Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Isolates
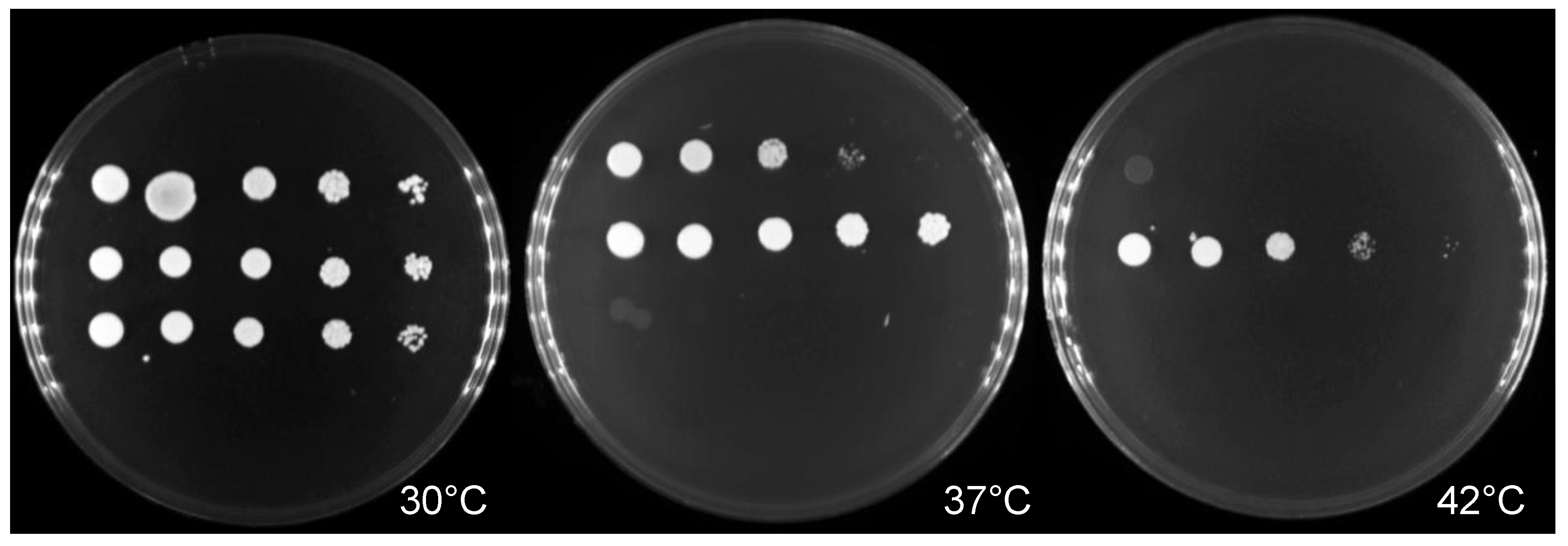
2.2. Thermotolerance and Antifungal Susceptibility Testing
2.3. Zebrafish Maintenance and Infection Experiment
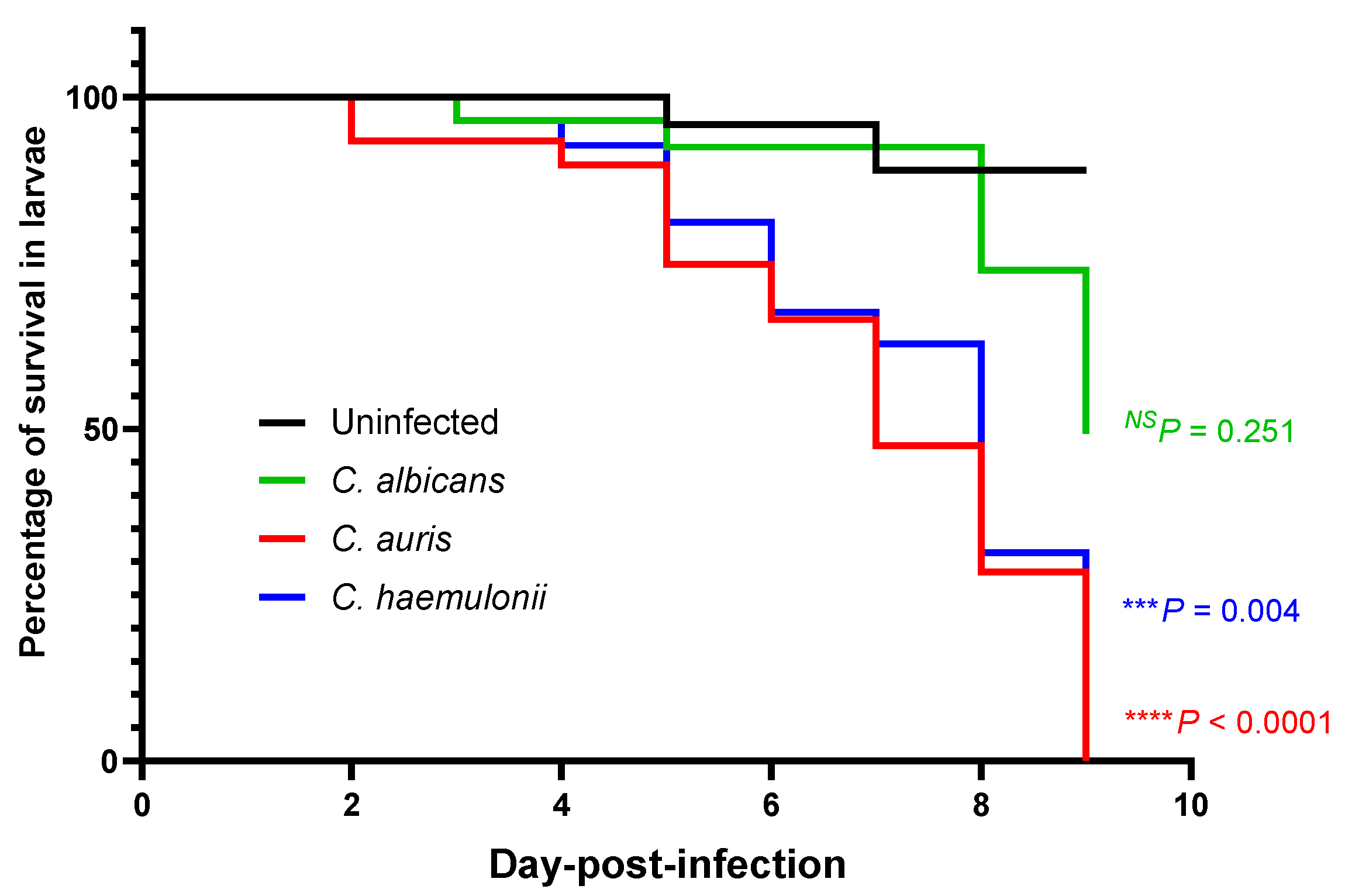
2.4. Survival Rate
2.5. Infectivity Assessment
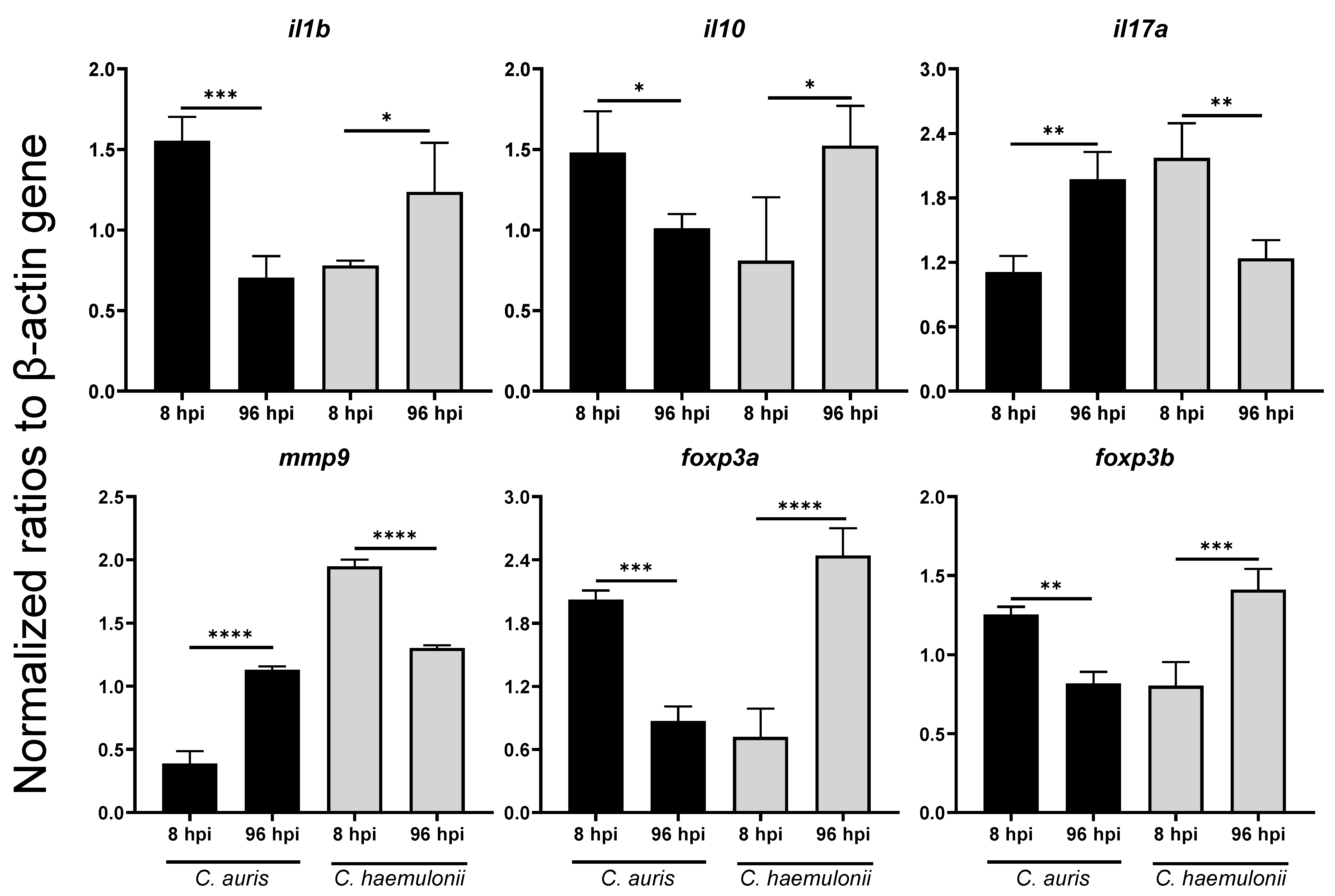
2.6. RNA Extraction and Expression Analysis
2.7. Statistical Analysis
3. Results
3.1. Antifungal Susceptibility and Thermotolerant Testing
3.2. Survival in a Zebrafish Model of a Mucosal Candida Infection at the Swim Bladder
3.3. Fungal Burden in Candidiasis Zebrafish Model
3.4. Dynamics of Immune Response Genes to Emerging Candida in Zebrafish Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [Green Version]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef]
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: A multi-institutional study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Playford, G.; Reboli, A.C.; Rex, J.H.; Sobel, J.D.; Pappas, P.G.; Kullberg, B.J. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: A patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012, 54, 1110–1122. [Google Scholar] [CrossRef]
- Castanheira, M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Antifungal susceptibility patterns of a global collection of fungal isolates: Results of the SENTRY Antifungal Surveillance Program (2013). Diagn. Microbiol. Infect. Dis. 2016, 85, 200–204. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Moet, G.J.; Messer, S.A.; Jones, R.N.; Castanheira, M. Geographic variations in species distribution and echinocandin and azole antifungal resistance rates among Candida bloodstream infection isolates: Report from the SENTRY Antimicrobial Surveillance Program (2008 to 2009). J. Clin. Microbiol. 2011, 49, 396–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Lee, W.G.; Shin, J.H.; Uh, Y.; Kang, M.G.; Kim, S.H.; Park, K.H.; Jang, H.C. First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 2011, 49, 3139–3142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kean, R.; Brown, J.; Gulmez, D.; Ware, A.; Ramage, G. Candida auris: A Decade of Understanding of an Enigmatic Pathogenic Yeast. J. Fungi 2020, 6, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, S.; Alfouzan, W. Candida auris: Epidemiology, Diagnosis, Pathogenesis, Antifungal Susceptibility, and Infection Control Measures to Combat the Spread of Infections in Healthcare Facilities. Microorganisms 2021, 9, 807. [Google Scholar] [CrossRef]
