Abstract
The cynomolgus macaque, Macaca fascicularis, is a non-human primate (NHP) widely used in biomedical research as its genetics, immunology and physiology are similar to those of humans. They may also be a useful model of the intestinal microbiome as their prokaryome resembles that of humans. However, beyond the prokaryome relatively little is known about other constituents of the macaque intestinal microbiome including the mycobiome. Here, we conducted a region-by-region taxonomic survey of the cynomolgus intestinal mycobiota, from duodenum to distal colon, of sixteen captive animals of differing age (from young to old). Using a high-throughput ITS1 amplicon sequencing-based approach, the cynomolgus gut mycobiome was dominated by fungi from the Ascomycota phylum. The budding yeast genus Kazachstania was most abundant, with the thermotolerant species K. pintolopesii highly prevalent, and the predominant species in both the small and large intestines. This is in marked contrast to humans, in which the intestinal mycobiota is characterised by other fungal genera including Candida and Saccharomyces, and Candida albicans. This study provides a comprehensive insight into the fungal communities present within the captive cynomolgus gut, and for the first time identifies K. pintolopesii as a candidate primate gut commensal.
1. Introduction
The cynomolgus macaque (Macaca fascicularis), also known as the long-tailed or crab-eating macaque, is a cercopithecine primate indigenous to mainland Southeast Asia, as well as the maritime islands of Borneo, Java, and Sumatra, and islands of the Philippines. Like its close relative the rhesus macaque (Macaca mulatta), the cynomolgus macaque shares behavioural, immunological, and physiological similarities, as well as a close evolutionary relationship with humans, making this non-human primate (NHP) an important animal model for biomedical research [1]. These similarities appear to extend to the intestinal microbiota, with some bacterial taxa being common to both humans and non-human primates. The phyla Bacteroidetes, Firmicutes and Proteobacteria are prominent members of the prokaryome of both the human [2,3,4], and macaque gastrointestinal tract (GIT) [5,6,7,8]. Furthermore, a recent comparative metagenomic survey has shown that the gastrointestinal (GI) microbiota of cynomolgus macaques is more like that of humans than that of either mice or pigs [6]. Hence, these non-human primates represent a more suitable animal model for studying biological processes such as human ageing, and how age shapes the composition (and function) of the GI microbiota, and how this in turn affects GI physiology and host health [9,10].
Beyond the GI prokaryome however, relatively little is known of other constituents of the cynomolgus macaque GI microbiome and in particular, the fungal microbiome (mycobiome). Whilst typically present in low abundance [11], enteric fungi nevertheless interact with both the prokaryome as well as host cells to alter host immunity, and can exacerbate the severity of several human diseases, including inflammatory bowel disease (IBD) and colorectal cancer [12,13,14,15,16,17,18,19]. A recent study characterising the oral and faecal mycobiomes of wild and captive Thai cynomolgus macaques, represents the first and only such study to date [20], revealing wild macaques have a significantly higher fungal alpha diversity than their captive counterparts. Overall, most fungi in the faecal (and oral) mycobiome, which is a surrogate of the intestinal mycobiome, of these primates belonged to the Ascomycota phylum, with the cynomolgus faecal mycobiota dominated by the budding yeast genus Kazachstania. Thermotolerant members of this genus and those belonging to the K. telluris species complex (incl. K. bovina, K. pintolopesii, K. slooffiae and K. telluris), are often found in the GIT of cows, pigs and rodents [21,22,23,24,25,26].
In the present study, we employed a high-throughput internal transcribed spacer region 1 (ITS1) amplicon sequencing approach, using an established ITS1 primer set [27,28], to conduct a comprehensive region-by-region taxonomic survey of the cynomolgus macaque intestinal mycobiota, from duodenum to distal colon, in a cohort of young, adult, and aged captive animals. Our goal was to gain a better insight into the composition and diversity of the fungal communities populating the GIT of this biomedically important NHP species, investigate how they change with age and identify candidate fungal GIT commensals.
2. Materials and Methods
2.1. Animals
Sixteen clinically healthy cynomolgus macaques were included in the study. The animals ranged from 4 to 20 years in age and were categorized into young (<7 years), adult (8 to 12 years), or aged (13 years or older) (see Table S1). All animals housed and bred at the UKHSA facility are derived from either Mauritian or South East Asia origin. No new animals have been introduced to these colonies since 2004. The colonies are licensed by the UK Home Office to breed, supply and use macaques for scientific research (Establishment license no. XBF9440B0). The breeding colonies are maintained to the highest standard in terms of animal welfare, health status, genetic profile, and behavioural compatibility, compliant with the UK Home Office Code of Practice for the Housing, and Care of Animals Bred, Supplied or Used for Scientific Purposes, 2014. This is achieved through facilities that provide an enriched and complex environment which meets the behavioural needs of the animals. The cynomolgus macaques are held in either harem breeding groups, or single sex, age matched holding groups. Their accommodation is a climate controlled, multiple room, solid floor caging system. Most groups also have access to an external ‘extension’ pen that is not climate controlled and open to the elements. All larger rooms have complex enrichment. Deep litter bedding is provided in the largest of these rooms. Water and a complete primate diet is provided ad lib. Fresh fruits, vegetables and pulses are provided daily as enrichment.
2.2. Sample Collection and DNA Extraction
All animals used in this study were required to be euthanized as part of normal colony management needs and requirements. Identified animals were initially sedated with ketamine hydrochloride at a dose of 10 mg/kg before exsanguination and euthanasia via intracardial injection with sodium pentobarbital at a dose of 80 mg/kg for elderly and 120–160 mg/kg for younger NHPs. All procedures were conducted under the authority and in compliance with a UK Homes Office project license. Luminal contents were collected from each GIT region of each animal and immediately frozen prior to transfer on dry ice to the laboratory for storage at −70 °C prior to processing. Total microbial DNA was extracted from ~200 mg of lumen content using the QIAamp PowerFecal Pro DNA kit (QIAGEN) and following the manufacturer’s protocol. In addition, all samples were homogenized using a FastPrep-24 benchtop tissue homogenizer (MP Bio) at 6.0 m/s for 1 min. This step was included to aid fungal cell wall disruption to improve fungal DNA recovery. Extracted DNA was quantified, and quality checked using the Qubit 3.0 fluorometer and associated Qubit dsDNA BR Assay Kit (Invitrogen). DNA samples were stored at −20 °C prior to further analysis.
