amplified by PCR with oligonucleotides that added sequences identical to the insertion site on the pETite™ N-His SUMO vector. The amplification reaction was carried out in a GeneAmp 9700 thermo-cycler (Applied Biosystems, Foster City, CA, USA). The 100 μL reactions contained 2X Phusion High-Fidelity PCR Master Mix with HF Buffer (Thermo Fisher Scientific, Waltham, MA, USA), 40 pmol of each oligonucleotide, and 25 ng plasmid DNA as template. The PCR reaction conditions were: an initial denaturation step at 98 °C for 30 s, followed by 30 cycles of 10 s denaturation at 98 °C, 30 s annealing at 59 °C, and 30 s extension at 72 °C. An additional final 5 min extension at 72 °C was performed.
After visualizing the PCR product on a 2% TBE agarose gel, 3 μL of the PCR product was mixed with 25 ng of pETite™ N-His SUMO vector and transformed directly into competent HI-Control 10G cells by heat shock in a 42 °C water bath for 45 s and returned to ice for 2 min. The transformed cells were incubated in Recovery Medium for 1 h at 37 °C with shaking, followed by overnight growth of cells on LB medium supplemented with 30 μg mL−1 kanamycin. Transformed bacterial colonies were screened for recombinant plasmids containing the Efe-afpA coding sequence inserts by using PCR with an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of 30 s denaturation at 94 °C, 30 s annealing at 55 °C, and 30 s extension at 72 °C. An additional final 7 min extension at 72 °C was performed. The PCR products were visualized on a 2% TBE agarose gel. Plasmids from selected positive bacterial colonies were isolated and sequenced (Genewiz, Inc., South Plainfield, NJ, USA) using the SUMO Forward primer. One hundred ng of a plasmid containing the correct sequence was transformed into SHuffle T7 Express Competent E. coli (C3029, New England BioLabs Inc.) by heat shock in a 42 °C water bath for 30 s and returned to ice for 5 min. The transformed cells were incubated in SOC medium for 1 h at 30 °C with shaking, followed by 2 d growth of cells on LB medium supplemented with 30 μg mL−1 kanamycin.
Expression and purification of the N-terminal Ala modified Efe-AfpA (described below) revealed that the SUMO-protease reaction was not efficient in cleaving the SUMO tag with this modified version of Efe-AfpA. The Expresso T7 SUMO Cloning and Expression System (Lucigen Corp.) manual suggests that if incomplete digestion persists to insert or substitute Gly and or Ser residues next to the SUMO tag. Therefore, three additional N-terminal modifications were generated, (1) the N-terminal Ala was changed to Gly, (2) a Gly was added to the N-terminal Ala, and (3) Gly-Ser was added to the N-terminal Ala. The modifications were generated by using the Q5-Site Directed Mutagenesis Kit according to the manufacturer’s instructions. The resulting modified pETite™ N-His SUMO plasmids were then cloned into SHuffle T7 Express Competent E. coli.
2.3. Recombinant N-Terminal Modified Efe-AfpA Protein Purification
For purification of modified Efe-AfpA proteins, a starter culture of the Shuffle T7 cells containing the appropriate plasmid in 50 mL LB supplemented with 30 µg mL−1 kanamycin was grown overnight at 30 °C with shaking. The following day this was subcultured into 1 L LB plus 30 µg mL−1 kanamycin and shaken at room temperature until an OD600 of 0.6 to 0.8 was reached. Efe-AfpA expression was then induced by the addition of 4 mL of 100 mM IPTG with overnight shaking at room temperature until an OD600 of about 1 was reached. Recombinant Efe-AfpA proteins were purified by using TALON ® Metal Affinity Resin (Takara Bio USA, Inc., San Jose, CA, USA). First, cells were collected by centrifugation followed by lysis using 100 mL 1× Fast Break Lysis Reagent (Promega Corporation, Madison, WI, USA) supplemented with 248 µL of 5 mg mL−1 DNase I (D4527, Sigma-Aldrich, St. Louis, MO, USA). The cells were rotated for 20 min to allow for complete lysis. The 6xHIS-SUMO tagged Efe-AfpA proteins were isolated by the addition of pre-equilibrated TALON resin and incubated for 20 min with rotation. The resin was collected by centrifugation and the supernatant decanted. The resin was then washed in TALON equilibration buffer, applied to a column, and bound SUMO-tagged Efe-AfpA was eluted using TALON Elution Buffer. High-molecular-weight proteins were removed from the eluate by centrifugation through a 30 kDa Amicon ® Ultra-15 Centrifugal Filter Unit (MilliporeSigma, Burlington, MA, USA). The protein in the flow-through was concentrated and the elution buffer was exchanged to 50 mM NaPO4, pH 7.0, 300 mM NaCl by using a 3 kDa Amicon ® Ultra-15 Centrifugal Filter Unit. To remove the 6xHis-SUMO tag from the modified forms of Efe-AfpA, 100 µg of purified 6xHis-SUMO-tagged protein was incubated overnight at 4 °C with 1 unit of SUMO protease (Lucigen Corp.). The buffer of the digested Efe-AfpA solution was exchanged to 50 mM NaPO4, pH 7.0, to reduce the salt concentration, by using a 3 kDa Amicon ® Ultra-15 Centrifugal Filter Unit. The released Efe-AfpA protein was then separated from the 6xHis-SUMO tag by batch ionic exchange purification by using carboxymethyl cellulose (CMC52) (Biophoretics, Sparks, NV, USA). CMC52 was pre-equilibrated with 50 mM NaPO4 pH 7.0 for 1 h, added to the digest solution, and incubated for 3 h at 25 °C. At pH 7.0, Efe-AfpA (pI = 8.9) is positively charged and binds to the CMC52. The CMC52 was applied to a column, washed with excess 50 mM NaPO4, pH 7.0, and Efe-AfpA was eluted with NaCl amended buffer ranging from 0.1 to 0.5 M. The A280 of the eluted fractions was monitored by using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Protein concentrations of the fractions were determined by using the molecular weight of Efe-AfpA (6.278 kDa) and the predicted extinction coefficient of 5220 M−1 cm−1 [30]. Protein containing fractions were then concentrated and the buffer exchanged to sterile water by using a 3 kDa Amicon ® Ultra-15 Centrifugal Filter. The purified Efe-AfpA solution was sterilized by filtering the protein through a 0.2 µm polyethersulfone syringe filter (Corning Inc., Corning, NY, USA).
2.4. Purification of Efe-AfpA from Culture Filtrates of Pichia pastoris
The cloning of Efe-AfpA in Pi. pastoris was described previously [20]. Expression and purification of Efe-AfpA is as described below. Pi. pastorisEfeAfpA was streaked onto solid YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) plates amended with 100 µg mL−1 Zeocin, and incubated at 30 °C until single colonies appeared. A single colony was grown in a starter culture of 50 mL BMGY (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 1% glycerol) at 30 °C with shaking at 200 rpm until an OD600 of at least 2 was reached. The culture was then subcultured into 1 L of fresh BMGY and grown at 30 °C with shaking at 200 rpm until the OD600 was at least 2. The cells were then pelleted by centrifugation and resuspended in 1 L BMMY (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 0.5% methanol) to induce expression of the Efe-AfpA protein. Expression of Efe-AfpA was induced with the addition of 5 mL of methanol daily for 5 days. The cells were then pelleted by centrifugation at 10,000 rpm for 10 min, and the culture supernatant transferred to SnakeSkinTM Dialysis Tubing (Thermo Fisher Scientific) with a molecular weight cut-off of 3.5 K and was dialyzed overnight in 8 L of 10 mM NaPO4 pH 6.6, 25 mM NaCl, 0.15 mM EDTA. Dialyzed culture supernatant was then applied to a CMC52 column pre-equilibrated in 10 mM NaPO4, pH 6.6, 25 mM NaCl, 0.15 mM EDTA buffer. The column was washed with excess buffer and eluted with increasing salt concentrations from 0.1 to 0.5 M NaCl. Protein concentration in the fractions was determined as described above and the protein containing fractions were then filtered through a 30 kDa Amicon ® Ultra-15 Centrifugal Filter to remove high molecular weight proteins. The flow-through containing Efe-AfpA was then concentrated and the buffer exchanged to sterile water by using a 3 kDa Amicon ® Ultra-15 Centrifugal Filter. The purified Efe-AfpA was then sterilized by filtering the protein through a 0.2 µm polyethersulfone syringe filter.
2.5. Cloning and Transformation of Efe-afpA into Penicillium chrysogenum
The pSK275paf plasmid and a Pe. chrysogenum isolate in which the paf gene was deleted were provided by Dr. Florentine Marx. The pSK275paf plasmid was developed for expressing the Pe. chrysogenum antifungal protein paf and paf variants in the Pe. chrysogenum strain in which the paf gene had been deleted, and includes the paf promoter and coding sequences [31]. The pSK275paf plasmid was modified by cloning the Efe-afpA coding sequence of the mature protein in place of the paf coding sequence of the mature protein. This generated a plasmid in which the Efe-afpA coding sequence of the mature protein was downstream of the Pe. chrysogenum paf promoter and paf signal peptide and propeptide.
Assembly of the pSK275:Efe-AfpA vector was completed using the NEBuilder® HiFi DNA Assembly Kit (New England Biolabs Inc.) The mature Efe-AfpA sequence was produced by PCR from the pETite:Efe-afpA vector (described above) with overhangs homologous to regions on the pSK275paf plasmid. The pSK275paf backbone fragments were also generated by PCR, and the final pSK275:Efe-afpA vector was constructed following the HiFi DNA Assembly Kit manual. Transformation of the Pe. chrysogenum Δpaf strain was completed as described previously [31]. Spores (2 × 108) of the Δpaf strain were inoculated in 200 mL complete medium [31] and shaken at 200 rpm at room temperature for 48 h. Fungal pellets were harvested by filtering through cheesecloth and converted to protoplasts by using 1.2 g Vinotaste Pro (Novozymes, Franklinton, NC, USA) in 30 mL lytic buffer (50 mM potassium phosphate, 0.7 M KCl, pH 5.8). Protoplasts were harvested by filtration through filter paper (Whatman Grade 1) and then pelleted by centrifugation (3000 rpm). The protoplasts were then washed twice with 0.7 M KCl, resuspended in a final volume of 300 µL 0.7 M KCl, and counted. Ten to fifteen µg of pSK275:Efe-afpA, linearized by digestion with Not1, were added to 100 µL of protoplasts (108) followed by 25 µL PCM (0.8% CaCl2, 2% MOPS, 50% PEG 6000, pH 5.8). Both the transformation solution and appropriate control solutions were incubated on ice for 30 min. PCM (250 µL) was then added to both the sample and controls, and they were further incubated for 20 min at room temperature. Lastly, 1 mL of 0.7 M KCl was added and both transformation and control solutions were mixed in melted PcMM (Pe. chrysogenum Minimal Media, 0.3% NaNO3, 0.05% MgSO4 × 7H2O, 0.05% KCl, 0.005% FeSO4 × 7H2O, 2% sucrose, 2.5% 1M potassium phosphate buffer pH 5.8, 0.1% trace elements solution A) top agar (1% agar) [31] prior to being plated on antibiotic amended (0.6 µg mL−1 pyrithiamine and 200 µg mL−1 noureseothricin) PcMM bottom agar (2% agar). Trace elements solution A was 0.1% FeSO4•7H2O, 0.9% ZnSO4, 0.04% CuSO4, 0.01% MnSO4, 0.01% H3BO3, 0.01% Na2MoO4.
Plates were incubated at 25 °C for up to two weeks to allow transformants to grow. Colonies were then subcultured onto fresh PcMMPyr0.6Nour200 and screened for gene integration by spore PCR using the primers Efe-AfpA Forward and Efe-AfpA Reverse with positive transformants yielding a band on a 2% agarose gel of 168 bp. Positive colonies were then subjected to single spore selection, allowing for the collection of a genetically homogeneous strain. PCR positive transformants from single spore isolates were then streaked onto PcMMPyr0.6Nour200. Spores were collected and inoculated into 25 mL liquid PcMM. After seven days of growth at room temperature with shaking at 200 rpm, the culture supernatant was collected, filtered through a 30 kDa Amicon ® Ultra-15 Centrifugal Filter Unit and concentrated on a 10 kDa Amicon ® Ultra-15 Centrifugal Filter Unit (Millipore). Supernatants were then quantitated by A280 (Nanodrop ND-1000, Thermo Fisher Scientific) and equal total protein amounts were compared by 16% SDS PAGE to visualize the secreted Efe-AfpA protein. Transformants with the highest levels of Efe-AfpA were chosen for further analysis. Primers pSK275:Efe-AfpA F and pSK275:Efe-AfpA R were used to amplify the Efe-AfpA gene region of the chosen transformants. The PCR fragment was gel purified using the QIAquick Gel Extraction Kit (Qiagen Inc, Germantown, MD, USA) and sequenced (Genewiz Inc., South Plainfield, NJ, USA).
2.6. Purification of Efe-AfpA from Culture Filtrates of Penicillium chrysogenum and of PAF from an Overexpression Strain of Pe. chrysogenum
A strain of Pe. chrysogenum that overexpressed PAF (designated Pe. chrysogenum paf) [31] was used to purify PAF for comparison of activity with Efe-AfpA purified from the different expression systems. Purification of PAF and Efe-AfpA from Pe. chrysogenum was as described below.
Pe. chrysogenumpaf conidia were streaked onto solid PcMM 2% agar plates supplemented with 200 µg mL−1 nourseothricin and 0.6 µg mL−1 pyrithiamine from freezer stocks and grown for 4 days at room temperature. Spores were then harvested in spore buffer (0.9% NaCl, 0.01% Tween 20), washed twice in spore buffer, and counted using a hemocytometer. Conidia (2 × 108) were inoculated into 200 mL of PcMM and grown at room temperature with shaking at 200 rpm for 72 h. The culture supernatant was filtered through cheesecloth to remove mycelia and any excess debris was pelleted by centrifugation at 10,000 rpm for 10 min. The culture supernatant was then applied to a CMC52 column pre-equilibrated in 10 mM NaPO4, 25 mM NaCl, 0.15 mM EDTA, pH 6.6 buffer. The column was washed with excess buffer and eluted with increasing salt concentrations from 0.1 to 0.5 M NaCl. Eluted fractions were then evaluated for the presence of protein at A280 (Nanodrop ND-1000) utilizing the molecular weight (6.25 kDa) and extinction coefficient (4845 M−1 cm−1) of PAF [32]. The protein-containing fractions were then filtered through a 30 kDa Amicon ® Ultra-15 Centrifugal Filter, concentrated and desalted using a 3 kDa Amicon ® Ultra-15 Centrifugal Filter and sterile distilled water, and sterilized by filtering the protein through a 0.2 µm polyethersulfone syringe filter.
Pe. chrysogenumEfe-AfpA required an altered cultivation method for protein expression due to the low production of Efe-AfpA when using the method described above for PAF production. This was likely due to Efe-AfpA activity against Pe. chrysogenum, as discussed below. Pe. chrysogenumEfe-AfpA conidia from freezer stocks were streaked onto solid PcMM 2% agar plates supplemented with 200 µg mL−1 nourseothricin and 0.6 µg mL−1 pyrithiamine and grown for 4 days at room temperature. Spores were then harvested in spore buffer, washed twice in spore buffer, and counted using a hemocytometer. Conidia (2 × 108) were inoculated into 200 mL of A. nidulans complete media [31] and grown at room temperature for 48 h with shaking at 200 rpm. Mycelia were harvested on cheesecloth, washed with sterile distilled water, and subcultured into 200 mL PcMM to induce expression of the Efe-AfpA protein for 72 h at room temperature with shaking at 200 rpm. Purification of Efe-AfpA from the culture filtrate was as described above for purification of PAF. For Efe-AfpA quantitation, the molecular weight 6.278 kDa and the theoretical extinction coefficient 5220 M−1 cm−1 were used [30].