- Lima, S.L.; Francisco, E.C.; de Almeida Júnior, J.N.; Santos, D.; Carlesse, F.; Queiroz-Telles, F.; Melo, A.S.A.; Colombo, A.L. Increasing Prevalence of Multidrug-Resistant Candida haemulonii Species Complex among All Yeast Cultures Collected by a Reference Laboratory over the Past 11 Years. J. Fungi 2020, 6, 110. [Google Scholar] [CrossRef]
- Lavarde, V. Peritonite mycosique a Torulopsis haemulonii. Bull. Soc. Fr. Mycol. Med. 1984, 13, 173–176. [Google Scholar]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gómez-López, A.; Boekhout, T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, S.-Y.; Kuo, Y.-W.; Huang, C.-T.; Hsiue, H.-C.; Hsueh, P.-R. Infections due to Candida haemulonii: Species identification, antifungal susceptibility and outcomes. Int. J. Antimicrob. Agents 2010, 35, 85–88. [Google Scholar] [CrossRef]
- Ramos, L.S.; Figueiredo-Carvalho, M.H.; Barbedo, L.S.; Ziccardi, M.; Chaves, A.L.; Zancopé-Oliveira, R.M.; Pinto, M.R.; Sgarbi, D.B.; Dornelas-Ribeiro, M.; Branquinha, M.H.; et al. Candida haemulonii complex: Species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J. Antimicrob. Chemother. 2015, 70, 111–115. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-ResistantCandida aurison 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Huffnagle, G.B.; Deepe, G.S. Innate and adaptive determinants of host susceptibility to medically important fungi. Curr. Opin. Microbiol. 2003, 6, 344–350. [Google Scholar] [CrossRef]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 4, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.P.; Moyes, D.L. Adaptive immune responses to Candida albicans infection. Virulence 2015, 6, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.J.; Davis, J.M.; Huttenlocher, A.; Kernien, J.F.; Nett, J.E. Emerging Fungal Pathogen Candida auris Evades Neutrophil Attack. mBio 2018, 9, e01403-18. [Google Scholar] [CrossRef] [Green Version]