2.3. ITS1 Amplification, Library Preparation and Sequencing
The fungal ITS1 region was amplified from 100 ng of template DNA by PCR using the ITS1F and ITS2 primer set [29,30], with each primer modified at the 5′ end to include an Illumina adapter tail using the following amplification conditions: 94 °C for 5 min; 35 cycles of 92 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; and a final extension of 72 °C for 5 min. Amplification reactions were set up in duplicate for each faecal DNA sample, and positive and negative controls were also included in each PCR run (see Section 2.5). Following ITS1 PCR, a 0.7× SPRI purification using KAPA Pure Beads (Roche, Wilmington, MA, USA) was performed and the purified DNA was eluted in 20 µL of EB buffer (10 mM Tris-HCl). In a second PCR, library index primers were added using a Nextera XT Index Kit v2 (Illumina, Cambridge, UK) and amplified using the following conditions: 95 °C for 5 min: 10 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; and a final extension of 72 °C for 5 min. Following PCR, libraries were quantified using the Invitrogen™ Quant-iT dsDNA high sensitivity assay kit (Thermo Fisher, Waltham, MA, USA) and run on a FLUOstar Optima plate reader (BMG Labtech, Aylesbury, UK). Libraries were pooled following quantification in equal quantities. The final pool was SPRI cleaned using 0.7× KAPA Pure Beads, quantified on a Qubit 3.0 fluorometer and run on a High Sensitivity D1000 ScreenTape (Agilent Inc, Santa Clara, CA, USA) using the Agilent Tapestation 4200 to calculate the final library pool molarity. The pool was then run at a final concentration of 8 pM, on an Illumina MiSeq instrument using the MiSeq® v3 (2× 300 bp) Kit (Illumina) at at the Quadram Institute Bioscience, Norwich. The raw data were analysed using MiSeq reporter. A mean sequence depth of 123,710 reads/sample was achieved; samples with fewer than 10,000 filtered sequences were excluded from further analysis (see Table S2).
2.4. Mycobiome Characterization
Illumina MiSeq reads were analysed using the automated pipeline Dadaist2, a dedicated workflow for ITS profiling [31]. The quality profile of the raw reads (in FASTQ format) was assessed using SeqFu 1.9.3 [32], followed by primer removal using Cutadapt 3.5 [33] and quality filtering via Fastp 0.20.0 [34]. Locus-specific primers and conserved flanking regions were removed using ITSxpress [35]. The identification of representative sequences was performed using DADA2 [36], to produce a set of amplicon sequence variants (ASVs), and their taxonomic assignment was determined using the UNITE Fungal ITS database (release 8.3) [37]. The multiple alignment of the representative sequences was performed using ClustalO [38] and the guide tree was produced using FastTree [39]. Data normalization and diversity were produced using the Rhea scripts [40]. The output feature table, taxonomic classification, phylogeny and metafiles were exported and further analysed using PhyloSeq [41], MicrobiomeAnalyst [42], and the built-in plotting provided by Dadaist2 (via MultiQC [43]).
The raw Illumina ITS1 sequence data produced by the present study have been deposited at the European Nucleotide Archive (EBI), under the Project accession number PRJEB54860. Metadata and supporting scripts are available from the GitHub repository https://github.com/quadram-institute-bioscience/nhp-gut (22 July 2022).
2.5. Inclusion of Controls
Controls were included at each stage of the study. During DNA extraction, an empty bead-beating tube was included and treated the same as tubes containing luminal content and was quantified similarly. This extraction control was included in the initial amplicon PCR to assess that no ITS1 amplicon was produced. Negative (microbial DNA-free H2O) and positive controls (50 ng K. telluris DNA) were included in each PCR run. Libraries were also prepared from the DNA extraction control and from single fungal species DNAs (C. albicans and K. telluris) and were used as pipeline controls in the downstream bioinformatic analyses.
3. Results
3.1. Ascomycetous Fungi Dominate the Captive Cynomolgus Gastrointestinal Tract (GIT) Mycobiome
Fungal community profiling of the luminal contents of the duodenum to distal colon of a cohort of 16 captive macaques (NHP1 to NHP16) was performed using high-throughput internal transcribed spacer 1 (ITS1) amplicon sequencing and a DNA extraction protocol we developed and optimised to characterise the preterm infant GIT mycobiome [44]. The macaques ranged from 4 to 20 years in age and categorized into young (<7 years), adult (8 to 12 years), or aged (≥13 years) (Table S1). A total of 6,927,777 quality trimmed ITS1 reads were obtained from 56 lumen samples, ranging from 14,599 (NHP 1, distal colon) to 248,538 (NHP7, caecum), with a sample average of 123,710 reads (Table S2). Over 700 unique amplicon sequence variants (ASVs) were identified, although only 134 ASVs had a relative abundance of 0.01% or more. Collectively, this set of ASVs accounted for 99% of all ITS1 reads and was selected for subsequent taxonomic analyses to determine the composition and relative abundance of the fungal microbiota (mycobiota) in the separate intestinal sites of each macaque. The number of ASVs detected in each macaque ranged from 12 (NHP9) to 68 (NHP2), with over a quarter of ASVs specific to a single animal (36/134; 26.9%).