2.7. Neurospora crassa Conidial Growth Assays
The antifungal activities of the purified forms of N-terminal modified Efe-AfpA expressed in E. coli were compared in a Neurospora crassa (wild type strain 74-OR23-IVA; Fungal Genetics Stock Center #2489) conidial growth assay carried out in 96-well microtiter plates. The assay was based on the method described previously [33]. Frozen N. crassa conidia were thawed and 50 μL were diluted in 1 mL of 2× low-cationic medium (LCM) (1× LCM is 2 g L−1 glucose, 0.1 g L−1 yeast extract, 0.05 g L−1 peptone). The sample was counted by using a hemocytometer and further diluted with 2X LCM to a final working concentration of 2 × 106 conidia mL−1. Solutions (100 µL) of varying amounts of the purified antifungal proteins in 20 mM Tris-Cl, pH 8.0, 150 mM NaCl buffer were mixed with 100 uL of 2 × 106 N. crassa conida for a final volume of 200 uL in each well. Each sample was assayed in triplicate. The untreated control sample was 100 μL conidia + 100 μL 20 mM Tris, pH 8.0, 150 mM NaCl. The optical density at 620 nm was measured immediately with a microtiter plate reader (Absorbance 96, Byonoy GmbH, Hamburg, Germany) and the plates incubated at room temperature. The optical density was measured again after 24 h incubation and the initial readings subtracted from the 24 h readings. The corrected optical density of the untreated control conidia was considered 100% growth (0% inhibition). The percentage inhibition of the Efe-AfpA treated samples was calculated by comparing the conidia growth in the absence of Efe-AfpA to the growth in the treated samples.
The antifungal activities of the purified Efe-AfpA proteins expressed in Pi. pastoris and Pe. chrysogenum and of PAF from Pe. chrysogenum paf were compared in a similar N. crassa wild type conidial growth assay carried out in 96-well microtiter plates.
2.8. Penicillium chrysogenum Δpaf Sensitivity to Efe-AfpA
The recipient strain P. chrysogenum Δpaf was cultured as described above on solid antibiotic amended PcMM. Spores were harvested in spore buffer, washed twice with excess spore buffer, washed once in 2× LCM, and resuspended in fresh 2× LCM. Spores were counted with a hemocytometer and diluted to a working concentration of 2 × 104 conidia mL−1. One hundred µL of spores were mixed with 100 µL of 10 µg mL−1 Efe-AfpA or PAF resulting in a final concentration of 1 × 104 conidia mL−1 spores and 5 µg mL−1 protein in a 96-well plate in triplicate. Sterile distilled water was used as the control. Growth was monitored by absorbance at 620 nm in a microtiter plate reader (Absorbance 96, Byonoy GmbH). Absorbances at 30 h were corrected by subtracting the initial absorbances at 0 h. The water treated wells were considered 100% growth (0% inhibition). Wells were also visualized microscopically (EVOS M5000, Invitrogen).
2.9. Clarireedia jacksonii Inhibition Assays
A C. jacksonii isolate recovered from the creeping bentgrass cultivar ‘Luminary’ was maintained on 1× potato dextrose agar (PDA). Since C. jacksonii does not produce spores, it was maintained by routinely transferring plugs from the growing edge of a plate to a new PDA plate.
The Evans blue staining assay was modified from that previously described [20,34]. One mL PDA was applied to a surface sterilized microscope slide making a 3 cm by 2.5 cm rectangle of PDA. A 5 mm plug of C. jacksonii was taken from a 4-day old PDA plate and placed on the left edge of the PDA rectangle on the slide. The microscope slides were incubated at room temperature for 24 h in a closed Petri dish. The next day, 10 µL containing 300 ng of Efe-AfpA or PAF, or water were placed at the growing edge of the C. jacksonii mycelium. The microscope slides were then incubated for another 24 h at room temperature to allow the mycelium to grow into the treatment. The mycelium was then stained in 1% Evan’s blue stain for 15 min and destained in distilled water. Mycelium was then visualized microscopically. Each treatment had 3 replicates, and the experiment was completed twice.
For plate assays, 35 mm Petri dish plates containing 8 mL of PDA amended with increasing concentrations of Efe-AfpA, PAF, or water were used. Concentrations of the antifungal proteins tested were 0, 0.5, 1, 10, 20, 30, 40, 50, 100 µg mL−1. A 5 mm plug of C. jacksonii was taken from a 4-day-old PDA plate and placed in the center of each 35 mm plate. Plates were incubated at room temperature for 4 days. Plates were photographed, and cross section mycelium diameters were measured daily. Each treatment had three replicates and the experiment was completed twice.
2.10. Strong Creeping Red Fescue and Creeping Bentgrass Greenhouse Infection Assay
Strong creeping red fescue and creeping bentgrass plants were maintained in a greenhouse in Pro-Mix BX potting mix (Premier Horticulture Inc., Quakertown, PA, USA).
The strong creeping red fescue plants used for the greenhouse assays were an endophyte-free line S1139E and the same genotype infected with the E. festucae Rose City isolate, S1139RC. The generation of these plants was described previously [35]. Plants were repotted into 14.6 cm diameter pots prior to infection and trimmed to even heights. Pots of similar density were chosen and inoculated with either an 8 mm plug of C. jacksonii grown for 5 days on PDA or an 8 mm plug of PDA. To ensure the high humidity required for infection, the plants were covered with a translucent plastic bag, and placed in a tray containing a shallow layer of water. The plants were sprayed daily for 10 days with either 1 mL of water containing 0.1% Tween 20 or 1 mL 100 µg mL−1 Efe-AfpA containing 0.1% Tween 20. Four replicates were used per treatment. Photographs were taken ten days post inoculation.
Creeping bentgrass ‘Crenshaw’ plugs (10 cm diameter × 6 cm depth) were taken from the Rutgers University Horticulture Farm 2 experimental research plots in North Brunswick, New Jersey, transferred to 10 cm × 10 cm pots, and maintained in the greenhouse. The plants were propagated by repotting as needed. For the disease assays, plants were repotted to 8 × 8 cm pots. Prior to inoculation, creeping bentgrass plants of similar density were chosen and trimmed to the same height. For the fungal inoculum, 8 mm plugs were harvested from the growing edge of 5-day-old C. jacksonii cultures on PDA plates, as well as 8 mm plugs of PDA for the controls. A plug of either C. jacksonii or PDA was placed in the center of a pot resting on the grass blades, the plants were covered with a translucent plastic bag, and placed in a tray containing a shallow layer of water. Each pot was then sprayed daily with either 1 mL of water containing 0.1% Tween 20 or 1 mL of Efe-AfpA at various concentrations with 0.1% Tween 20. Six replicates were used per treatment. Photographs were taken 7 days post inoculation.
The timing of applications was also tested. Pots of creeping bentgrass plants and C. jacksonii inoculum were prepared as described above. Either 1 mL of water containing 0.1% Tween 20 or 1 mL of 100 µg mL−1 Efe-AfpA containing 0.1% Tween 20 was sprayed onto the pots at 0 h, 0 and 48 h, or daily for a total of one, two, or seven applications, respectively. Six replicates were used per treatment. Photographs were taken 7 days post inoculation.
2.11. Evaluation of the Effect of Neurospora crassa Glucosylceramides on Efe-AfpA Activity
To compare the activity of Efe-AfpA to PAF and evaluate the effect of N. crasssa glucosylceramides on Efe-AfpA activity, the assay was carried out on conidia of the glucosylceramide synthase [36] and 2-hydroxy fatty N-acyl-Δ3(E)-desaturase [37] deletion mutants, Fungal Genetics Stock Center #13794 and FGSC #22453, respectively [38,39].
N. crassa wild type, Δgcs, and Δdtd strains were grown on Vogel’s solid media (supplemented with 200 µg mL−1 hygromycin for the two knockout strains). The three strains were streaked onto fresh Vogel’s media from freezer stocks and incubated at room temperature for four to five days to allow for spore generation. Spores were harvested and washed as described above, diluted in 2× LCM to a working concentration of 2 × 104 conidia mL−1. One hundred µL of spores were incubated with 100 µL of water or increasing concentrations of either Efe-AfpA or PAF to final concentrations of 0.3, 0.6, 1.2, 5, 10, 20, 30, 40, 50, and 100 µg mL−1. Growth was monitored by absorbance at 620 nm at 0 and 24 h as described above. Untreated spores were considered 100% growth as compared to treated spores. All treatments were done in triplicate and the experiments were completed at least three times. Minimal inhibitory concentration (MIC) was determined from the absorbance data. MIC is considered the concentration of protein required for ≥ 90% inhibition of growth. Samples were also visualized microscopically (EVOS M5000, Invitrogen).
To further determine the susceptibility of the three N. crassa strains to Efe-AfpA, spores of each were cultured on solid Vogel’s media supplemented with increasing concentrations of either Efe-AfpA or PAF (0–100 µg mL−1) in 24-well plates [36]. Five µL of each strain at a concentration of 2 × 105 conidia mL−1 were plated on 500 µL Vogel’s media and incubated at room temperate for 96 h and photographed at 72 h. The experiment was done in duplicate per protein and completed twice.
Efe-AfpA’s effect on conidia germination of the three N. crassa strains was determined by modifying the method previously described [40]. One hundred µL of N. crassa conidia (1 × 105 mL−1) were incubated with 100 µL of 20 µg mL−1 Efe-AfpA or PAF, or water for 6 h in a 96 well plate at room temperature. Samples were also visualized microscopically, and 100 conidia were counted from each replicate per treatment. Germination efficiency was calculated as the percent of the 100 conidia that germinated. Each treatment was done in triplicate and the experiment was completed three times.
2.12. Phylogenetic Analysis
The Clustal-X program [41] was used to align the full antifungal protein amino acid sequences, including the signal peptides. The phylogenetic analysis was performed with the Paup* program, version 4.0b10 for Macintosh. The phylogenetic analysis was done by using the maximum parsimony full heuristic search option set to random sequence addition, tree-bisection-reconnection (TBR) branch swapping, and Multrees on, with 1000 bootstrap replications. Gaps were treated as missing.
2.13. Protein Gel Electrophoresis
For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, protein samples were mixed with 5× SDS sample buffer (5:1, v/v), [42], then boiled for 5 min and subjected to electrophoresis in 16% polyacrylamide gels. Gels were stained with Coomassie Blue to visualize protein bands. Gels were destained and then dried as described previously [43].
3. Results
3.1. Production of Efe-AfpA in E. coli
Bacterial expression of mature Efe-AfpA was achieved from a plasmid that adds a cleavable N-terminal 6xHis-SUMO (small ubiquitin-like modifier) tag [44] in the engineered E. coli strain SHuffle, which maintains an oxidative cytoplasmic environment allowing disulfide bond formation in the recombinant protein [45]. Modification of the mature N-terminus of Efe-AfpA was required since the native N-terminal isoleucine is known to inhibit cleavage of the SUMO tag by the SUMO protease. Four modified N-terminal coding sequences of the mature form of Efe-AfpA were cloned into the E. coli expression vector pETite™ N-His-SUMO and the resulting plasmids cloned into SHuffle competent cells. The modifications and the resulting protein designations are presented in Table 1.

Table 1.
N-terminal modifications of mature Efe-AfpA expressed in E. coli.
In all cases, the 6xHis-SUMO-tagged Efe-AfpA was a major soluble protein after induction by IPTG and could be purified by binding to TALON resin. The 6xHis-SUMO tag could be cleaved by digestion with an engineered 6xHis-tagged SUMO protease, releasing Efe-AfpA from the SUMO tag. Efe-AfpA was separated from the released tag and the SUMO protease by binding to CMC52. Three of the four N-terminal modified Efe-AfpA proteins were efficiently cleaved by the SUMO-protease (Figure S1). The A-Efe-AfpA-modified protein was not efficiently cleaved by the protease, even on extended incubation, and was therefore dropped from further characterization.
3.2. Activity of the Modified Efe-AfpA Proteins
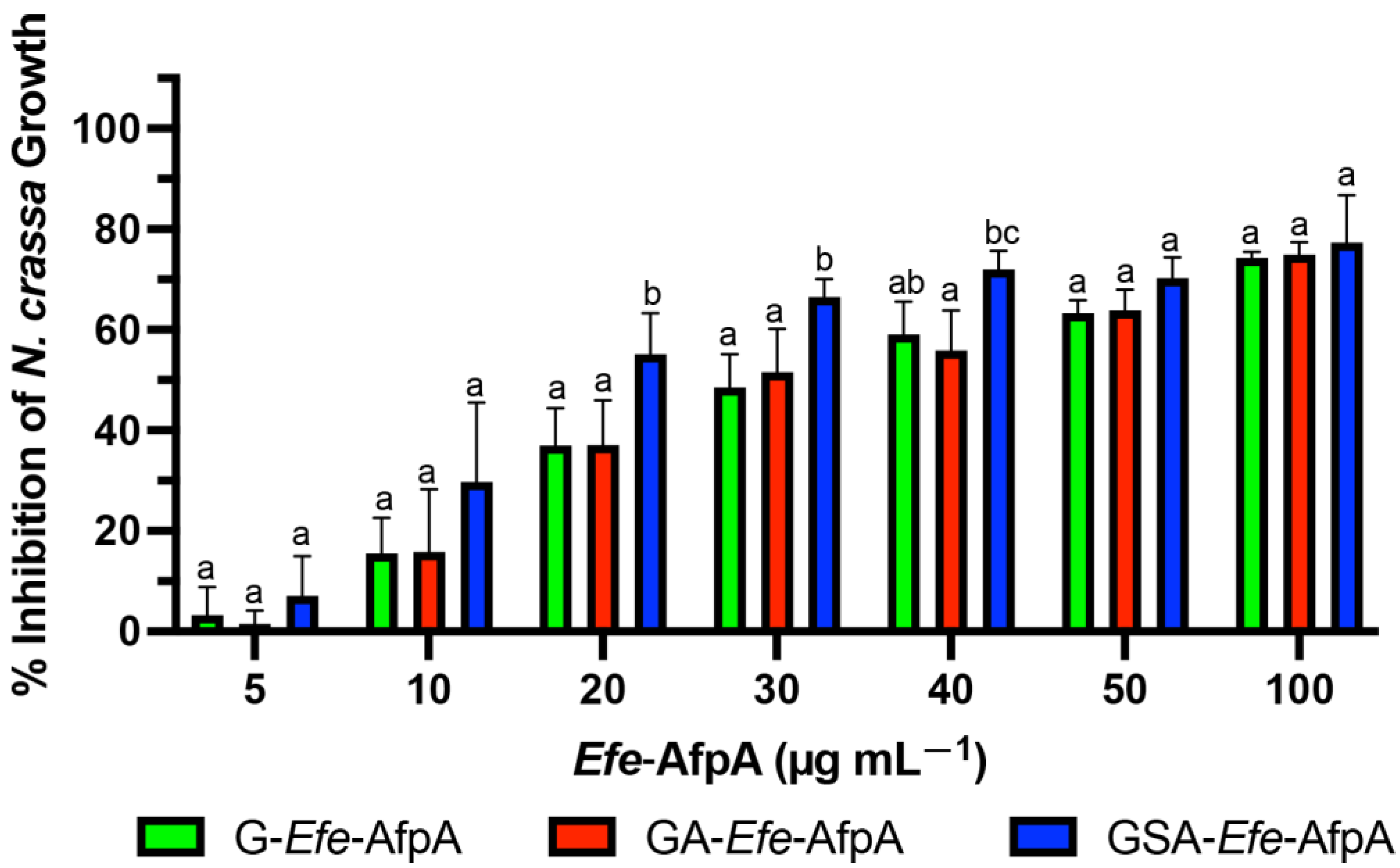
The antifungal activity of the N-terminal-modified Efe-AfpA proteins produced in E. coli was quantitatively assessed against N. crassa conidia germination in a microtiter plate assay. N. crassa has been established as a sensitive model system for testing the antifungal activity of the similar Pe. chrysogenum protein, PAF [46]. All three of the tested N-terminal modified forms of Efe-AfpA had activity at all the concentrations tested, with the GSA-Efe-AfpA form having statistically significant higher inhibition at 20, 30 and 40 µg mL−1 (Figure 1). At 100 µg mL−1, the inhibition of N. crassa growth was similar with all modified forms, ranging from 74 to 77%.
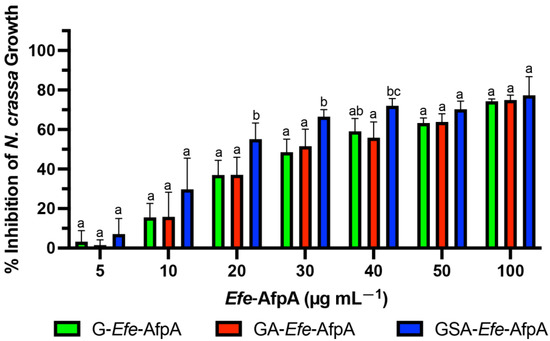
Figure 1.
Comparison of activity of bacterially produced Efe-AfpA with varying N-terminal amino acids. Increasing concentrations of the modified Efe-AfpA proteins were assayed for activity in the N. crassa growth assay with 1 × 106 conidia mL −1. The data presented are the means and standard deviations of three replicates. For each concentration, columns with different letters indicate a significant difference in activity (p ≤ 0.05, two-way ANOVA).