- Bruno, M.; Kersten, S.; Bain, J.M.; Jaeger, M.; Rosati, D.; Kruppa, M.D.; Lowman, D.W.; Rice, P.J.; Graves, B.; Ma, Z.; et al. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat. Microbiol. 2020, 5, 1516–1531. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.; Ellett, F.; Murray, G.L.; Kostoulias, X.; Cerqueira, G.M.; Schulze, K.E.; Mahamad Maifiah, M.H.; Li, J.; Creek, D.J.; Lieschke, G.J.; et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA 2016, 113, 9599–9604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Merseguel, K.B.; Nishikaku, A.S.; Rodrigues, A.M.; Padovan, A.C.; e Ferreira, R.C.; de Azevedo Melo, A.S.; Briones, M.R.; Colombo, A.L. Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect. Dis. 2015, 15, 57. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.M.; Huang, M.; Botts, M.R.; Hull, C.M.; Huttenlocher, A. A Zebrafish Model of Cryptococcal Infection Reveals Roles for Macrophages, Endothelial Cells, and Neutrophils in the Establishment and Control of Sustained Fungemia. Infect. Immun. 2016, 84, 3047–3062. [Google Scholar] [CrossRef] [Green Version]
- Spaink, H.P.; Cui, C.; Wiweger, M.I.; Jansen, H.J.; Veneman, W.J.; Marin-Juez, R.; de Sonneville, J.; Ordas, A.; Torraca, V.; van der Ent, W.; et al. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 2013, 62, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Hachicho, N.; Reithel, S.; Miltner, A.; Heipieper, H.J.; Küster, E.; Luckenbach, T. Body Mass Parameters, Lipid Profiles and Protein Contents of Zebrafish Embryos and Effects of 2,4-Dinitrophenol Exposure. PLoS ONE 2015, 10, e0134755. [Google Scholar] [CrossRef] [Green Version]
- Bergeron, A.C.; Seman, B.G.; Hammond, J.H.; Archambault, L.S.; Hogan, D.A.; Wheeler, R.T. Candida and Pseudomonas interact to enhance virulence of mucosal infection in transparent zebrafish. Infect. Immun. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef]