One hundred and thirty fungal taxa were classified to phylum level (97%), with the majority (95%) resolved to the genus level, and seventy-six of these (57%) to the species level. Four taxa could not be assigned at the phylum level or below and were classified as ‘Unidentified’ (Table S3). At the phylum level, >99% of fungi belonged to either Ascomycota or Basidiomycota, with a single taxon assigned to the Mucormycota subphylum (Figure 1a), with Ascomycota the predominant phylum accounting for 83% of all fungal reads (Figure 1a). In all, 85 taxa were ascomycetes, 44 were basidiomycetes, and one was identified as a mucormycete (Mucor saturninus) (Table S3).
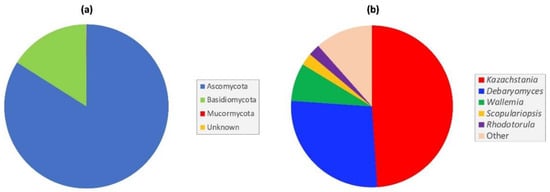
Figure 1.
Most abundant fungi in the captive cynomolgus macaque GIT at (a) phylum, and (b) genus level.
At the genus level, Kazachstania and Debaryomyces were the dominant genera, accounting for 76% of all ITS1 reads (Figure 1b). Both ascomycetous genera were detected, with varying abundance, in all macaque samples, irrespective of age. Overall, Kazachstania was the predominant genus, accounting for 49.0% of all ITS1 reads, and was present in varying abundance in all intestinal samples. Other notable, but less abundant genera included Wallemia (7.6%), Scopulariopsis (2.4%), and Rhodotorula (2.3%) (Figure 1b). Among the ten most abundant genera found in the macaque GIT, seven were yeast genera (Candida, Cystobasidium, Debaryomyces, Filobasidium, Kazachstania, Rhodotorula and Symmetrospora) (Table S4).
3.2. Fungal Community Analysis and Identification of a Core NHP Gut Mycobiome
The 134 ASVs were also used to conduct a community analysis of the fungi present in each NHP age group. Specifically, to identify taxa (ASVs) that were specific to or found predominantly in a particular macaque age group, as well as those shared between two or more age groups. The analysis was restricted to those ASVs detected in at least 50% of animals in a particular age group (51/134; Table S5). A Venn diagram was produced from the resulting dataset (Figure 2). In total, seventeen ASVs were found predominantly in the young macaques, seven in the adults, and one in the aged animals. Interestingly, a core set of thirteen ASVs, representing twelve different fungal taxa, were shared between all three age groups (Figure 2). Using type strain ITS1 sequences, ten taxa were resolved to species level and were identified as Candida albicans, C. parapsilosis, Cutaneotrichosporon cutaneum, Debaryomyces hansenii, Filobasidium uniguttulatum, Kazachstania pintolopesii, Pichia fermentans, Rhodotorula mucilaginosa, Vishniacozyma carnescens and Wallemia muriae (Table S5). Amongst these fungi, only C. albicans, C. parapsilosis, K. pintolopesii and P. fermentans are known to grow at 37 °C (or above), and thus represent candidate cynomolgus gut commensals. Overall, two species, namely D. hansenii and K. pintolopesii, were found in every single macaque, irrespective of animal age (Table S5).
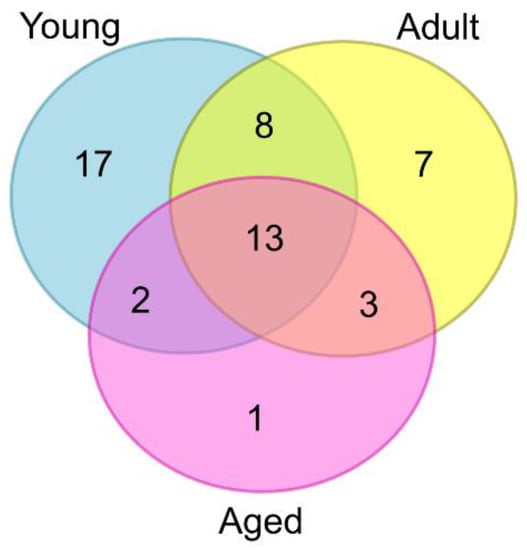
Figure 2.
Venn diagram showing the number of fungal taxa (ASVs) found predominantly in each macaque age group (diagram produced using web-based software at: https://www.vanderpeerlab.org/?q=tools/venn-diagram (16 September 2022).
3.3. Kazachstania pintolopesii and Debaryomyces hansenii Are Prevalent throughout the Cynomolgus GIT
Within the enteric fungal communities, two yeast species, Kazachstania pintolopesii and Debaryomyces hansenii (Candida famata) dominated and were present throughout the macaque GIT. Both species were present in all macaque samples irrespective of age, and with varying abundance (Figure 3). Overall, K. pintolopesii, a thermotolerant yeast species [22] dominated most GIT samples of adult and aged macaques (72.7%), and caecum of one young macaque (NHP9) (Figure 3). In contrast, D. hansenii, a yeast widespread in nature [45], dominated the GIT of two young macaques (NHP11 and NHP12) and most intestinal samples (9/11) of three aged animals (NHP1, NHP7 and NHP10) (Figure 3).

Figure 3.
Prevalence and abundance of K. pintolopesii (Kaz) and D. hansenii (Dhans) in macaque GIT samples.
Although luminal samples could not be obtained from each intestinal region of every animal, samples (40) from nine animals (2 young, 3 adult and 4 aged) were used to conduct a comparative analysis of the most abundant fungi in the small and large intestine. Duodenum-, jejunum- and ileum-derived samples were pooled for taxonomic profiling of the small intestine, while the caecal and colon content samples were likewise pooled for the large intestine analysis. Overall, the two fungal profiles were broadly similar, with K. pintolopesii and D. hansenii being the most abundant species in both intestinal regions (Figure 4). Two other fungi present in both intestinal regions, but in lower abundance, were the basidiomycetes Wallemia muriae and Rhodotorula mucilaginosa. Notable differences between the two intestinal regions included the presence of Scopulariopis brevicaulis, a soil saprotroph, in the small intestine (Figure 4a), and presence of an Aspergillus piperis-like species in the large intestine (Figure 4b).