3.3. Expression of Efe-AfpA in Penicillium chrysogenum
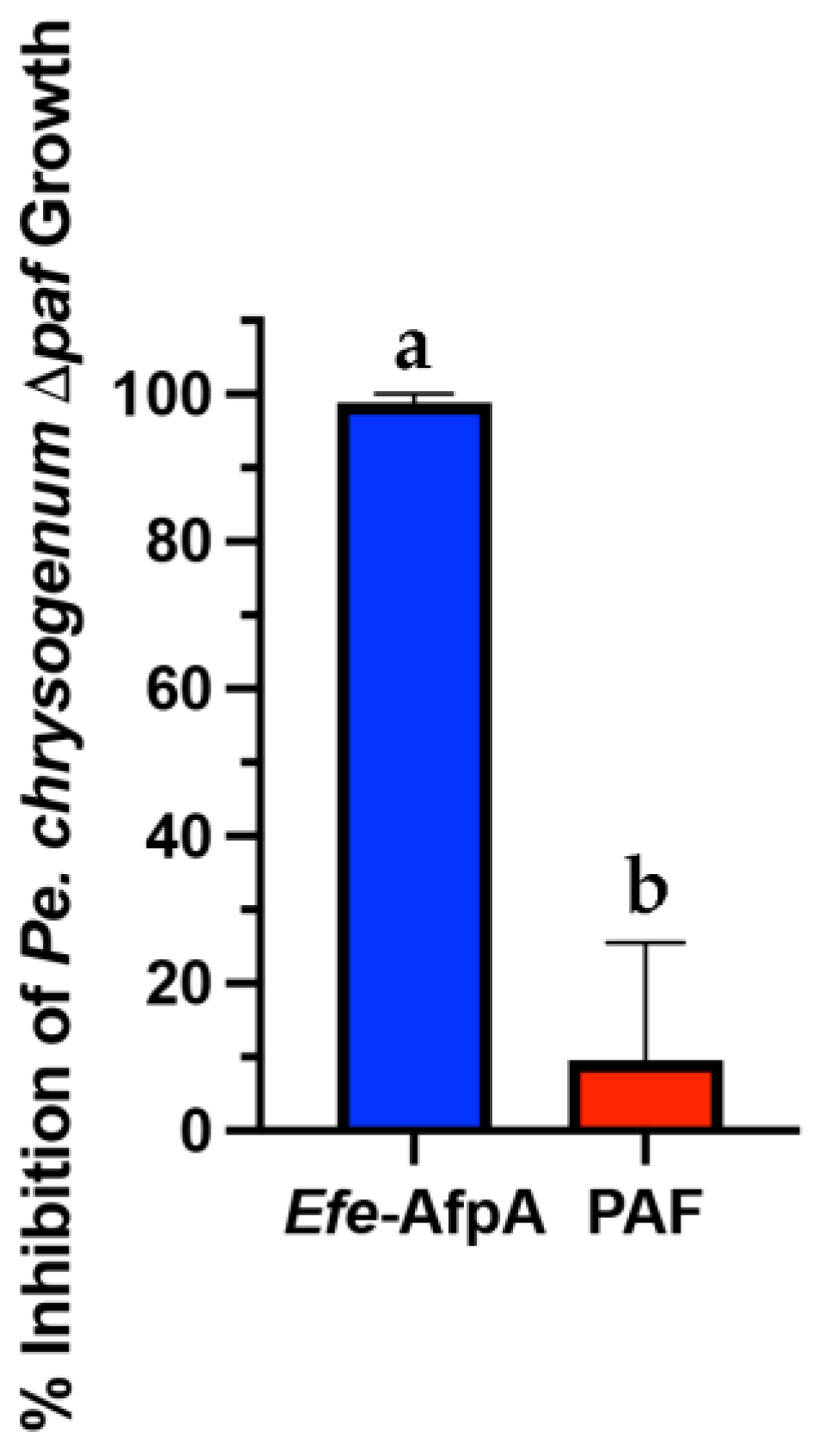
Pe. Chrysogenum, the producer of the antifungal protein PAF, has been developed as an efficient expression system for other antifungal proteins [31]. In this system the expression of the target protein is driven by the native paf promoter in a strain in which the paf gene has been deleted. Although active Efe-AfpA could be recovered from the Pe. Chrysogenum expression system when using the growth conditions used for PAF [31], yields were variable and in some cases the Pe. Chrysogenum cells appeared to lyse suggesting that Efe-AfpA may have activity against Pe. Chrysogenum. To test this possibility, the activity of Efe-AfpA on Pe. Chrysogenum conidial growth was assayed as for N. crassa. Efe-AfpA nearly completely inhibited Pe. Chrysogenum conidial growth even at the low concentration of 5 µg mL−1, whereas PAF was not inhibitory (Figure 2).
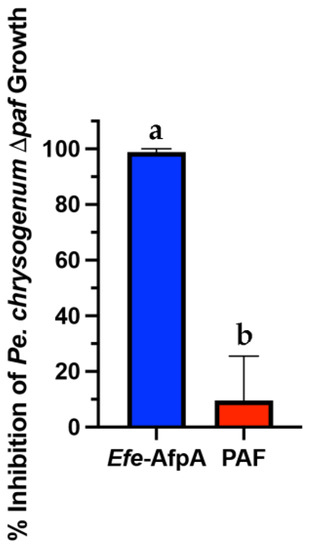
Figure 2.
Comparison of activity of Efe-AfpA and PAF against Pe. chrysogenum Δpaf conidial growth. Five µg mL−1 of either Efe-AfpA or PAF was assayed for activity against 1 × 104 conidia mL−1. The data presented are the means and standard deviations of three replicates. Columns with different letters indicate a significant difference in activity (p ≤ 0.05, one-way ANOVA).
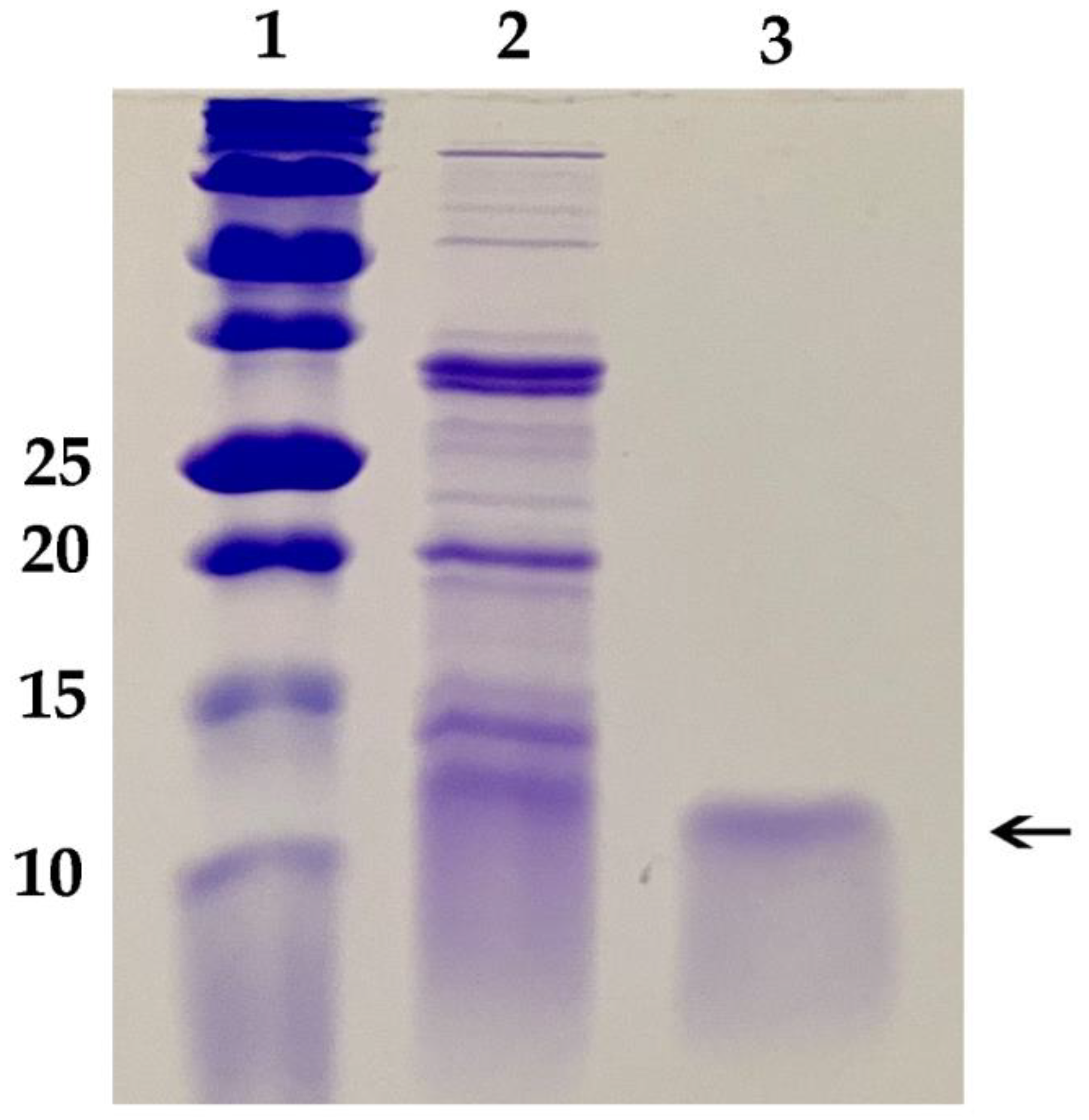
These results necessitated development of a new protocol for growth of P. chrysogenum expressing Efe-AfpA to avoid the toxic effects of the Efe-AfpA on Pe. chrysogenum. A successful protocol was developed that paired growth of the fungus for 48 h in a high nutrient medium that did not induce expression of Efe-AfpA, followed by transfer of the fungal mycelium to a low nutrient medium for 48 to 72 h. In the high nutrient medium, the fungus grew rapidly generating a large biomass of mycelium. In the low nutrient medium Efe-AfpA was expressed and secreted to the surrounding medium from the large biomass, resulting in high yields of active protein. Efe-AfpA was purified from the culture medium using a combination of cation exchange and size exclusion filtration (Figure 3). Although the calculated size of the mature protein is 6278 Daltons, it ran on a gel at a position similar to the 10 kDa marker.
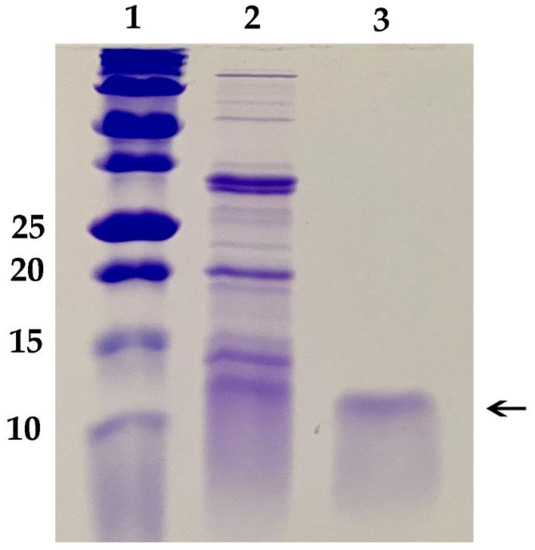
Figure 3.
SDS polyacrylamide gel of purification of the E. festucae antifungal protein produced in Pe. chrysogenum. Lane 1, Bio-Rad Precision Plus Protein Dual Xtra Standards, size of markers in kD given on left; Lane 2, Crude culture filtrate of Pe. chrysogenum expressing the antifungal protein; Lane 3, 1 µg purified Efe-AfpA, indicated by the arrow.
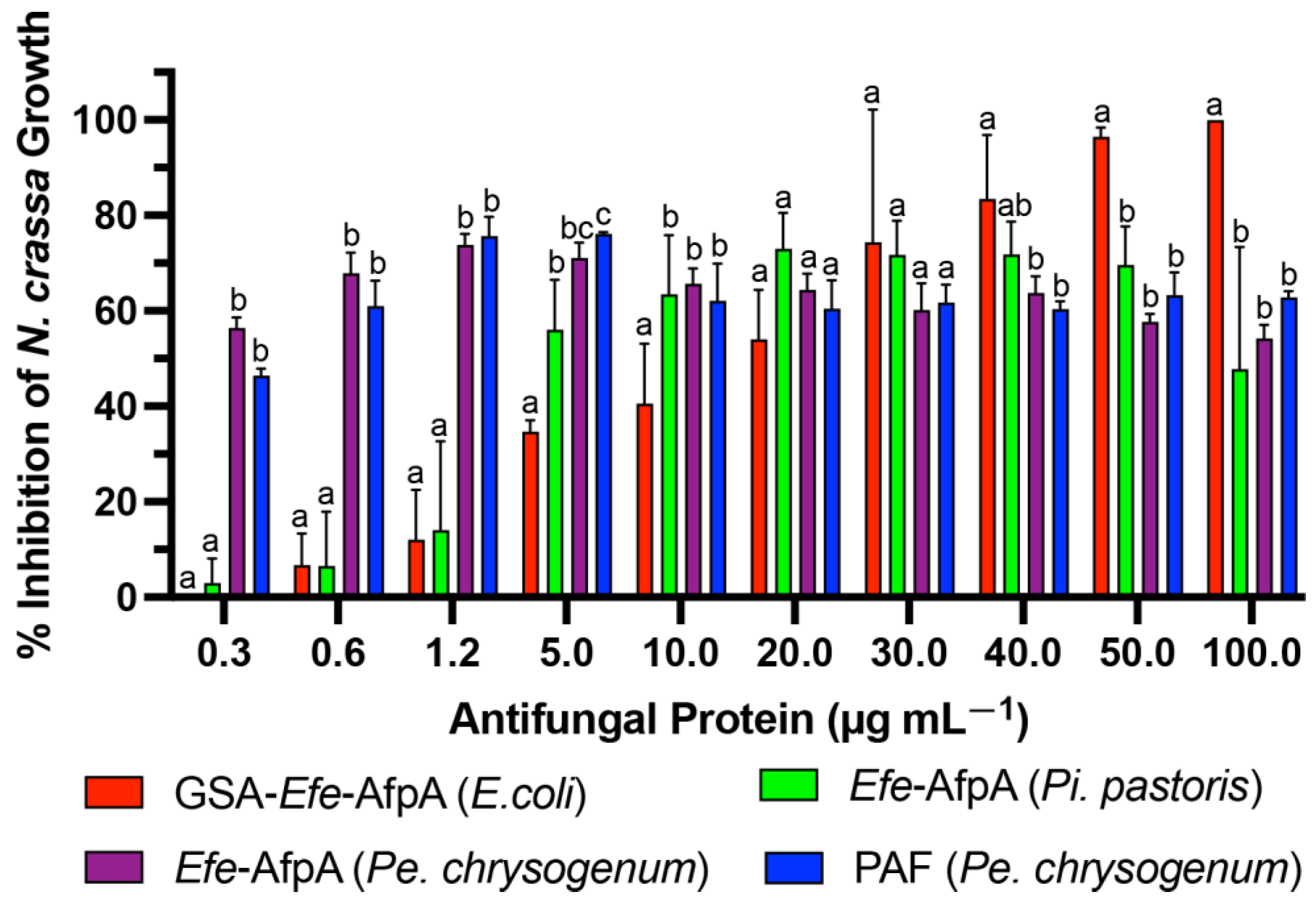
Efe-AfpA was expressed in the Pe. chrysogenum system and the activity of the purified protein was compared with the activity when expressed in Pi. pastoris and in E. coli, as well as with PAF, in the N. crassa conidial growth assay (Figure 4). The activity of Efe-AfpA expressed in Pe. chrysogenum was similar to the activity of PAF at all concentrations tested. At lower concentrations, Efe-AfpA expressed in Pe. chrysogenum had significantly higher activity than when produced in Pi. pastoris and E. coli. At 20 and 30 µg mL−1 the activity of Efe-AfpA expressed in all three systems was similar. At the higher concentrations, 40, 50, and 100 µg mL−1, Efe-AfpA produced in E. coli had significantly higher activity than the protein expressed in Pi. pastoris or Pe. chrysogenum. However, the significantly lower yields of pure protein from the E. coli system makes that system impractical for large-scale production of the protein (Table 2). The Pe. chrysogenum system was therefore chosen as the optimal system for further characterization of Efe-AfpA since it produced the highest quantity of Efe-AfpA and was the most convenient for purification. The difference in activity in Efe-AfpA produced in the different expression systems may be due to a difference in proper folding of the protein, which is known to affect the activity of the similar protein PAF [33].