- Xu, S.; Webb, S.E.; Lau, T.C.K.; Cheng, S.H. Matrix metalloproteinases (MMPs) mediate leukocyte recruitment during the inflammatory phase of zebrafish heart regeneration. Sci. Rep. 2018, 8, 7199. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Chowdhary, A.; Anil Kumar, V.; Sharma, C.; Prakash, A.; Agarwal, K.; Babu, R.; Dinesh, K.R.; Karim, S.; Singh, S.K.; Hagen, F.; et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Sharma, C.; Duggal, S.; Agarwal, K.; Prakash, A.; Singh, P.K.; Jain, S.; Kathuria, S.; Randhawa, H.S.; Hagen, F.; et al. New clonal strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013, 19, 1670–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef] [Green Version]
- Arendrup, M.C.; Prakash, A.; Meletiadis, J.; Sharma, C.; Chowdhary, A. Comparison of EUCAST and CLSI Reference Microdilution MICs of Eight Antifungal Compounds for Candida auris and Associated Tentative Epidemiological Cutoff Values. Antimicrob. Agents Chemother. 2017, 61, e00485-17. [Google Scholar] [CrossRef] [Green Version]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef] [Green Version]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.U.; Al-Sweih, N.A.; Ahmad, S.; Al-Kazemi, N.; Khan, S.; Joseph, L.; Chandy, R. Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin B, itraconazole, and fluconazole. J. Clin. Microbiol. 2007, 45, 2025–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodero, L.; Cuenca-Estrella, M.; Córdoba, S.; Cahn, P.; Davel, G.; Kaufman, S.; Guelfand, L.; Rodríguez-Tudela, J.L. Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J. Clin. Microbiol. 2002, 40, 2266–2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Ko, K.S.; Moon, S.Y.; Lee, M.S.; Lee, M.Y.; Son, J.S. Catheter-related candidemia caused by Candida haemulonii in a patient in long-term hospital care. J. Korean Med. Sci. 2011, 26, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Reséndiz-Sánchez, J.; Ortiz-Álvarez, J.; Casimiro-Ramos, A.; Hernández-Rodríguez, C.; Villa-Tanaca, L. First report of a catheter-related bloodstream infection by Candida haemulonii in a children’s hospital in Mexico City. Int. J. Infect. Dis. 2020, 92, 123–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandra, R.M.; McCarron, P.; Viganor, L.; Fernandes, M.F.; Kavanagh, K.; McCann, M.; Branquinha, M.H.; Santos, A.L.S.; Howe, O.; Devereux, M. In vivo Activity of Copper(II), Manganese(II), and Silver(I) 1,10-Phenanthroline Chelates Against Candida haemulonii Using the Galleria mellonella Model. Front. Microbiol. 