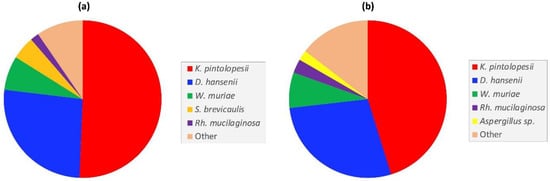
Figure 4.
Comparison of the most abundant fungal taxa in the captive macaque (a) small- and, (b) large intestines.
The caecum provided the most samples (13/16 macaques) (Table S2), enabling a detailed comparative characterization of the fungal communities of this section of the macaque GIT. Three distinct profiles (C1–3) were identified based upon the presence and relative abundance of D. hansenii and K. pintolopesii (Figure 5). In the majority (C1; 7/13 macaques), K. pintolopesii was the predominant species. The second commonest (C2; 5/13) was D. hansenii, with a third profile (C3) characterized by the presence of basidiomycetous taxa (e.g., Cystobasidium pallidum and Rh. mucilaginosa). The C3 profile was atypical and restricted to a single adult female (NHP2) (Figure 5). In this animal, this species profile was caecum-specific, and was not replicated in either of the other two intestinal sites analysed, D. hansenii being predominant in the duodenum and K. pintolopesii in the ileum (Figure 3).
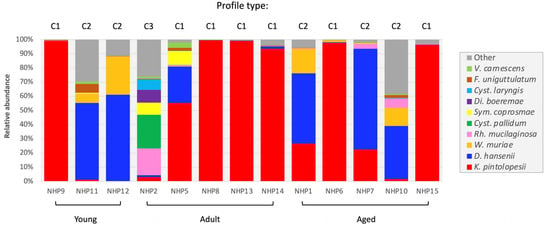
Figure 5.
Comparison of the caecal mycobiome of 13 captive macaques of differing age (young, adult or aged).
3.4. Prevalence of Human-Associated Fungi
Candida such as C. albicans and C. parapsilosis are frequent members of the human GIT mycobiota [44,46,47,48]. In our captive macaque cohort this genus accounted for <1% of all fungal reads (Table S4). A total of eight Candida species were identified, including C. albicans, C. parapsilosis and C. tropicalis, all of which are fungal pathobionts [49]. In contrast, C. anglica, C. freidrichii, C. oleophila, C. saitoana and C. sake were considered GI transients based on their inability to grow above 30 °C [49].
Candida parapsilosis was the most prevalent Candida sp. and except for NHP8 was detected in all animals. C. albicans, while less prevalent (12/16) was present in all age groups (Figure 6). For those animals for which multiple intestinal samples were available (Table S6) these two Candida were not restricted to one region of the GIT (e.g., C. albicans in NHP14; C. parapsilosis in NHP12) (Figure 6). In contrast, C. tropicalis was detected in only 4 GIT samples of three aged animals (NHP7, NHP15 and NHP16; Figure 6).
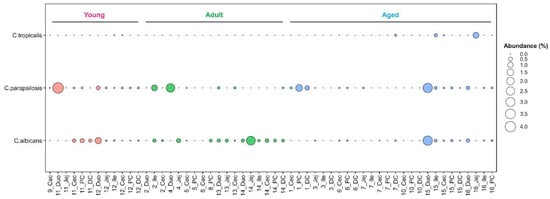
Figure 6.
Prevalence and abundance of Candida pathobionts in the captive macaque GIT.
Saccharomyces cerevisiae frequently found in the human GIT [47], acquired via diet rather than by vertical transmission [50], accounted for less than 0.2% of all ITS1 reads in NHP samples. This species was detected in varying abundance (0.02 to 4.2%; Table S6) in only seven macaques; two young (NHP9 and NHP12), two adult (NHP2 and NHP13) and three aged (NHP7, NHP10 and NHP15). Like Candida spp., S. cerevisiae was not restricted to one GI region and in NHP12 was detected in all three small intestinal sites, as well as the caecum.
Debaryomyces hansenii, a food-borne (dairy) yeast commonly detected in the human GIT [45,47] was the most prevalent and abundant human-associated fungus identified in this study. This yeast was detected in all macaques and was present in moderate to high abundance in five macaques; two young (NHP11 and NHP12) and three aged (NHP1, NHP7 and NHP 10) (Figure 3; Table S6).
4. Discussion
Non-human primates, such as cynomolgus macaques represent important animal models in microbiome research, not least for the unprecedented opportunity they offer for gaining better insights into the biological processes (e.g., ageing) and factors (e.g., diet) that influence and shape the human microbiome. To date, much of this research has concentrated on the NHP prokaryome, with relatively little attention given to the other constituents, including intestinal fungi which are typically present in the GIT in much lower abundance than their bacterial counterparts [11]. To begin addressing this shortfall, we used a high-throughput ITS1 amplicon sequencing approach, that we first developed and used to profile the preterm infant GIT mycobiome [44], to characterize the enteric mycobiota in lumen contents collected from six intestinal sites of a cohort of captive macaques of differing age (i.e., from young to old). The results revealed that the cynomolgus GIT, from duodenum to distal colon, is populated by more than 50 genera, almost exclusively from the Ascomycota and Basidiomycota phyla, with Ascomycota the predominant phylum. This dominance, in both the macaque small- and large intestine, was largely due to the presence of taxa from the budding yeast genera Debaryomyces and Kazachstania, which together accounted for >70% of fungal reads in each intestinal region.