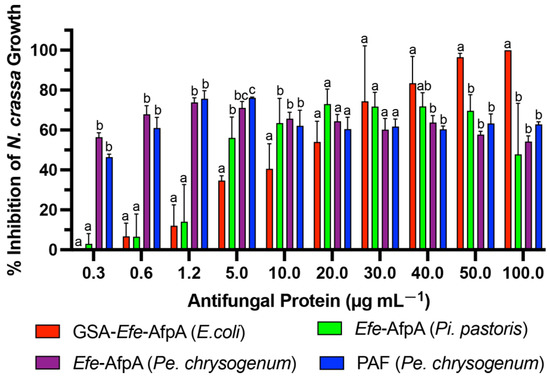
Figure 4.
Comparison of activities of PAF purified from Pe. chrysogenum and Efe-AfpA purified from E. coli, Pi. pastoris, and Pe. chrysogenum. Increasing concentrations of the antifungal proteins were assayed for activity in the N. crassa growth assay with 1 × 106 conidia mL−1. The data presented are the means and standard deviations of three replicates. For each concentration, columns with different letters indicate a significant difference in activity (p ≤ 0.05, two-way ANOVA).

Table 2.
Yield of purified Efe-AfpA from the different expression systems.
3.4. Activity of Efe-AfpA and PAF against Clarireedia jacksonii in Culture
Previously, Efe-AfpA produced in Pi. pastoris was reported to have activity in culture against C. jacksonii, the dollar spot pathogen of turfgrasses [20]. Since the activity of Efe-AfpA produced in Pe. chrysogenum was similar to that of PAF in the N. crassa assay, the activity of the two antifungal proteins against C. jacksonii was compared in culture (Figure 5). Efe-AfpA clearly inhibited the growth of the C. jacksonii mycelial plug, whereas PAF did not (Figure 5A). The dye Evans blue enters cells that have damaged cell membranes [47]. Treatment of C. jacksonii with the E. festucae antifungal protein resulted in damage to the cell membranes as evidenced by Evans blue staining, whereas treatment with PAF did not (Figure 5B).
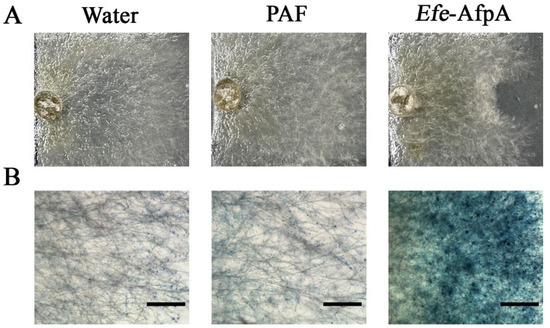
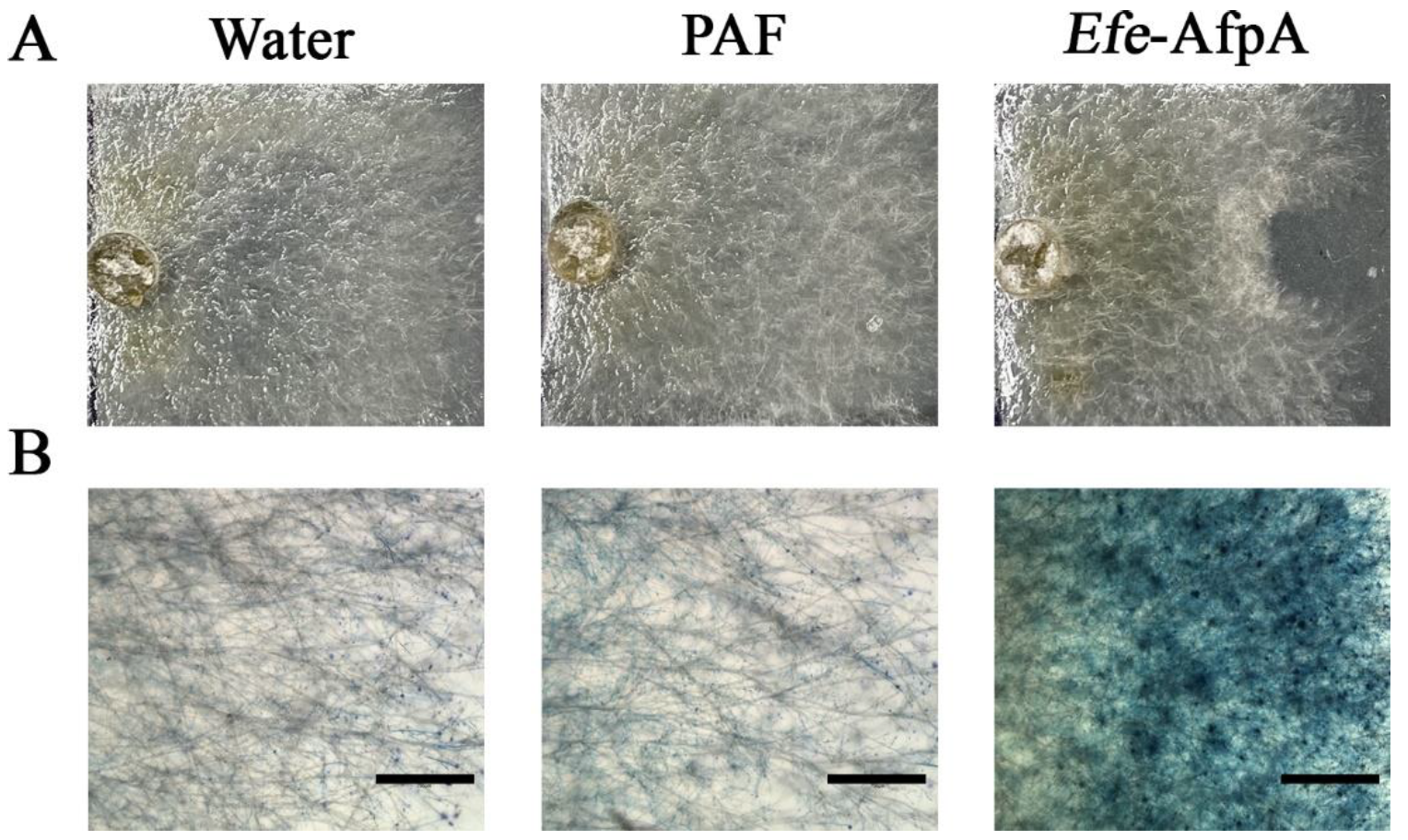
Figure 5.
Comparison of the activity of Efe-AfpA expressed in Pe. chrysogenum with PAF, a similar antifungal protein from Pe. chrysogenum, against C. jacksonii in culture. In the upper panels (A) water (10 µL), PAF (300 ng), or purified Efe-AfpA (300 ng), was placed on the right side of a plug of C. jacksonii. The lower panels (B) show the C. jacksonii hyphae from the upper panels treated with Evans blue. Bars in the lower panels are 750 μm.
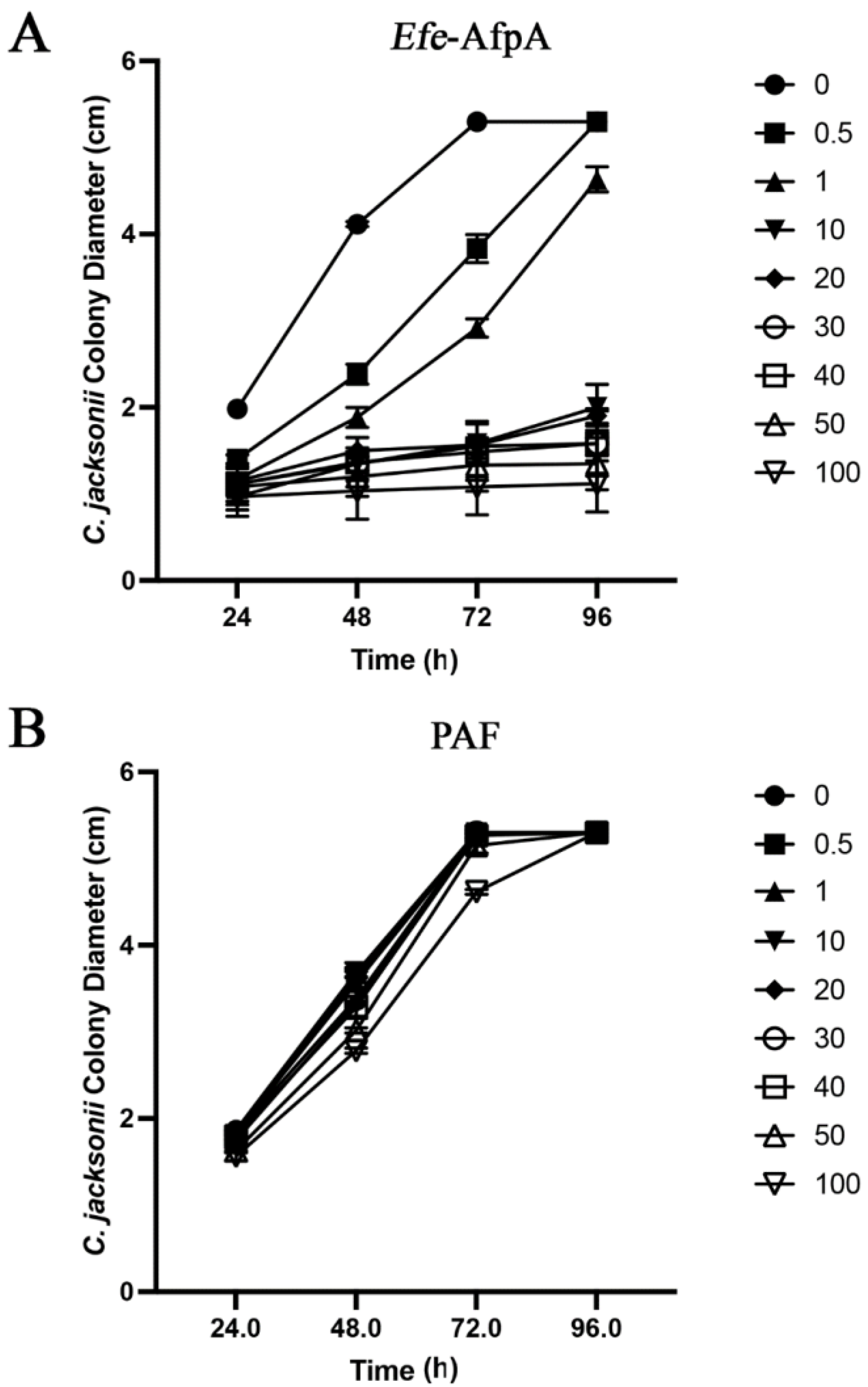
In agar plate assays in which either purified Efe-AfpA or PAF were incorporated into the agar, Efe-AfpA inhibited C. jacksonii growth at concentrations of 0.5 μg mL−1 and higher, whereas PAF was not inhibitory at any concentration (Figure 6, Figure S1 and Figure S2). Although PAF is similar to Efe-AfpA in amino acid sequence and in activity against N. crassa, it did not have activity against C. jacksonii in culture.
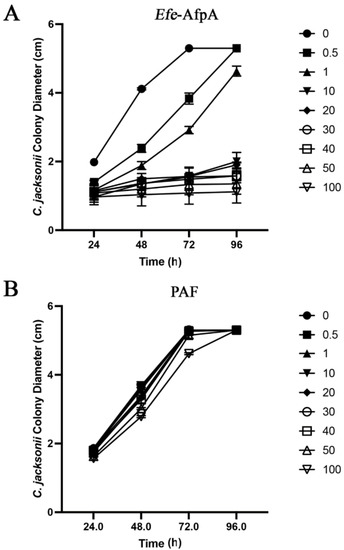
Figure 6.
Effect of Efe-AfpA (A) or PAF (B) on C. jacksonii mycelial growth. C. jacksonii mycelial plugs were subcultured onto PDA plates amended with increasing concentrations of either Efe-AfpA or PAF. The colony diameters were measured daily. The data presented are the means and standard deviations of three replicates. Photographs of the plates are shown in Figures S2 and S3.