2020, 11, 470. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.E.; Ramirez, L.M.; Dias, L.D.S.; Rivas, L.A.; Ramos, L.S.; Santos, A.L.S.; Taborda, C.P.; Parra-Giraldo, C.M. Pathogenicity Levels of Colombian Strains of Candida auris and Brazilian Strains of Candida haemulonii Species Complex in Both Murine and Galleria mellonella Experimental Models. J. Fungi 2020, 6, 104. [Google Scholar] [CrossRef]
- Borman, A.M.; Szekely, A.; Johnson, E.M. Comparative Pathogenicity of United Kingdom Isolates of the Emerging Pathogen Candida auris and Other Key Pathogenic Candida Species. mSphere 2016, 1, e00189-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Alampalli, S.V.; Nageshan, R.K.; Chettiar, S.T.; Joshi, S.; Tatu, U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom. 2015, 16, 686. [Google Scholar] [CrossRef] [Green Version]
- Fakhim, H.; Vaezi, A.; Dannaoui, E.; Chowdhary, A.; Nasiry, D.; Faeli, L.; Meis, J.F.; Badali, H. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 2018, 61, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.H.; Bhuiyan, M.S.; Shen, H.H.; Cameron, D.R.; Rupasinghe, T.W.T.; Wu, C.M.; Le Brun, A.P.; Kostoulias, X.; Domene, C.; Fulcher, A.J.; et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 3722–3727. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Gong, Q.Y.; Lai, S.S.; Cheng, Z.X.; Chen, Z.G.; Zheng, J.; Peng, B. Phenylalanine enhances innate immune response to clear ceftazidime-resistant Vibrio alginolyticus in Danio rerio. Fish Shellfish. Immunol. 2019, 84, 912–919. [Google Scholar] [CrossRef]
- Chin, V.K.; Foong, K.J.; Maha, A.; Rusliza, B.; Norhafizah, M.; Chong, P.P. Early expression of local cytokines during systemic Candida albicans infection in a murine intravenous challenge model. Biomed. Rep. 2014, 2, 869–874. [Google Scholar] [CrossRef]
- Wagener, J.; Malireddi, R.K.; Lenardon, M.D.; Köberle, M.; Vautier, S.; MacCallum, D.M.; Biedermann, T.; Schaller, M.; Netea, M.G.; Kanneganti, T.D.; et al. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 2014, 10, e1004050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Sutmuller, R.; Hermann, C.; Van der Graaf, C.A.; Van der Meer, J.W.; van Krieken, J.H.; Hartung, T.; Adema, G.; Kullberg, B.J. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J. Immunol. 2004, 172, 3712–3718. [Google Scholar] [CrossRef] [Green Version]