Kazachstania is the predominant genus in our captive macaque cohort, accounting for >72% of all fungal reads, and was found with varying abundance in every animal, and across all age groups. The dominance of this ascomycetous genus was due almost exclusively to one species, K. pintolopesii. While K. telluris, a close relative, was found in a limited number of macaques from each age group, it was typically present in very low abundance (<1.0%). In contrast, K. pintolopesii was highly prevalent throughout the small- and large intestines of each macaque. Furthermore, it was frequently the predominant fungus, and in a third of all GIT samples, K. pintolopesii abundancy exceeds 90%.
Kazachstania is a large and diverse yeast genus comprising more than 40 species [51]. Within the genus, K. pintolopesii is closely related to K. bovina, K. heterogenica, K. slooffiae and K. telluris. Collectively, these five comprise the K. telluris species complex, a phylogenetically distinct group of yeasts characterized by their ability to grow at elevated temperature (i.e., 37 °C) [22]. Some representatives of K. pintolopesii can survive and grow at temperatures as high as 42 °C [22], a physiological trait rare in yeasts. To date, most strains from this thermotolerant species complex have been isolated from the nasal passages and GIT of birds and mammals [21,22,23,24,25]. Prior to this study, the principal hosts of K. pintolopesii appeared to be mice (captive and wild) and rats [21,22,25] with the only member of the K. telluris species complex previously found in NHPs being K. heterogenica, which was limited to a single strain from a young female white-handed gibbon (Hylobates lar) [52].
Debaryomyces hansenii is also highly prevalent in the captive macaque cohort. Distributed throughout the cynomolgus GIT it was the predominant fungus in both the young and elderly animals, albeit to a lesser extent than K. pintolopesii. Despite having a lower optimum growth temperature than K. pintolopesii [22,45], this halotolerant food-borne (dairy) yeast is a frequent member of the human GI mycobiome [44,48,53,54] and can be cultured from human faeces, and is associated with ulcerative colitis (UC), Crohn’s Disease (CD) and colorectal cancer [19,55,56]. D. hansenii produces mycocins that kill C. albicans [57] and it was interesting to note that in three macaques where this yeast was in high abundance, C. albicans was absent. However, given the low abundance (<1.0%) of C. albicans in the macaque GIT it is difficult to draw any significance from these observations.
The human GIT like that of the cynomolgus macaque, is largely dominated by fungi from the Ascomycota and Basidiomycota phyla [20,46,48,53,58,59]. However, despite this similarity, our study has revealed distinct differences at both the genus and species levels between the intestinal mycobiota in humans and cynomolgus macaques. Most notable is the predominance of the Kazachstania genus in the cynomolgus gut, with K. pintolopesii, a fungus rarely found in humans, the dominant species throughout the cynomolgus GIT (this study; [20]). In contrast, Candida and Saccharomyces are prominent genera in the human intestinal mycobiome [46,48,53,58,59], often attributed to the presence of the fungal pathobiont C. albicans and the food-borne yeast S. cerevisiae [46,48,53,58,59]. Despite being rare in the environment [60], C. albicans is a common commensal of both the human GIT and oral cavity, and of the vaginal mycobiome [44,48,53,61,62,63]. Although human-associated fungi were detected in the cynomolgus GIT, most in low abundance (e.g., C. albicans, C. parapsilosis and S. cerevisiae) with the exception of D. hansenii, which was highly prevalent in the macaque cohort, and the predominant intestinal fungus in some of the young and aged animals. In addition to its use as a dairy yeast [45], D. hansenii is ubiquitous in nature, and frequently isolated from soil [45,64]. Thus, the presence of this ascomycete in the cynomolgus GIT may be of environmental origin but not from the primate diet. In contrast, the presence of P. fermentans, which was detected in many of the macaques, albeit in low abundance, is most likely due to diet. This yeast is frequently found in food and fruit juices [65], and so was most likely acquired from the daily supplement of fresh fruits and vegetables given to the animals. Two basidiomycetous yeasts that were also frequently detected were Cutaneotrichosporon cutaneum and Filobasidium uniguttulatum. Cut. cutaneum, like other members of this genus, is a common colonizer of animal skin [66], and its presence and prevalence could be as the result of skin-oral contact (i.e., grooming) within the colony. Indeed, in Thai cynomolgus macaque, Sawaswong and colleagues found Cutaneotrichosporon to be the prominent fungal genus of the oral microbiome of captive macaques [20], providing further support for social grooming as an additional route of fungal acquisition and transmission between primates. F. uniguttulatum, like Cut. cutaneum, is unable to grow at elevated temperature (i.e., 37 °C), and although its natural habitat remains unknown, it has been isolated previously from animal bedding [67]. Thus, given that the macaques are provided with deep litter bedding this may explain why this basidiomycete was detected in many of the animals. In addition to diet and environment, other factors that may contribute to help shape the enteric fungal communities in cynomolgus macaques and humans, include differing GI physiology and normal core body temperature (i.e., humans, 37.0 °C; macaques, 37.0 to 39.5 °C) [68].
The persistence of K. pintolopesii throughout the GIT in young, adult as well as elderly cynomolgus macaques coupled with an innate ability to grow at and above 37 °C [21], suggests that K. pintolopesii represents a plausible primate GI commensal. If proven then this raises the question as to what role it performs in the cynomolgus GIT. Insights into its potential role(s) may come from K. slooffiae, a close relative, and the predominant fungus in the post-weaning porcine gut [21,23,24,69]. This member of the K. telluris species complex [21,22] provides amino acids as an energy source for microbial and piglet growth and is an important source of health promoting micronutrients including vitamin C and formic acid [23,26,69]. Furthermore, a strong (positive) correlation has been identified between K. slooffiae and beneficial intestinal bacteria, including Lactobacillus and Prevotella [24]. In the human GIT, C. albicans can interact directly with Lactobacillus spp. [70,71], leading to the proposal that K. slooffiae may behave similarly to commensal Candida spp. in humans [24]. Given the paucity of human-associated Candida (e.g., C. albicans and C. parapsilosis) in these macaques, coupled with the prevalence and abundance of Prevotella and Lactobacillus spp., including L. acidophilus and L. reuteri, in the cynomolgus GIT [6], it is conceivable that K. pintolopesii performs an equivalent role to that of K. slooffiae in pigs. However, this remains to be established in future cynomolgus microbiome studies encompassing both the mycobiome and bacteriome.