3.5. Activity of Applied Efe-AfpA on Expression of Dollar Spot Symptoms When Strong Creeping Red Fescue and Creeping Bentgrass Plants Were Inoculated with Clarireedia jacksonii in a Greenhouse Assay
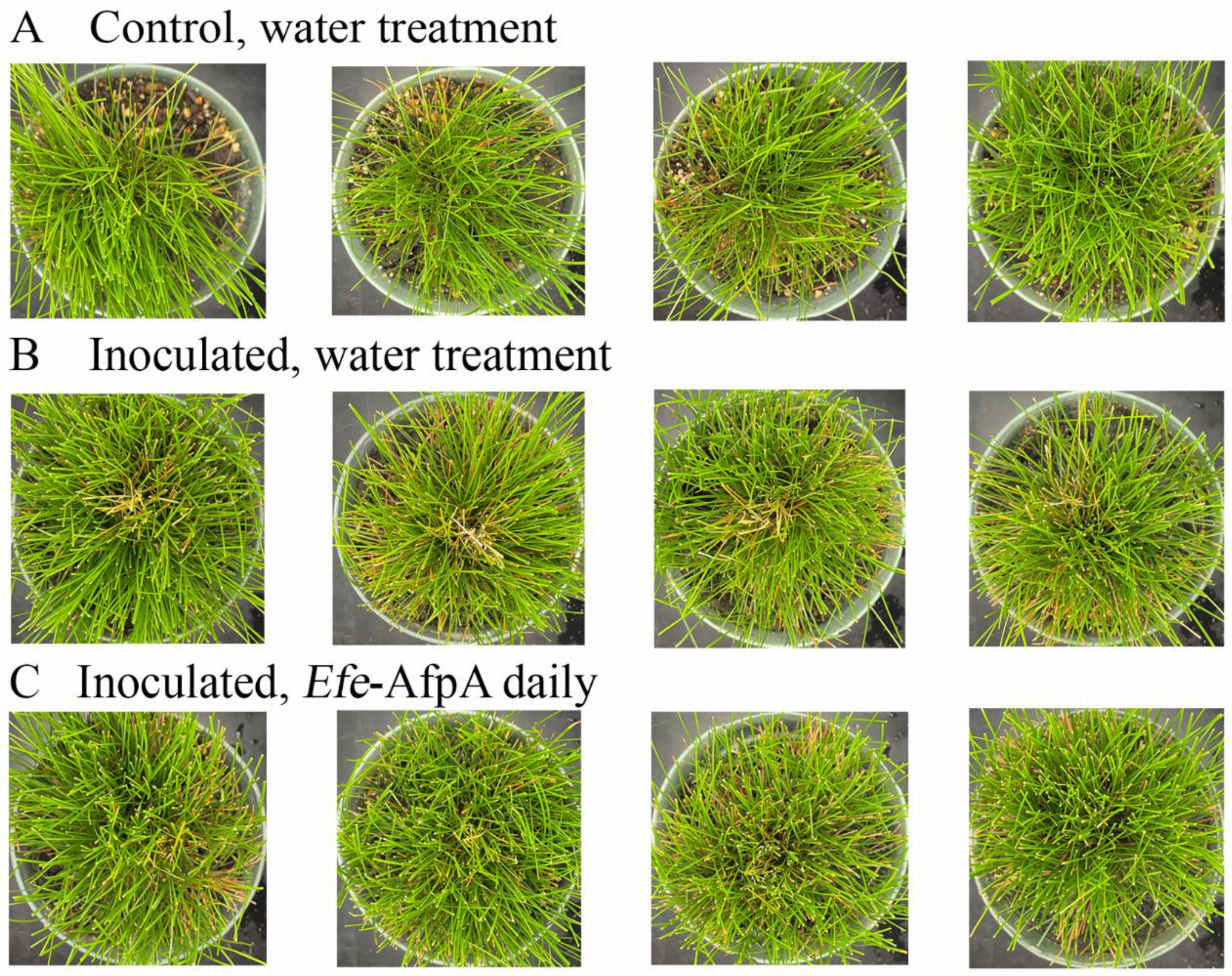
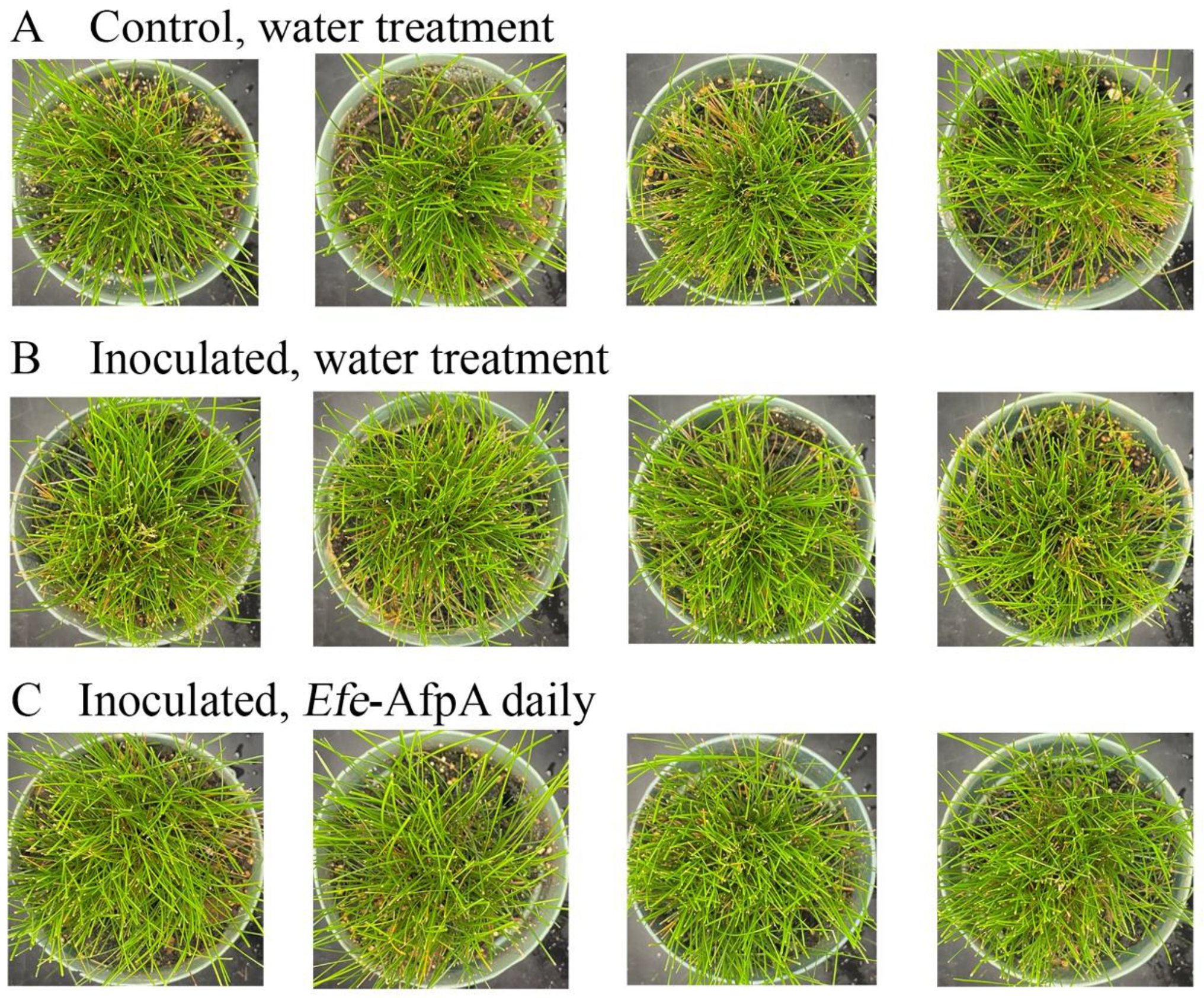
As described above, in field studies, endophyte-infected strong creeping red fescue plants were resistant to dollar spot infection, whereas endophyte-free plants were susceptible to the disease [19]. Efe-AfpA is an abundant E. festucae protein in endophyte-infected plants and may be the reason for the disease resistance seen in endophyte-infected plants. it was therefore of interest to determine if application of the protein could protect endophyte-free plants from dollar spot caused by C. jacksonii. Purified Efe-AfpA was tested against C. jacksonii on endophyte-free and endophyte-infected strong creeping red fescue plants in greenhouse assays where an agar plug of C. jacksonii was used to inoculate the plants. Plants were sprayed with either water or a 100 μg mL−1 Efe-AfpA solution and the symptom expression of the controls and treated plants was compared. As seen in Figure 7, the C. jacksonii inoculated endophyte-free plants had severe disease symptoms, observed as necrotic tillers surrounding the point of inoculation in the center of plants. In contrast, the Efe-AfpA treated inoculated endophyte-free plants had only minor symptoms of dollar spot disease. The endophyte-infected plants inoculated with C. jacksonii had only minor disease symptoms (Figure 8), when compared with the endophyte-free plants, similar to what is seen in field studies [19]. These results support the hypothesis that Efe-AfpA is a major contributor to the dollar spot resistance seen in E. festucae-infected strong creeping red fescue plants.
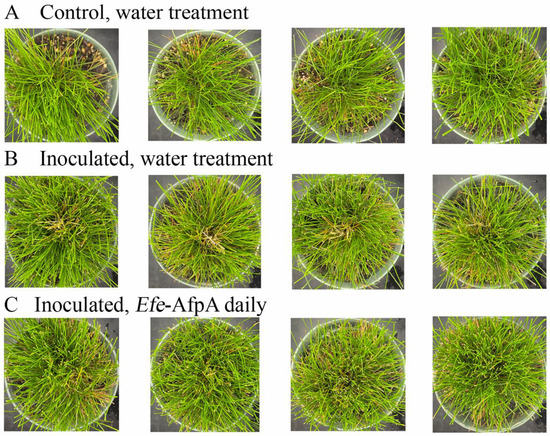
Figure 7.
Efe-AfpA prevented severe symptoms of dollar spot disease when endophyte-free strong creeping red fescue plants were inoculated with an 8 mm plug of C. jacksonii. Plants were sprayed daily for 10 days with either water (A,B) or 100 μg mL−1 of Efe-AfpA (C). Photos within a row are replicates of the labeled treatment.
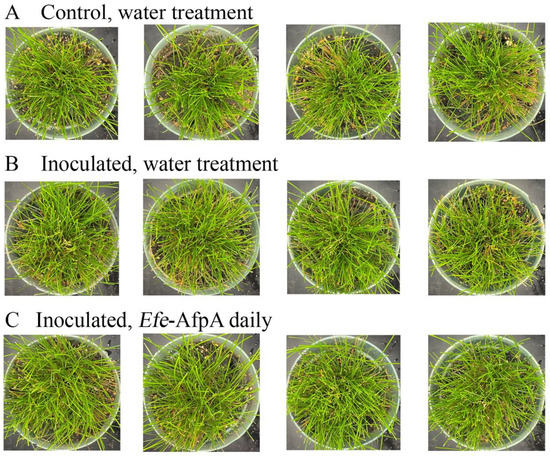
Figure 8.
Endophyte infected strong creeping red fescue plants exhibited no dollar spot disease symptoms when inoculated with an 8 mm plug of C. jacksonii. Plants were sprayed daily for 10 days with either water (A,B) or 100 μg mL−1 of Efe-AfpA (C). Photos within a row are replicates of the labeled treatment.
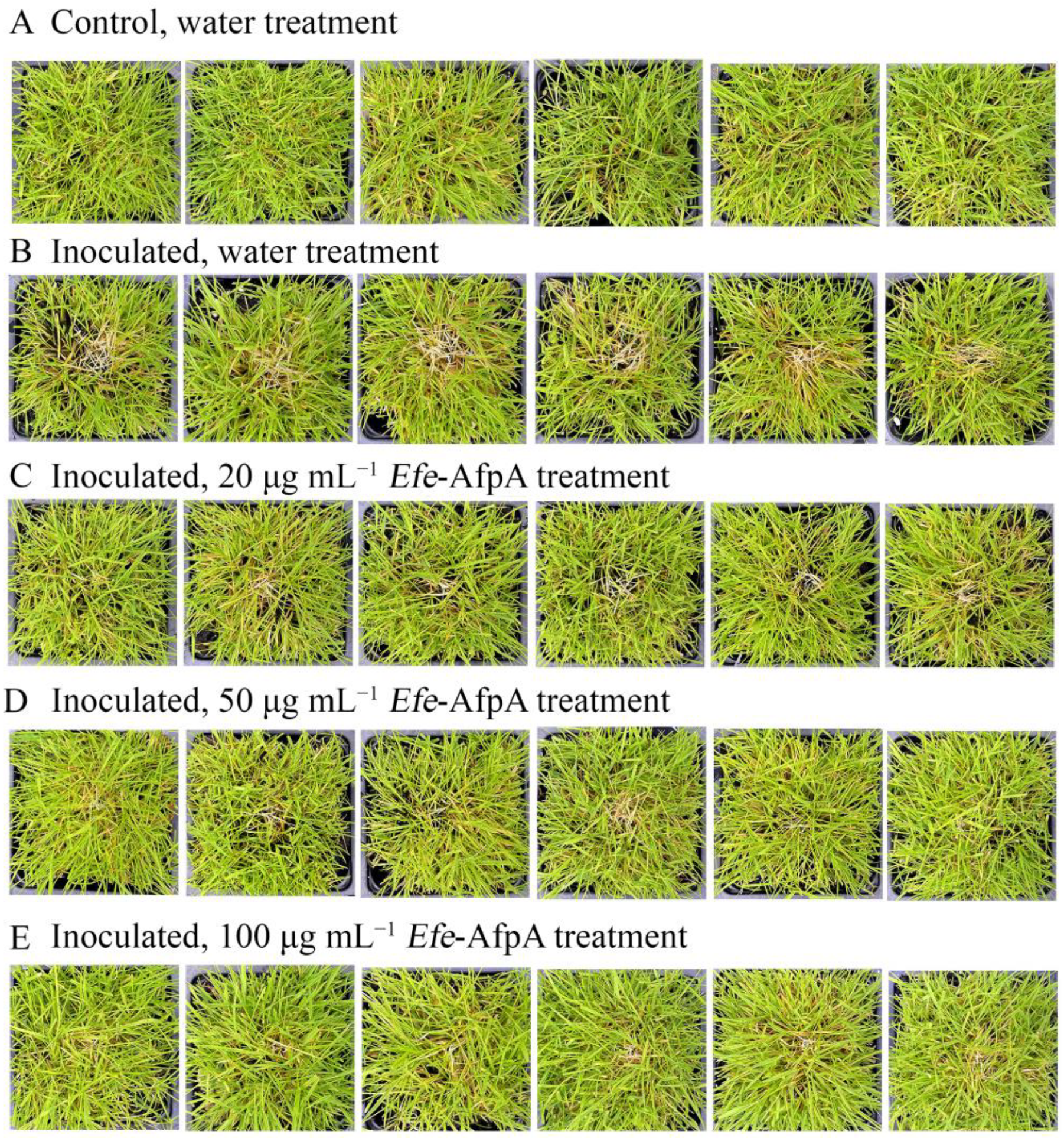
Since dollar spot disease is a serious problem on creeping bentgrass, the ability of Efe-AfpA to protect creeping bentgrass plants from dollar spot caused by C. jacksonii was also determined in greenhouse assays. Plants were sprayed with either water or Efe-AfpA solutions and symptom expression of the control and treated plants was compared. Dollar spot symptoms on creeping bentgrass are characterized by necotic sunken areas of the turf [26] as seen in the inoculated water-treated plants (Figure 9). When sprayed daily for 7 days with either 20, 50 or 100 μg mL−1 Efe-AfpA, the plants had less severe disease symptoms at all concentrations, with 100 μg mL−1 providing the best protection.
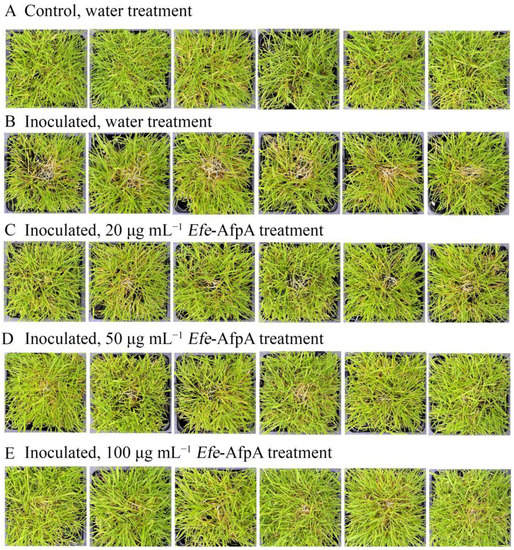
Figure 9.
Effect of Efe-AfpA concentration on expression of dollar spot disease symptoms when creeping bentgrass cv. ‘Crenshaw’ plants were inoculated with an 8 mm plug of C. jacksonii. Plants were sprayed daily for 7 days with either water (A,B) or different concentrations of Efe-AfpA (C–E). Photos within a row are replicates of the labeled treatment.
To determine the effect of sequential applications on dollar spot severity, plants sprayed daily with 100 μg mL−1 Efe-AfpA were compared with plants sprayed only at the time of inoculation or at the time of inoculation and again at 48 h post-inoculation (Figure 10). The protection provided by two sprays was similar to that provided by seven daily applications.

Figure 10.
Effect of the number of treatments of Efe-AfpA applications on expression of dollar spot disease symptoms after creeping bentgrass cv. ‘Crenshaw’ plants were inoculated with an 8 mm plug of C. jacksonii. Plants were treated water (A,B) or with 100 μg mL −1 of Efe-AfpA as indicated (C–E) and photographed 7 days post inoculation. Photos within a row are replicates of the labeled treatment.
3.6. Efe-AfpA Activity against Neurospora crassa Glucosylceramide Mutant Strains
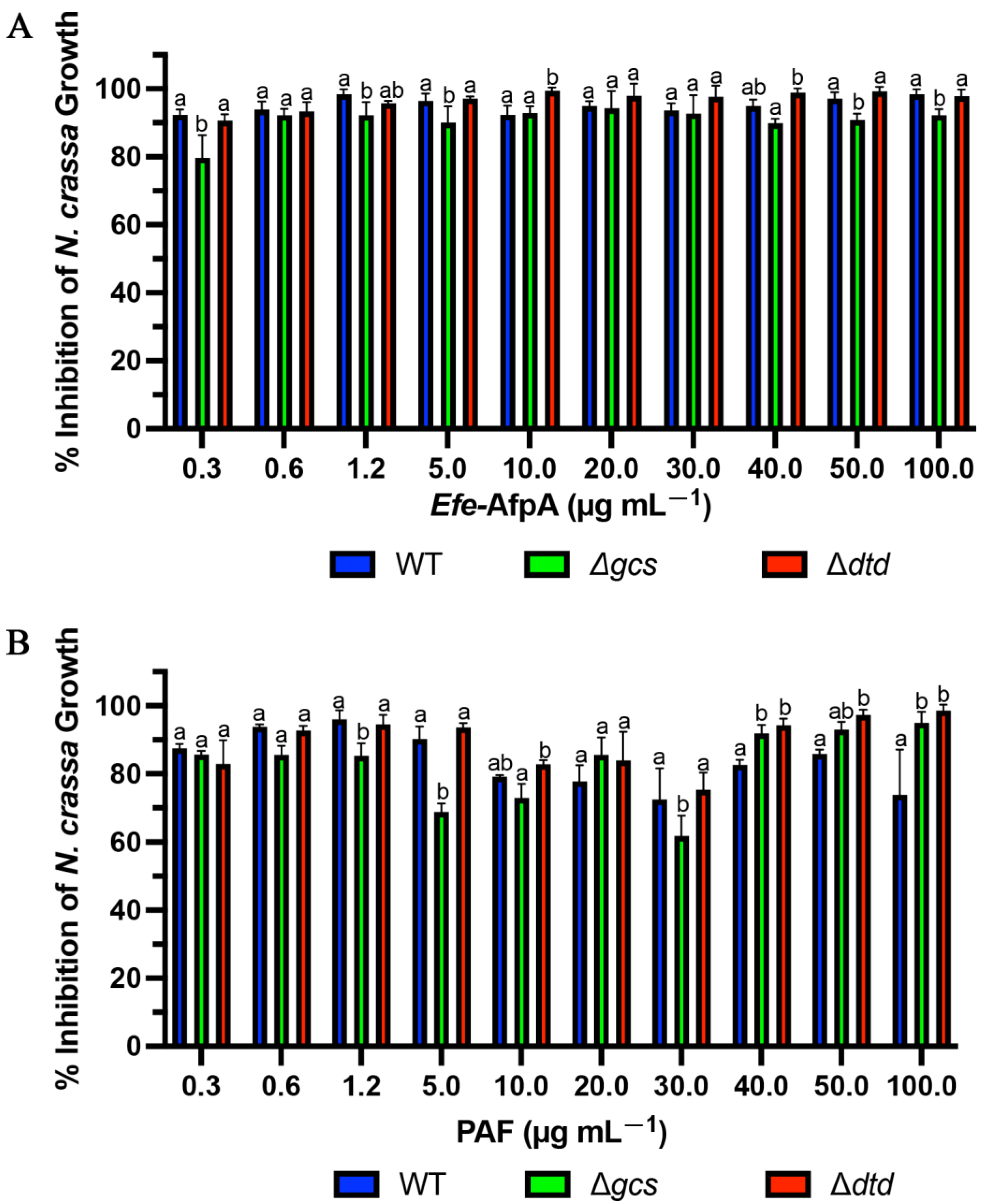
PAF from Pe. chrysogenum and AFP from A. giganteus have been reported to require presence of the membrane sphingolipids glucosylceramide or Δ3-unsaturated glucosylceramide, respectively (Figure S4), in the susceptible target fungi for full antifungal activity [36,48]. Since Efe-AfpA and PAF have both similarities and differences in their activity against N. crassa and C. jacksonii, respectively, we compared their activities against some N. crassa membrane glucosylceramide mutants. The activities of PAF and Efe-AfpA were compared in the conidial growth assay with N. crassa glucosylceramide synthase (gcs) and 2-hydroxy fatty N-acyl-Δ3(E)-desaturase (sphingolipid Δ3(E)-desaturase), here designated dtd (delta three desaturase), deletion mutants
In plate assays against wild type N. crassa, PAF and Efe-AfpA had similar levels of inhibition at concentrations of 3 μg mL−1 and above. At 25 μg mL−1 and above, there appeared to be complete inhibition of wild type N. crassa growth with both antifungal proteins. At the two lowest concentrations tested, PAF had higher levels of inhibition than Efe-AfpA (Figure S5). In plate assays, Huber et al. [36] reported reduced activity of PAF against the N. crassa Δgcs mutant deficient in glucosylceramide relative to activity against the wild-type strain. The level of inhibitory activity was similar from 0.5 μM to 16 μM (3 μg mL−1 to 100 μg mL−1) [36]. Our results with PAF are similar, with similar levels of inhibition of the Δgcs strain from 1.5 to 100 μg mL−1 (Figure S5). In contrast, the inhibitory activity of Efe-AfpA against the Δgcs mutant was similar to its activity against the wild type strain (Figure S5), where Efe-AfpA exhibited complete inhibition at the higher concentrations. The activities of both PAF and Efe-AfpA against the Δdtd mutant strain were similar to their activities against the wild-type strain (Figure S5).