Finally, C. albicans is a common member of the normal human microbiome, and in healthy individuals, it can remain a lifelong benign commensal. However, under certain circumstances (e.g., immunosuppression, broad-spectrum antibiotic treatment) it can cause infections ranging from superficial infections of mucosal surfaces to life-threatening systemic candidiasis [62,72,73]. C. albicans outgrowth in the human GIT can also compound pre-existing CD and UC [18,74]. Thus, given the pathobiont nature of C. albicans (in humans), future research is needed to investigate the pathogenic potential of K. pintolopesii in the cynomolgus macaque, and factors that may trigger a transitional shift from harmless commensal to pathogen. This is especially pertinent given that K. pintolopesii causes gastric infections in laboratory mice, which can prove fatal [21]. Moreover, K. heterogenica, another member of the K. telluris species complex and close relative of K. pintolopesii [22], can exacerbate Helicobacter suis-associated gastric infection in Mongolian gerbils [75], and has been linked to a fatal infection in a young female white-handed gibbon, the first documented case of its kind [52].
5. Conclusions
Our study identified a diverse array of fungi throughout the cynomolgus GIT, with Ascomycota and Basidiomycota the dominant phyla. A characteristic feature is the prominence of the ascomycetous yeast K. pintolopesii, a member of the K. telluris species complex, which we propose represents a credible primate intestinal commensal. This study paves the way for further investigations, firstly to confirm that K. pintolopesii is a primate GIT commensal, and if proven then establish what function it performs in the cynomolgus macaque GIT.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8101054/s1, Table S1: Details of the captive cynomolgus macaques included in this study, Table S2: Quality-trimmed fungal ITS1 reads per GIT luminal sample, Table S3: Fungal ASVs with a relative abundance of 0.01% or more, Table S4: Most abundant fungal genera in the captive macaque GIT, Table S5: Fungal community analysis. ASVs (fungal taxa) specific/predominantly found in one or more NHP age groups, Table S6:Prevalence and abundance of human-associated fungi in the captive macaque GIT.
Author Contributions
Conceptualization, S.R.C., S.G.P.F., A.P. and S.A.J.; methodology, S.A.J., A.P., C.P., A.T., S.H., J.D. and D.B.; software, A.T.; validation, S.A.J. and A.T.; formal analysis, A.T. and S.A.J.; resources, S.H., J.D. and S.G.P.F.; data curation, A.T.; writing—original draft preparation, S.A.J.; writing—review and editing, S.R.C. and S.A.J.; supervision, S.R.C.; project administration, S.R.C.; funding acquisition, S.R.C. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC), this research was funded by BBSRC Core Capability Grant BB/CCG1860/1, and BBSRC Institute Strategic Programme Grant Gut Microbes and Health (BB/R012490/1) and constituent project BBS/E/F/000PR10353. Additional support was provided by a BBSRC NRP DTP Studentship Grant BB/T008717/1 (C.P.).
Institutional Review Board Statement
All procedures were conducted under the authority and in compliance with UK Home Office project license number PD28B8ED5 and were approved by Porton Committee for the Animal Welfare and Ethical Review Body.
Data Availability Statement
The raw Illumina ITS1 sequence data used in this study have been deposited at the European Nucleotide Archive (EBI), under the Project accession number PRJEB54860. Metadata and supporting scripts are available from the GitHub repository https://github.com/quadram-institute-bioscience/nhp-gut, accessed on 22 July 2022.
Acknowledgments
The authors would like to thank Sumeet Tiwari from the Quadram Institute Bioscience for his rendering of Figure 5. We thank all staff of the specialized UK Health Security Agency (UKHSA) primate facility at Porton and in particular, Caroline Cruttwell, for animal husbandry, and ensuring animal welfare and for maximizing the use of tissues that would otherwise have been discarded. UKHSA also acknowledges funding from the UK Department of Health for funding for the maintenance and operation of the primate facility. Bioinformatics analyses were performed on CLIMB-BIG-DATA computing infrastructure (funded by the UK’s Medical Research Council through grant MR/T030062/1) and the Norwich BioScience Institute Partnership “Research Computing” team (for High Performance Computing resources).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Carlsson, H.E.; Schapiro, S.J.; Farah, I.; Hau, J. Use of primates in research: A global overview. Am. J. Primatol. 2004, 63, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.J.; Li, R.Q.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, R.; Gundamaraju, R.; Shastri, M.D.; Shukla, S.D.; Kalpurath, K.; Ball, M.; Tristram, S.; Shankar, E.M.; Ahuja, K.; Eri, R. Gut Microbial Changes, Interactions, and Their Implications on Human Lifecycle: An Ageing Perspective. Biomed Res. Int. 2018, 2018, 4178607. [Google Scholar] [CrossRef]
- Yasuda, K.; Oh, K.; Ren, B.Y.; Tickle, T.L.; Franzosa, E.A.; Wachtman, L.M.; Miller, A.D.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J.; et al. Biogeography of the Intestinal Mucosal and Lumenal Microbiome in the Rhesus Macaque. Cell Host Microbe 2015, 17, 385–391. [Google Scholar] [CrossRef]