In the quantitative 96-well microtitre plate liquid culture assays the inhibitory activity of PAF was reduced against the Δgcs mutant strain relative to the wild-type strain at some, but not all, concentrations tested (Figure 11). The higher level of inhibition seen in Figure 11 relative to Figure 4 is because fewer N. crassa spores were used in Figure 11. Huber et al. [36] reported a lack of PAF inhibition of the growth of the Δgcs mutant at 0.06 and 1 μM (approximately 0.37 and 6.2 µg mL−1). Here, at the similar PAF concentrations of 0.3 and 5 μg mL−1, there was a reduction in inhibition of growth of the Δgcs mutant only at the higher concentration of 5 μg mL−1, but not the apparent complete loss of inhibition seen by Huber et al. [36]. Interestingly, at the higher concentrations of PAF, the Δgcs mutant was more inhibited by PAF than the wild-type strain. At 1.2 and 5 μg mL−1, the Δgcs mutant was also less inhibited by Efe-AfpA, but to a lesser extent than with PAF (Figure 11). There was no reduction in inhibition, relative to the wild-type strain, of the Δdtd mutant by either PAF or Efe-AfpA.
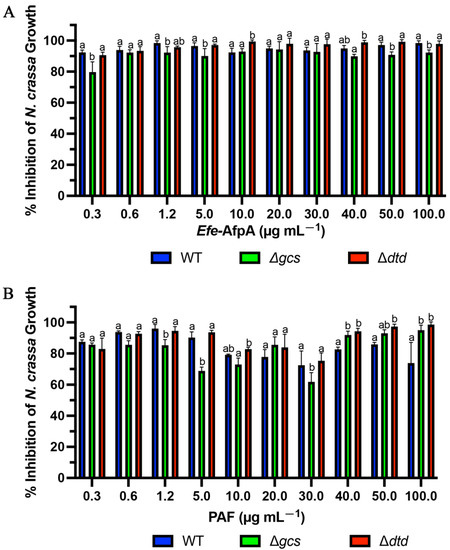
Figure 11.
Efe-AfpA (A) and PAF (B) activity against N. crassa wild type and glucosylceramide pathway mutants. Increasing concentrations of the proteins were assayed for activity in the N. crassa growth assay with 1 × 104 conidia mL−1. The data presented are the means and standard deviations of three replicates. For each concentration, columns with different letters indicate a significant difference in activity (p ≤ 0.05, two-way ANOVA).
With PAF, but not Efe-AfpA, the activity against the wild-type strain dropped between the concentrations 10 and 30 μg mL−1 and then rose at the higher concentrations. This drop in inhibitory activity of PAF against the wild-type N. crassa strain was reproducible, as it was seen in three independent assays. The explanation for this is unknown but does suggest there may be different modes of action at different concentrations. This decrease in inhibition (increased growth) can be seen in the accompanying microscopy (Figure S6).
From the 96 well liquid assays, the minimum inhibitory concentration (MIC) for Efe-AfpA and PAF against the wild-type and mutant N. crassa strains was determined (Table 3). The MIC is defined as the concentration needed for ≥90% inhibition. The low concentrations of 0.3 and 0.6 µg mL−1 for Efe-AfpA for all strains and for PAF for the wild type and Δdtd strains are characteristic of antifungal proteins, as one of their hallmarks is high activity at low concentrations. The MIC of PAF for the Δgcs mutant was much higher, at 40 µg mL−1. Although PAF does have inhibitory activity against the Δgcs mutant, its reduced activity relative to the wild-type strain both in quantitative assays and in plate assays at some concentrations does support the proposal by Huber et al. [36] that membrane glucosylceramide in the target fungus is an important factor in PAF activity. The presence of glucosylceramides does not appear to be a major factor for Efe-AfpA activity.

Table 3.
Minimum inhibitory concentrations of Efe-AfpA and PAF against wild type and glucosylceramide mutant strains of Neurospora crassa.
In contrast to PAF, PAFB, a similar antifungal protein from Pe. chrysogenum, was found to have similar inhibition of the N. crassa wild type and glucosylceramide synthase mutant strains [36]). This difference was ascribed to a higher electrostatic affinity of PAFB to the anionic membrane surfaces [36]. Efe-AfpA also has a predicted higher net charge than PAF (Table 4), which may explain the differences in observed inhibition of the Δgcs mutant strain relative to PAF.

Table 4.
Characteristics of the mature amino acid sequences of PAF, PAFB, and Efe-AfpA The numbers of charged amino acids in each protein are given.
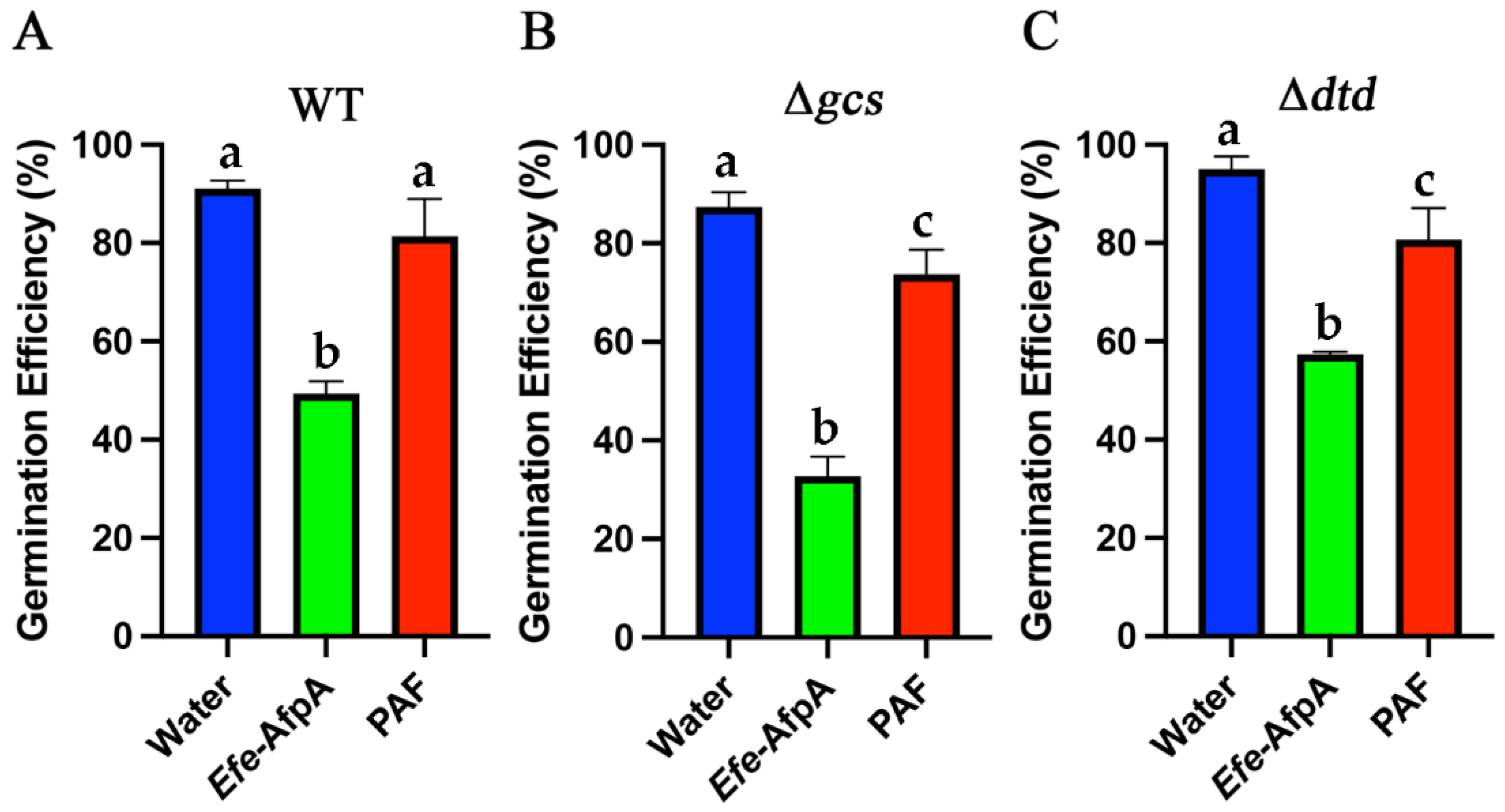
The comparison between PAF and Efe-AfpA for the inhibition of wild-type N. crassa growth in the 96-well liquid assay revealed that at concentrations between 10 and 100 μg mL−1 Efe-AfpA had significantly greater inhibitory activity (Figure S7). This effect was also seen with the two mutant strains at some concentrations. This phenomenon was further investigated by comparing conidial germination rates in the presence of 10 μg mL−1 PAF or Efe-AfpA (Figure 12). After 6 h of incubation, all three N. crassa strains had significantly fewer germinated conidia in the Efe-AfpA treated samples relative to the PAF treated samples when compared to the water treated control. The differences in activities of the two proteins seen in the 96-well liquid assay appears to be, in part, due to their differential effect on conidial germination.
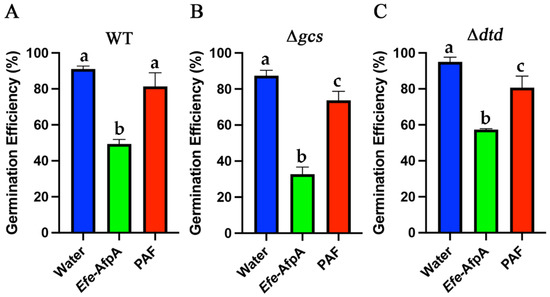
Figure 12.
Effect of Efe-AfpA and PAF on conidia germination of N. crassa wild type (A) and glucosylceramide pathway mutants (B,C). Conidia (100 μL of 1 × 105 conidia mL−1) were treated for six hours with either water or an antifungal protein at 10 μg mL−1. One-hundred conidia were counted as either germinated or ungerminated. The data presented are the means and standard deviations of three replicates. For each N. crassa strain, columns with different letters indicate a significant difference in activity (p ≤ 0.05, one-way ANOVA).
3.7. Relationship of Efe-AfpA to Other Antifungal Proteins from Filamentous Fungi
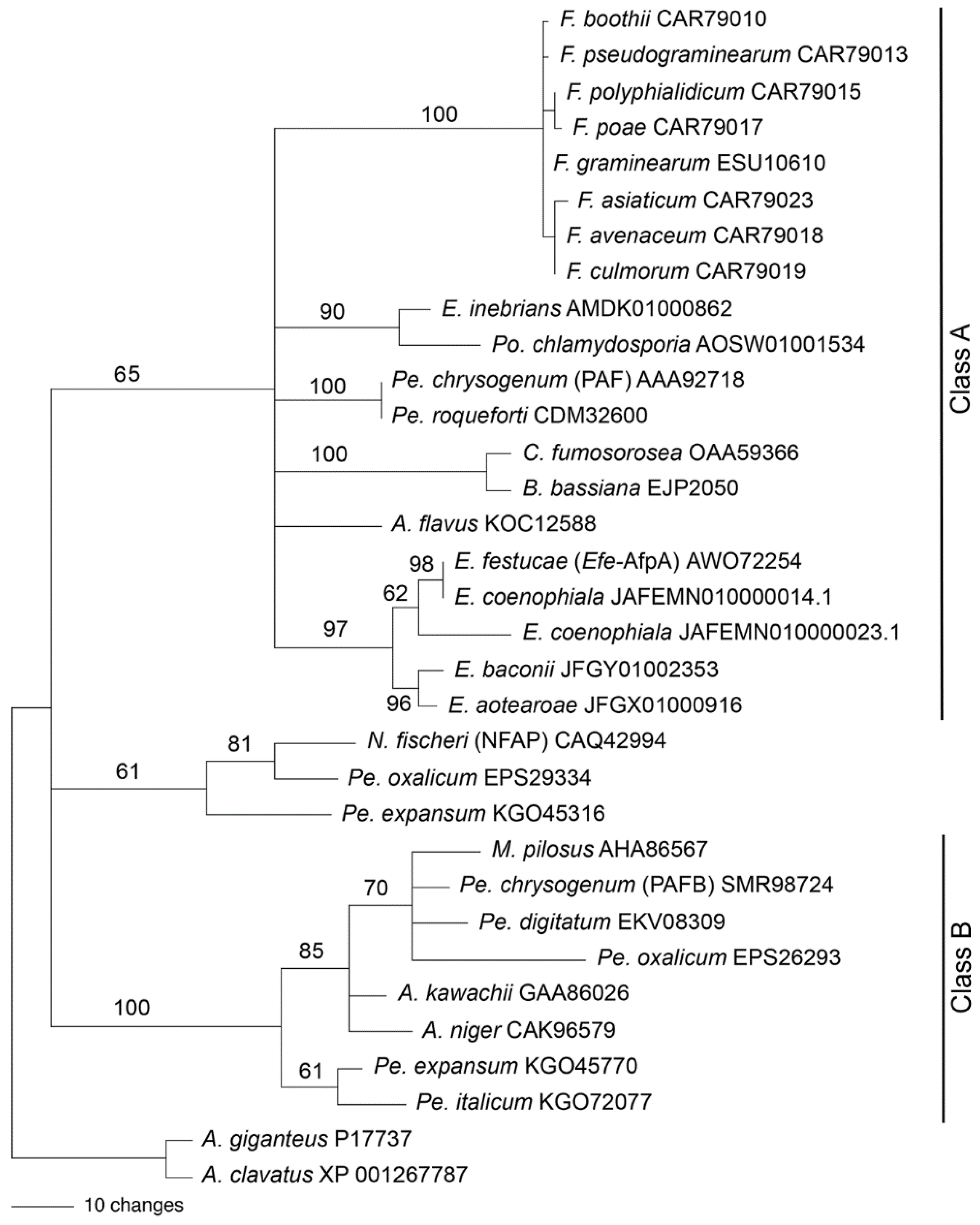
We previously reported a phylogenetic analysis of antifungal proteins including Efe-AfpA [22]. Garrigues et al. [49] proposed a phylogenetic classification of the antifungal proteins. We therefore generated a new phylogenetic analysis to place Efe-AfpA into the antifungal protein classification scheme (Figure 13). The phylogeny reported here includes the sequences from Class A and Class B used previously [49], as well as some additional sequences. The Class C proteins were not included here since they are distinctly different than those of Classes A and B. Efe-AfpA, as well as the similar sequences from the other four Epichloë spp. that have antifungal protein genes, grouped with the Class A sequences.