- Li, X.P.; Liang, S.S.; Xia, Z.K.; Qu, J.; Liu, H.; Liu, C.; Yang, H.M.; Wang, J.; Madsen, L.; Hou, Y.; et al. Establishment of a Macaca fascicularis gut microbiome gene catalog and comparison with the human, pig, and mouse gut microbiomes. Gigascience 2018, 7, giy100. [Google Scholar] [CrossRef]
- Cui, Y.F.; Wang, F.J.; Yu, L.; Ye, H.H.; Yang, G.B. Metagenomic comparison of the rectal microbiota between rhesus macaques (Macaca mulatta) and cynomolgus macaques (Macaca fascicularis). Zool. Res. 2019, 40, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.G.; Yeoh, Y.K.; Hui, M.M.; Wong, P.Y.; Chan, M.C.W.; Ip, M.; Yu, J.; Burk, R.D.; Chan, F.K.L.; Chan, P.K.S. Diversity of macaque microbiota compared to the human counterparts. Sci. Rep. 2018, 8, 15573. [Google Scholar] [CrossRef]
- Didier, E.S.; MacLean, A.G.; Mohan, M.; Didier, P.J.; Lackner, A.A.; Kuroda, M.J. Contributions of Nonhuman Primates to Research on Aging. Vet. Pathol. 2016, 53, 277–290. [Google Scholar] [CrossRef]
- Duan, J.J.; Yin, B.M.; Li, W.; Chai, T.J.; Liang, W.W.; Huang, Y.; Tan, X.M.; Zheng, P.; Wu, J.; Li, Y.F.; et al. Age-related changes in microbial composition and function in cynomolgus macaques. Aging 2019, 11, 12080–12096. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Noverr, M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Lliev, L.D. The mycobiota: Interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014, 14, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.G.; Xie, L.L.; Yang, X.; Miao, H.F.; Lv, N.; Zhang, R.F.; Xiao, X.; Hu, Y.F.; Liu, Y.L.; Wu, N.; et al. Dysbiosis of Fungal Microbiota in the Intestinal Mucosa of Patients with Colorectal Adenomas. Sci. Rep. 2015, 5, 7980. [Google Scholar] [CrossRef]
- Suhr, M.J.; Hallen-Adams, H.E. The human gut mycobiome: Pitfalls and potentials-a mycologist’s perspective. Mycologia 2015, 107, 1057–1073. [Google Scholar] [CrossRef]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. Mbio 2016, 7, e01250-16. [Google Scholar] [CrossRef]
- Iliev, I.D.; Leonardi, I. Fungal dysbiosis: Immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 2017, 17, 635–646. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Jain, U.; Ver Heul, A.M.; Xiong, S.S.; Gregory, M.H.; Demers, E.G.; Kern, J.T.; Lai, C.W.; Muegge, B.D.; Barisas, D.A.G.; Leal-Ekman, J.S.; et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 2021, 371, 1154–1159. [Google Scholar] [CrossRef]
- Sawaswong, V.; Chanchaem, P.; Khamwut, A.; Praianantathavorn, K.; Kemthong, T.; Malaivijitnond, S.; Payungporn, S. Oral-fecal mycobiome in wild and captive cynomolgus macaques (Macaca fascicularis). Fungal Genet. Biol. 2020, 144, 103468. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Ward, J.M.; Brayton, C.; Gorelick, P.; Walsh, T.J. Multigene phylogenetic analysis of pathogenic Candida species in the Kazachstania (Arxiozyma) telluris complex and description of their ascosporic states as Kazachstania bovina sp nov., K-heterogenica sp nov., K-pintolopesii sp nov., and K-slooffiae sp nov. J. Clin. Microbiol. 2005, 43, 101–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vaughan-Martini, A.; Lachance, M.-A.; Kurtzman, C.P. Kazachstania Zubkova (1971). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, pp. 439–470. [Google Scholar]
- Urubschurov, V.; Busing, K.; Freyer, G.; Herlemann, D.P.R.; Souffrant, W.B.; Zeyner, A. New insights into the role of the porcine intestinal yeast, Kazachstania slooffiae, in intestinal environment of weaned piglets. FEMS Microbiol. Ecol. 2017, 93, fiw245. [Google Scholar] [CrossRef] [PubMed]
- Arfken, A.M.; Frey, J.F.; Ramsay, T.G.; Summers, K.L. Yeasts of Burden: Exploring the Mycobiome-Bacteriome of the Piglet GI Tract. Front. Microbiol. 2019, 10, 2286. [Google Scholar] [CrossRef] [PubMed]
- Bendova, B.; Pialek, J.; Dureje, L.; Schmiedova, L.; Cizkova, D.; Martin, J.F.; Kreisinger, J. How being synanthropic affects the gut bacteriome and mycobiome: Comparison of two mouse species with contrasting ecologies. BMC Microbiol. 2020, 20, 194. [Google Scholar] [CrossRef]
- Summers, K.L.; Frey, J.F.; Arfken, A.M. Characterization of Kazachstania slooffiae, a Proposed Commensal in the Porcine Gut. J. Fungi 2021, 7, 146. [Google Scholar] [CrossRef]
- Huseyin, C.E.; Rubio, R.C.; O’Sullivan, O.; Cotter, P.D.; Scanlan, P.D. The Fungal Frontier: A Comparative Analysis of Methods Used in the Study of the Human Gut Mycobiome. Front. Microbiol. 2017, 8, 1432. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.L.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Ansorge, R.; Birolo, G.; James, S.A.; Telatin, A. Dadaist2: A Toolkit to Automate and Simplify Statistical Analysis and Plotting of Metabarcoding Experiments. Int. J. Mol. Sci. 2021, 22, 5309. [Google Scholar] [CrossRef]