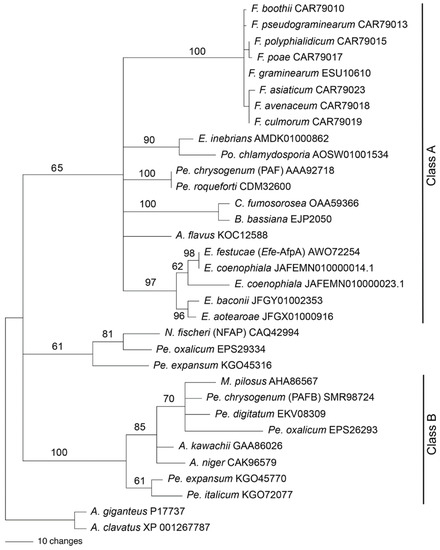
Figure 13.
Rooted 50% majority rule maximum parsimony phylogenetic tree of the antifungal protein amino acid sequences. The A. giganteus and A. clavatus sequences were designated as the outgroups for rooting the tree. The numbers at the nodes are the bootstrap percentages based on 1000 replications. The tree was based upon 101 total characters, of which 14 were constant, 6 variable characters were parsimony uninformative, and 81 variable characters were parsimony informative. The NCBI accession numbers are given following the species names. The Efe-AfpA, PAF, NFAP, and PAFB sequences are identified in parentheses. The clades previously designated as Class A and Class B [49] are indicated. Genera abbreviations are F., Fusarium; E., Epichloë; Po., Pochonia; Pe., Penicillium; C., Cordyceps; B., Beauveria; A., Aspergillus; N., Neosartorya; M., Monascus.
Rooted phylogenetic trees can be informative regarding the evolutionary history of a protein, but can be problematic when the ancestral sequence is unknown or when there is horizontal gene transfer between unrelated species. Both of these issues are relevant with antifungal proteins. PAF from Pe. chrysogenum was placed in a clade of antifungal proteins from the Hypocreales rather than the Eurotiales, suggesting the possibility of horizontal gene transfer [22]. Large genomic regions containing the PAF gene sequence were found to be horizontally transferred among several cheese-associated Penicillium spp. [50,51]. The antifungal protein gene sequence from E. inebrians was more similar to that from Pochonia chlamydosporia, a parasite of nematode eggs, than to antifungal protein sequences from other Epichloë spp., suggesting its origin from horizontal gene transfer [20]. These issues make it difficult to choose a sequence with which to root the tree based on species phylogeny. The phylogenetic tree in Figure 13 was therefore rooted with the A. giganteus and A. clavatus sequences since those antifungal proteins are distinct from the rest, in that they have eight cysteines involved in four disulfide bonds [52,53] in contrast to six cysteines involved in three disulfide bonds in the other antifungal proteins used in the analysis.
The phylogenetic tree presented in Figure 13 is similar to that of Garrigues et al. [49] except in the placement of the sequences from Neosartorya fischeri, Pe. oxalicum, and Pe. expansum, which were previously grouped with the Class A proteins but here are not grouped with either of the two major clades. Hajdu et al. [54] also found that the NFAP-type antifungal proteins formed a clade separate from the Class A and Class B proteins. Here, all the Class B proteins are from fungal species within the Eurotiales whereas Class A contains species from both the Eurotiales and the Hypocreales.
3.8. Afp-A Genes in Epichloë spp.
We previously reported that most Epichloë spp. for which sequence data is available do not have a gene similar to Efe-afpA [20,22]. Since then, genome sequence data have become publicly available for two commercially important Epichloë spp., E. festucae var. lolii and E. coenophiala, which are endophytes of the turf and forage grasses perennial ryegrass and tall fescue, respectively. The E. festucae var. lolii AR5 genome (NCBI accession SRX1531627) [55] does not have a gene similar to Efe-afpA but the E. coenophiala genome does (NCBI accession JAFEMN010000000) [56].
Tall fescue is an important cool-season turf and forage grass used extensively worldwide. It is often infected with the endophyte E. coenophiala, which is a triparental hybrid fungus derived from the haploid species E. festucae, E. typhina subsp. poae, and a member of the Lolium-associated endophyte clade possibly derived from E. baconii [57,58]. The E. coenophiala genome sequence has two sequences similar to Efe-afpA, and these are included in the phylogenetic tree in Figure 13. At the amino acid level, the E. coenophiala AfpA sequence on scaffold 14 is 100% identical to that of Efe-AfpA and therefore likely derives from the E. festucae component of the E. coenophiala genome. The E. coenophiala AfpA sequence on scaffold 23 is more similar to that of E. festucae than to the AfpA sequence found in E. baconii and so cannot be unambiguously attributed to the Lolium-associated endophyte progenitor of E. coenophiala. However, the sequenced isolate of the other ancestral parent of E. coenophiala, E. typhina subsp. poae, does not have an AfpA gene in its genome [22].
Dinkins et al. [59] reported an extensive Illumina-based transcriptome comparison of endophyte-infected versus endophyte-free tall fescue. E. coenophiala Efe-afpA-like transcripts were represented, but at a low level. There were a total of 42 Efe-afpA-like reads in the endophyte-infected pseudostem samples out of a total of 1.3 million E. coenophiala reads, and 1 Efe-afpA-like read in the endophyte-infected leaf samples out of a total of 10,084 reads. Both E. coenophiala genes were represented among the reads. The low level of expression of the E. coenophiala Efe-afpA-like genes in endophyte-infected tall fescue is in stark contrast to the high level of expression in strong creeping red fescue infected with E. festucae, where Efe-afpA was among the most abundant fungal transcripts [22,60]. Tall fescue infected with E. coenophiala has not been reported to exhibit enhanced disease resistance in the field [19,20]. Therefore, it seems likely a high expression level of afpA in planta, as seen in strong creeping red fescue infected with E. festucae, is required for disease resistance. How expression of Efe-afpA in planta is regulated is unknown, but in strong creeping red fescue is 700-fold higher in planta than in culture [28].
4. Discussion
Here, we compared the activity of Efe-AfpA expressed in the prokaryote E. coli and in two eukaryotic systems, Pi. pastoris and Pe. chrysogenum. Active Efe-AfpA was obtained from all systems, with the Pe. chrysogenum system chosen as the most convenient. Purified Efe-AfpA clearly has antifungal activity, as shown here against N. crassa and C. jacksonii, and presumably also has antifungal activity in planta. Since application of purified Efe-AfpA was shown to protect endophyte-free strong creeping red fescue inoculated with C. jacksonii from developing severe symptoms of dollar spot, it seems likely that Efe-AfpA is the main factor in the observed field level resistance seen in endophyte-infected strong creeping red fescue. Efe-afpA knockout isolates of E. festucae were previously produced with the objective of directly assessing this presumption by inoculating the knockouts into endophyte-free strong creeping red fescue and determining if resistance to C. jacksonii was diminished relative to the wild type E. festucae strain [28]. However, the Efe-afpA knockout isolates were unable to infect the grass, whereas wild type and complemented isolates were able to infect. These results suggested that Efe-AfpA also affects the interaction between E. festucae and its host in addition to having antifungal activity. Similarly, additional roles for PAF in Pe. chrysogenum including conidiation and autophagy have been proposed [61,62].
Efe-AfpA expressed in Pe. chrysogenum was tested on creeping bentgrass plants inoculated with the dollar spot pathogen and was shown to be effective in reducing the severity of disease symptoms. Sapkota et al. [26] reviewed the economic importance of dollar spot disease on both cool-season and warm-season turfgrass species. Control of dollar spot disease on creeping bentgrass is a major problem for golf course managers and currently relies heavily on fungicide applications. Additionally, the dollar spot pathogen has developed reduced sensitivity or increased tolerance to many fungicide classes. Ongoing efforts to address this problem have focused on breeding tolerant cultivars and on developing and implementing improved best management protocols. The results presented here offer a potential additional/complementary approach for the management of dollar spot disease, the application of purified Efe-AfpA for use as an antifungal agent against this economically important plant pathogen.
The direct comparison of bacterial and eukaryotic expression systems reported here revealed the superiority of the Pe. chrysogenum system for producing Efe-AfpA, based on the quantity of protein produced and the ease of purification. However, the E. coli system was also effective at producing active Efe-AfpA. Bacterial expression systems are often used for eukaryotic proteins because of their ease of purification through affinity tags. Attempts at expression of antifungal proteins in E. coli have rarely been reported. An attempt at producing functional PAF from P. chrysogenum in bacteria was reported as unsuccessful, yielding only inactive protein, which was attributed to improper folding of the protein [63]. Similarly, expression of the antifungal protein from Monascus pilosus in E. coli resulted in a protein with 100-fold less activity than the native protein [64].
A factor that is likely important in expression of active Efe-AfpA in bacteria is the correct folding of the recombinant protein. Class A antifungal proteins, such as Efe-AfpA, all have six conserved cysteine residues involved in intramolecular disulfide bonds that likely contribute to the compact structure and stability of the proteins. The cysteine pairing in PAF from P. chrysogenum was determined by mass spectrometry of disulfide-bonded peptides [33]. The structure of PAF that is maintained by proper disulfide bonding is critical for the antifungal activity of the protein. The presence of glutathione, which resulted in a reduction in disulfide bonds, caused PAF to be maintained in the linear form and was inactive [33]. Production of disulfide-bonded proteins in E. coli has proved to be problematic because of the reducing environment of the cytoplasm. Here, Efe-AfpA was expressed in SHuffle cells, which are from an E. coli strain that has been engineered to constitutively express disulfide bond isomerase (DsbC) in the cytoplasm, which corrects disulfide bonding in incorrectly oxidized proteins, and in which thioredoxin reductase and glutathione reductase are deleted [45]. SHuffle cells were used previously to successfully produce functional small cysteine-rich effector proteins from fungal plant pathogens [65].
Another factor likely to be important in the production of active Efe-AfpA in bacteria is the use of the vector pETite, which adds a 6XHis-Sumo tag to the recombinant protein and allows the protein to be purified by metal affinity chromatography. The 107 amino acid 6XHIS-SUMO tag can then be cleaved precisely at the junction between the tag and the recombinant protein by SUMO protease, thereby leaving no extra amino acids on the target protein. Many bacterial expression systems inherently result in extra amino acids on the protein that often have no effect on protein activity. However, we found that even just six extra amino acids on the Efe-AfpA protein reduced the activity of the protein expressed in yeast [20]. A longer form of AFP from A. giganteus with six additional amino acids at the N-terminus had less activity than the fully mature form [66]. The reported previous attempts at expressing a similar antifungal protein in bacteria used systems that added 11 or 30 amino acids to the protein [63,64], which is common in many bacterial expression systems, and is presumably one reason those proteins had no or reduced activity. Apparently, the antifungal proteins cannot tolerate excessive extra amino acids, which results in loss of activity. Here, the N-terminal modified forms of Efe-AfpA, which added only one or two extra amino acids, were active. The combined use of the pETite vector, which has the precisely cleavable SUMO tag, and the SHuffle cells, which were designed to facilitate disulfide bonding, resulted in active Efe-AfpA. However, higher concentrations of the protein were required for the same level of activity, relative to the proteins expressed in Pi. pastoris or Pe. chrysogenum. Whether the lower activity when expressed in E. coli is due to the modified N-terminal amino acids or to less efficient disulfide bonding is not known. For Efe-AfpA the eukaryotic expression systems were clearly preferable for recovery of high levels of active protein.
The use of antifungal proteins as alternatives or complements to fungicides warrants further exploration. Here, we demonstrated the effectiveness of Efe-AfpA in protecting endophyte-free strong creeping red fescue and creeping bentgrass from developing severe symptoms of the important turfgrass disease dollar spot. Other antifungal proteins have also been reported to be effective in protecting plants from specific fungal pathogens. Direct application of an antifungal protein from Pe. expansum, PeAfpA, was successful in protecting orange fruit from infection by the post-harvest pathogen Pe. digitatum and tomato leaves from infection by Botrytis cinerea [67]. PAF applied to detached tomato leaves was protective against B. cinerea [68]. Gandia et al. [69] compared the effectiveness of three antifungal proteins from Penicillium spp. and NFAP2 from N. fischeri in delaying symptoms of Penicillium postharvest fruit decay. They reported some species-specific reductions in infections. Here, we also observed some species-specific effects. Although PAF and Efe-AfpA are similar proteins, their activity against C. jacksonii was quite different. Additionally, the comparison of PAF and Efe-AfpA in activity against the N. crassa Δgcs mutant revealed an apparent difference in the requirement for membrane glucosylceramide in the target fungus.
A challenge for the commercialization of antifungal proteins as alternatives or complements to fungicides is an efficient large-scale production system for the proteins. The Pe. chrysogenum system [31], as used here for Efe-AfpA, can yield 12 μg L−1 of culture and could be a viable system. Additionally, a plant-based transient expression system using tobacco mosaic virus has been developed and was successful in producing the Pe. digitatum antifungal protein AfpB in Nicotiana benthamiana [70]. Overall, the future development of Efe-AfpA, as well as other antifungal proteins, for use in combatting specific plant pathogenic fungi could provide new approaches to plant disease management and potentially help reduce fungicide inputs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8101097/s1, Table S1: Sequences of oligonucleotide primers used in this study; Figure S1: SDS-PAGE of N-terminal modified Efe-AfpA proteins expressed in E. coli; Figure S2: Effect of Efe-AfpA on C. jacksonii mycelial growth; Figure S3: Effect of PAF on C. jacksonii mycelial growth; Figure S4: Final steps in the fungal sphingolipid biosynthetic pathway; Figure S5: Plate assays of effect of Efe-AfpA and PAF on growth of N. crassa wild type and glucosylceramide mutants; Figure S6: Microscopy of effect of Efe-AfpA and PAF on N. crassa wild type and glucosylceramide pathway mutants; Figure S7: Quantitative comparison of activity of Efe-AfpA and PAF against N. crassa wild type and the glucosylceramide mutants Δgcs and Δdtd.
Author Contributions
Conceptualization, P.A.F., Z.T., B.B.C. and F.C.B.; methodology, P.A.F., Z.T., B.B.C. and F.C.B.; investigation, P.A.F. and Z.T.; data curation, P.A.F.; writing—original draft preparation, P.A.F. and F.C.B.; writing—review and editing, P.A.F., Z.T., B.B.C. and F.C.B.; visualization, P.A.F.; supervision, F.C.B.; project administration, F.C.B.; funding acquisition, B.B.C. and F.C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported with funds provided by the United States Golf Association Davis Program, the Rutgers Center for Turfgrass Science, and the USDA National Institute of Food and Agriculture Hatch project accession number 1024790 through the New Jersey Agricultural Experiment Station, Hatch project NJ12108.
Data Availability Statement
All data supporting the findings of this study are available within the paper and within itsSupplementary Materials published online.
Acknowledgments
We thank Florentine Marx for providing the Penicillium chrysogenum paf knockout and overexpressing isolates and the pSK275paf plasmid. We thank William Beldon for providing the Neurospora crassa conidia.