- Telatin, A.; Fariselli, P.; Birolo, G. SeqFu: A Suite of Utilities for the Robust and Reproducible Manipulation of Sequence Files. Bioengineering 2021, 8, 59. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glockner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. The Clustal Omega Multiple Alignment Package. In Multiple Sequence Alignment: Methods and Protocols; Katoh, K.W., Walker, J.M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2021; Volume 2231, pp. 3–16. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Fischer, S.; Kumar, N.; Clavel, T. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. Peerj 2017, 5, e2836. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J.G. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- James, S.A.; Phillips, S.; Telatin, A.; Baker, D.; Ansorge, R.; Clarke, P.; Hall, L.J.; Carding, S.R. Preterm Infants Harbour a Rapidly Changing Mycobiota That Includes Candida Pathobionts. J. Fungi 2020, 6, 273. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Prasad, G.S.; Kurtzman, C.P. Debaryomyces Lodder & Kreger-van Rij (1952). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, pp. 361–372. [Google Scholar]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.Z.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Kachman, S.D.; Kim, J.; Legge, R.M.; Martinez, I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015, 15, 9–17. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Lachance, M.-A.; Boekhout, T.; Scorzetti, G.; Fell, J.W.; Kurtzman, C.P. Candida Berkhout (1923). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, pp. 987–1278. [Google Scholar]
- Bliss, J.M.; Basavegowda, K.P.; Watson, W.J.; Sheikh, A.U.; Ryan, R.M. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr. Infect. Dis. J. 2008, 27, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Ke, T.; Zhai, Y.C.; Yan, Z.L.; Hui, F.L. Kazachstania jinghongensis sp. nov. and Kazachstania menglunensis f.a., sp. nov., two yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 2019, 69, 3623–3628. [Google Scholar] [CrossRef]
- Alvarez-Perez, S.; Mateos, A.; Dominguez, L.; Martinez-Nevado, E.; Rodriguez-Bertos, A.; Blanco, J.L.; Garcia, M.E. First isolation of the anamorph of Kazachstania heterogenica from a fatal infection in a primate host. Med. Mycol. 2012, 50, 193–196. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358. [Google Scholar] [CrossRef]
- Schei, K.; Avershina, E.; Øien, T.; Rudi, K.; Follestad, T.; Salamati, S.; Ødegard, R.A. Early gut mycobiota and mother-offspring transfer. Microbiome 2017, 5, 107. [Google Scholar] [CrossRef]
- Mar, J.S.; LaMere, B.J.; Lin, D.L.; Levan, S.; Nazareth, M.; Mahadevan, U.; Lynch, S.V. Disease Severity and Immune Activity Relate to Distinct Interkingdom Gut Microbiome States in Ethnically Distinct Ulcerative Colitis Patients. Mbio 2016, 7, e01072-16. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Banjara, N.; Nickerson, K.W.; Suhr, M.J.; Hallen-Adams, H.E. Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int. J. Food Microbiol. 2016, 222, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sam, Q.H.; Chang, M.W.; Chai, L.Y.A. The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. Int. J. Mol. Sci. 2017, 18, 330. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Amaretti, A.; Gozzoli, C.; Simone, M.; Righini, L.; Candeliere, F.; Brun, P.; Ardizzoni, A.; Colombari, B.; Paulone, S.; et al. Longitudinal Survey of Fungi in the Human Gut: ITS Profiling, Phenotyping, and Colonization. Front. Microbiol. 2019, 10, 1575. [Google Scholar] [CrossRef]
- Bensasson, D.; Dicks, J.; Ludwig, J.M.; Bond, C.J.; Elliston, A.; Roberts, I.N.; James, S.A. Diverse Lineages of Candida albicans Live on Old Oaks. Genetics 2019, 211, 277–288. [Google Scholar] [CrossRef]
- Sobel, J.D. Vulvovaginal candidosis. Lancet 2007, 369, 1961–1971. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Moran, G.; Coleman, D.; Sullivan, D. An introduction to the medically important Candida species. In Candida and Candidiasis, 2nd ed.; Calderone, R.A., Clancy, C.J., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 11–25. [Google Scholar]
- Breuer, U.; Harms, H. Debaryomyces hansenii—An extremophilic yeast with biotech nological potential. Yeast 2006, 23, 415–437. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Pichia, E.C. Hansen (1904). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, pp. 685–707. [Google Scholar]
- Sugita, T. Trichosporon Behrend (1890). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 2015–2061. [Google Scholar]
- Kwon-Chung, K. Filobasidium Olive (1968). In The Yeasts: A Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 1457–1465. [Google Scholar]
- Laffins, M.M.; Mellal, N.; Almlie, C.L.; Regalia, D.E. Evaluation of Infrared Thermometry in Cynomolgus Macaques (Macaca fascicularis). J. Am. Assoc. Lab. Anim. Sci. 2017, 56, 84–89. [Google Scholar]
- Summers, K.L.; Frey, J.F.; Ramsay, T.G.; Arfken, A.M. The piglet mycobiome during the weaning transition: A pilot study. J. Anim. Sci. 2019, 97, 2889–2900. [Google Scholar] [CrossRef]
- Mason, K.L.; Downward, J.R.E.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and Bacterial Microbiota Interactions in the Cecum during Recolonization following Broad-Spectrum Antibiotic Therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [PubMed]
- Allonsius, C.N.; van den Broek, M.F.L.; De Boeck, I.; Kiekens, S.; Oerlemans, E.F.M.; Kiekens, F.; Foubert, K.; Vandenheuvel, D.; Cos, P.; Delputte, P.; et al. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb. Biotechnol. 2017, 10, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Opin. Microbiol. 2020, 56, 7–15. [Google Scholar] [CrossRef]
- Richard, M.L.; Lamas, B.; Liguori, G.; Hoffmann, T.W.; Sokol, H. Gut Fungal Microbiota: The Yin and Yang of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 656–665. [Google Scholar] [CrossRef]
- Flahou, B.; De Baere, T.; Chiers, K.; Pasmans, F.; Haesebrouck, F.; Ducatelle, R. Gastric Infection with Kazachstania heterogenica Influences the Outcome of a Helicobacter suis Infection in Mongolian Gerbils. Helicobacter 2010, 15, 67–75. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).