Conflicts of Interest
Rutgers University has filed a provisional patent concerning the purification and use of Efe-AfpA for fungal pathogen disease control. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Marx, F. Small, basic antifungal proteins secreted from filamentous ascomycetes: A comparative study regarding expression, structure, function and potential application. App. Microbiol. Biotechnol. 2004, 65, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, N.; Marx, F. Antifungal proteins: More than antimicrobials? Fungal Biol. Rev. 2013, 26, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Marx, F.; Haas, H.; Reindl, M.; Stoffler, G.; Lottspeich, F.; Redl, B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum paf gene encoding an abundantly secreted protein with antifungal activity. Gene 1995, 167, 167–171. [Google Scholar] [CrossRef]
- Hegedus, N.; Leiter, E.; Kovacs, B.; Tomori, V.; Kwon, N.-J.; Emri, T.; Marx, F.; Batta, G.; Csernoch, L.; Haas, H. The small molecular mass antifungal protein of Penicillium chrysogenum—A mechanism of action oriented review. J. Basic Microbiol. 2011, 51, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Galgoczy, L.; Varadi, G.; Holzknecht, J.; Kakar, A.; Malanovic, N.; Leber, R.; Koch, J.; Keller, M.A.; Batta, G.; et al. Two small, cysteine-rich and cationic antifungal proteins from Penicillium chrysogenum: A comparative study of PAF and PAFB. Biochim. Biophys. Acta—Biomembr. 2020, 1862, 183246. [Google Scholar] [CrossRef]
- Whendt, S.; Ulbrich, N.; Stahl, U. Molecular cloning, sequence analysis and expression of the gene encoding an antifungal-protein from Aspergillus giganteus. Curr. Genet. 1994, 25, 519–523. [Google Scholar] [CrossRef]
- Meyer, V. A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Appl. Microbiol. Biotechnol. 2008, 78, 17–28. [Google Scholar] [CrossRef]
- Kovacs, L.; Viragh, M.; Tako, M.; Papp, T.; Vagvolgyi, C.; Galgoczy, L. Isolation and characterization of Neosartorya fischeri antifungal protein (NFAP). Peptides 2011, 32, 1724–1731. [Google Scholar] [CrossRef]
- Delgado, J.; Owens, R.A.; Doyle, S.; Asensio, M.A.; Nunez, F. Antifungal proteins from moulds: Analytical tools and potential application to dry-ripened foods. Appl. Microbiol. Biotechnol. 2016, 100, 6991–7000. [Google Scholar] [CrossRef]
- Leiter, E.; Gall, T.; Csernoch, L.; Pocsi, I. Biofungicide utilizations of antifungal proteins of filamentous ascomycetes: Current and foreseeable future developments. BioControl 2017, 62, 125–138. [Google Scholar] [CrossRef]
- Palicz, Z.; Gall, T.; Leiter, E.; Kollar, S.; Kovacs, I.; Miszti-Blasius, K.; Pocsi, I.; Csernoch, L.; Szentesi, P. Application of a low molecular weight antifungal protein from Penicillium chrysogenum (PAF) to treat pulmonary aspergillosis in mice. Emerg. Microbes Infect. 2016, 5, e114. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Culebras, P.V.; Gandia, M.; Boronat, A.; Marcos, J.F.; Manzanares, P. Differential susceptibility of mycotoxin-producing fungi to distinct antifungal proteins (AFPs). Food Microbiol. 2021, 97, 103760. [Google Scholar] [CrossRef] [PubMed]
- Ruemmele, B.A.; Wipff, J.K.; Brilman, L.; Hignight, K.W. Fine-leaved Festuca species. In Turfgrass Biology, Genetics, and Breeding; Cassler, M.D., Duncan, R.R., Eds.; Wiley: Hoboken, NJ, USA, 2003; pp. 129–174. [Google Scholar]
- Braun, R.C.; Patton, A.J.; Watkins, E.; Koch, P.L.; Anderson, N.P.; Bonos, S.A.; Brilman, L.A. Fine fescues: A review of the species, their improvement, production, establishment, and management. Crop Sci. 2020, 60, 1142–1187. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef] [PubMed]
- Caradus, J.R.; Johnson, L.J. Epichloë fungal endophytes—From a biological curiosity in wild grasses to an essential component of resilient high performing ryegrass and fescue pastures. J. Fungi 2020, 6, 322. [Google Scholar] [CrossRef]
- Funk, C.R.; White, R.H.; Breen, J.P. Importance of Acremonium endophytes in turfgrass breeding and management. Agric. Ecosyst. Environ. 1993, 44, 215–232. [Google Scholar] [CrossRef]
- Bonos, S.A.; Wilson, M.M.; Meyer, W.A.; Funk, C.R. Suppression of red thread in fine fescues through endophyte-mediated resistance. Appl. Turfgrass Sci. 2005, 10, 1094. [Google Scholar] [CrossRef]
- Clarke, B.B.; White, J.F., Jr.; Hurley, R.H.; Torres, M.S.; Sun, S.; Huff, D.R. Endophyte-mediated suppression of dollar spot disease in fine fescues. Plant Dis. 2006, 90, 994–998. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, R.; Ambrose, K.V.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae antifungal protein has activity against the plant pathogen Sclerotinia homoeocarpa, the causal agent of dollar spot disease. Sci. Rep. 2017, 7, 5643. [Google Scholar] [CrossRef]
- Heineck, G.C.; Qiu, Y.; Ehlke, N.J.; Watkins, E. The fungal endophyte Epichloë festucae var. lolii plays a limited role in mediating crown rust severity in perennial ryegrass. Crop Sci. 2020, 60, 1090–1104. [Google Scholar]
- Ambrose, K.V.; Belanger, F.C. SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS ONE 2012, 7, e53214. [Google Scholar] [CrossRef]
- Card, S.D.; Bastias, D.A.; Caradus, J.R. Antagonism to plant pathogens by Epichloë fungal endophytes—A review. Plants 2021, 10, 1997. [Google Scholar] [CrossRef]
- Fernando, K.; Reddy, P.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. Metabolic potential of Epichloë endophytes for host grass fungal disease resistance. Microorganisms 2022, 10, 64. [Google Scholar] [CrossRef]
- Salgado-Salazar, C.; Beirn, L.A.; Ismaiel, A.; Boehm, M.J.; Carbone, I.; Putman, A.I.; Tredway, L.P.; Clarke, B.B.; Crouch, J.A. Clarireedia: A new fungal genus comprising four pathogenic species responsible for dollar spot disease of turfgrass. Fungal Biol. 2018, 122, 761–773. [Google Scholar] [CrossRef]
- Sapkota, S.; Catching, K.E.; Raymer, P.L.; Martinez-Espinoza, A.D.; Bahri, B.A. New approaches to an old problem: Dollar spot of turfgrass. Phytopathology 2022, 112, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Walsh, B.; Ikeda, S.S.; Boland, G.J. Biology and management of dollar spot (Sclerotinia homoeocarpa); an important disease of turfgrass. HortScience 1999, 34, 13–21. [Google Scholar] [CrossRef]
- Wang, R.; Luo, S.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae antifungal protein Efe-AfpA is also a possible effector protein required for the interaction of the fungus with its host grass Festuca rubra subsp. rubra. Microorganisms 2021, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Scott, B. Recommendations for gene nomenclature for Epichloë species and related Clavicipitaceae. In Epichloae, Endophytes of Cool Season Grasses: Implications, Utilization and Biology; Young, C.A., Aiken, G.E., McCulley, R.L., Strickland, J.R., Schardl, C.L., Eds.; The Samuel Roberts Noble Foundation: Ardmore, OK, USA, 2012; pp. 84–87. [Google Scholar]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Sonderegger, C.; Galgoczy, L.; Garrigues, S.; Fizil, A.; Borics, A.; Manzanares, P.; Hededus, N.; Huber, A.; Marcos, J.F.; Batta, G.; et al. A Penicillium chrysogenum-based expression system for the production of small, cysteine-rich antifungal proteins for structural and functional analyses. Microb. Cell Factories 2016, 15, 192. [Google Scholar] [CrossRef]
- Marx, F.; Binder, U.; Leiter, E.; Pocsi, I. The Penicillium chrysogenum antifungal protein PAF, a promising tool for the development of new antifungal therapies and fungal cell biology studies. Cell. Mol. Life Sci. 2008, 65, 445–454. [Google Scholar] [CrossRef]
- Varadi, G.; Toth, G.; Kele, Z.; Galgoczy, L.; Fizil, A.; Batta, G. Synthesis of PAF, an antifungal protein from P. chrysogenum, by native chemical ligation: Native disulfide pattern and fold obtained upon oxidative refolding. Chemistry 2013, 19, 12684–12692. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.R.; Lyons, E.M.; Jordan, K.S.; Raizada, M.N. Bacterial endophytes from wild and ancient maize are able to suppress the fungal pathogen Sclerotinia homoeocarpa. J. Appl. Microbiol. 2016, 120, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Cicalese, J.; Secks, M.E.; Lam, C.K.; Meyer, W.A.; Murphy, J.A.; Belanger, F.C. Cross species inoculation of Chewings and strong creeping red fescues with fungal endophytes. Crop Sci. 2000, 40, 1485–1489. [Google Scholar] [CrossRef]
- Huber, A.; Oemer, G.; Malanovic, N.; Lohner, K.; Kovacs, L.; Salvenmoser, W.; Zschocke, J.; Keller, M.A.; Marx, F. Membrane sphingolipids regulate the fitness and antifungal protein susceptibility of Neurospora crassa. Front. Microbiol. 2019, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Zauner, S.; Zahringer, U.; Lindner, B.; Warnecke, D.; Sperling, P. Identification and functional characterization of the 2-hydroxy fatty N-acyl-Δ3(E)-desaturase from Fusarium graminearum. J. Biol. Chem. 2008, 283, 36734–36742. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Dunlap, J.C.; Borkovich, K.A.; Henn, M.R.; Turner, G.E.; Sachs, M.S.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W.; et al. Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv. Genet. 2007, 57, 49–96. [Google Scholar]
- Sonderegger, C.; Fizil, A.; Burtscher, L.B.; Hajdu, D.; Munoz, A.; Gaspari, Z.; Read, N.D.; Batta, G.; Marx, F. D19S mutation of the cationic, cysteine-rich protein PAF: Novel insights into its structral dynamics, thermal unfolding and antifungal function. PLoS ONE 2017, 12, e0169920. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ambrose, K.V.; Tian, Z.; Wang, Y.; Smith, J.; Zylstra, G.; Huang, B.; Belanger, F.C. Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci. Rep. 2015, 5, 10939. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.Z.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. Shuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef]
- Binder, U.; Chu, M.; Read, N.D.; Marx, F. The antifungal activity of the Penicillium chrysogenum protein PAF disrupts calcium homeostasis in Neurospora crassa. Eukaryot. Cell 2010, 9, 1374–1382. [Google Scholar] [CrossRef]
- Gaff, D.F.; Okong’O-Ogola, O. The use of non-permeating pigments for testing the survival of cells. J. Exp. Bot. 1971, 22, 756–758. [Google Scholar] [CrossRef]
- Paege, N.; Warnecke, D.; Zauner, S.; Hagen, S.; Rodrigues, A.; Baumann, B.; Thiess, M.; Jung, S.; Meyer, V. Species-specific differences in the susceptibility of fungi to the antifungal protein AFP depend on C-3 saturation of glycosylceramides. MSphere 2019, 4, e00741-19. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Gandia, M.; Marcos, J.F. Occurrence and function of fungal antifungal proteins: A case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 2016, 100, 2243–2256. [Google Scholar] [CrossRef]
- Cheeseman, K.; Ropars, J.; Renault, P.; Dupont, J.; Gouzy, J.; Branca, A.; Abraham, A.-L.; Ceppi, M.; Conseiller, E.; Debuchy, R.; et al. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat. Commun. 2013, 5, 2876. [Google Scholar] [CrossRef]
- Ropars, J.; Rodriguez de la Vega, R.; Lopez-Villavicencio, M.; Gouzy, J.; Sallet, E.; Dumas, E.; Lacoste, S.; Debuchy, R.; Dupont, J.; Branca, A.; et al. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr. Biol. 2015, 25, 2562–2569. [Google Scholar] [CrossRef]
- Campos-Olivas, R.; Bruix, M.; Santoro, J.; Lacadena, J.; del Pozo, A.M.; Gavilanes, J.G.; Rico, M. NMR solution structure of the antifungal protein from Aspergillus giganteus: Evidence for cysteine pairing isomerism. Biochemistry 1995, 34, 3009–3021. [Google Scholar] [CrossRef]
- Skouri-Gargouri, H.; Ali, M.B.; Gargouri, A. Molecular cloning, structural analysis and modeling of the AcAFP antifungal peptide from Aspergillus clavatus. Peptides 2009, 30, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, D.; Huber, A.; Czajlik, A.; Toth, L.; Kele, Z.; Kocsube, S.; Fizil, A.; Marx, F.; Galgloczy, L.; Batta, G. Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP). Int. J. Biol. Macromol. 2019, 129, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Tapper, B.A.; Simpson, W.R.; Johnson, R.D.; Mace, W.; Ram, A.; Lukito, Y.; Dupont, P.-Y.; Johnson, L.J.; Scott, D.B.; et al. Epichloë hybrida, sp. nov., an emerging model system for investigating fungal allopolyploidy. Mycologia 2017, 109, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Florea, S.; Jaromczyk, J.; Schardl, C.L. Non-transgenic CRISPR-mediated knockout of entire ergot alkaloid gene clusters in slow-growing asexual polyploid fungi. Toxins 2021, 13, 153. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-F.; Liu, J.-S.; Staben, C.; Christensen, M.J.; Latch, G.C.M.; Siegel, M.R.; Schardl, C.L. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc. Natl. Acad. Sci. USA 1994, 91, 2542–2546. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.D.; Craven, K.D.; Leuchtmann, A.; Clement, S.L.; Schardl, C.L. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol. Ecol. 2004, 13, 1455–1467. [Google Scholar] [CrossRef]
- Dinkins, R.D.; Nagabhyru, P.; Graham, M.A.; Boykin, D.; Schardl, C.L. Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala. New Phytol. 2016, 213, 324–337. [Google Scholar] [CrossRef]
- Wang, R.; Clarke, B.B.; Belanger, F.C. Transcriptome analysis of choke stroma and asymptomatic inflorescence tissues reveals changes in gene expression in both Epichloë festucae and its host plant Festuca rubra subsp. rubra. Microorganisms 2019, 7, 567. [Google Scholar] [CrossRef]
- Hegedus, N.; Sigl, C.; Zadra, I.; Pocsi, I.; Marx, F. The paf gene product modulates asexual development in Penicillium chrysogenum. J. Basic Microbiol. 2011, 51, 253–262. [Google Scholar] [CrossRef]
- Kovacs, B.; Hegedus, N.; Balint, M.; Szabo, Z.; Emri, T.; Kiss, G.; Antal, M.; Pocsi, I.; Leiter, E. Penicillium antifungal protein (PAF) is involved in the apoptotic and autophagic processes of the producer Penicillium chrysogenum. Acta Microbiol. Immunol. Hung. 2014, 61, 379–388. [Google Scholar] [CrossRef]
- Marx, F.; Salvenmoser, W.; Kaiserer, L.; Graessle, S.; Weiler-Gorz, R.; Zadra, I.; Oberparleiter, C. Proper folding of the antifungal protein PAF is required for optimal activity. Res. Microbiol. 2005, 156, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.-Y.; Chen, Y.-P.; Yu, M.-C.; Hwang, I.-E.; Wu, D.-Y.; Liaw, L.-L. Characterization and expression of the antifungal protein from Monascus pilosus and its distribution among various Monascus species. J. Biosci. Bioeng. 2016, 122, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nguyen, N.; Breen, S.; Outram, M.A.; Dodds, P.N.; Kobe, B.; Solomon, P.S.; Williams, S.J. Production of small cysteine-rich effector proteins in Escherichia coli for structural and functional studies. Mol. Plant Pathol. 2017, 18, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ruiz, A.; Martinez del Pozo, A.; Lacadena, J.; Mancheno, J.M.; Onaderra, M.; Gavilanes, J.G. Characterization of a natural larger form of the antifungal protein (AFP) from Aspergillus giganteus. Biochim. Biophys. Acta 1997, 1340, 81–87. [Google Scholar] [CrossRef]
- Garrigues, S.; Gandia, M.; Castillo, L.; Coca, M.; Marx, F.; Marcos, J.F.; Manzanares, P. Three antifungal proteins from Penicillium expansum: Different patterns of production an antifungal activity. Front. Microbiol. 2018, 9, 2370. [Google Scholar] [CrossRef]
- Toth, L.; Boros, E.; Poor, P.; Ordog, A.; Kele, Z.; Varadi, G.; Holzknecht, J.; Bratschun-Khan, D.; Nagy, I.; Toth, G.K.; et al. The potential use of the Penicillium chrysogenum antifungal protein PAF, the designed variant PAFopt and its γ-core peptide Pγopt in plant protection. Microb. Biotechnol. 2020, 13, 1403–1414. [Google Scholar] [CrossRef]
- Gandia, M.; Kakar, A.; Giner-Llorca, M.; Holzknecht, J.; Martinez-Culebras, P.; Galgoczy, L.; Marx, F.; Marcos, J.F.; Manzanares, P. Potential of antifungal proteins (AFPs) to control Penicillium postharvest fruit decay. J. Fungi 2021, 7, 449. [Google Scholar] [CrossRef]
- Shi, X.; Cordero, T.; Garrigues, S.; Marcos, J.F.; Daros, J.-A.; Coca, M. Efficient production of antifungal proteins in plants using a new transient expression vector derived from tobacco mosaic virus. Plant Biotechnol. J. 2019, 17, 1069–1080. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).