- Zenobia, C.; Hajishengallis, G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontol. 2000 2015, 69, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Elkington, P.T.; O’Kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef]
- McQuibban, G.A.; Gong, J.H.; Wong, J.P.; Wallace, J.L.; Clark-Lewis, I.; Overall, C.M. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood 2002, 100, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273, Table of Contents. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, V.; Viemann, D.; Schmidt, M.; Endres, N.; Ludwig, S.; Leverkus, M.; Roth, J.; Goebeler, M. Candida albicans triggers activation of distinct signaling pathways to establish a proinflammatory gene expression program in primary human endothelial cells. J. Immunol. 2007, 179, 8435–8445. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Cai, J.P.; Luo, Y.L.; Chen, C.; Zhang, S. The Specific Roles of JAK/STAT Signaling Pathway in Sepsis. Inflammation 2015, 38, 1599–1608. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [Green Version]
- Josefowicz, S.Z.; Lu, L.F.; Rudensky, A.Y. Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 2012, 30, 531–564. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Hui, S.P.; Sheng, D.Z.; Nakayama, M.; Kikuchi, K. Zebrafish FOXP3 is required for the maintenance of immune tolerance. Dev. Comp. Immunol. 2017, 73, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Whibley, N.; Maccallum, D.M.; Vickers, M.A.; Zafreen, S.; Waldmann, H.; Hori, S.; Gaffen, S.L.; Gow, N.A.; Barker, R.N.; Hall, A.M. Expansion of Foxp3(+) T-cell populations by Candida albicans enhances both Th17-cell responses and fungal dissemination after intravenous challenge. Eur. J. Immunol. 2014, 44, 1069–1083. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Yang, L.; Chen, Z.G.; Lai, S.S.; Zheng, J.; Peng, B. Exogenous maltose enhances Zebrafish immunity to levofloxacin-resistant Vibrio alginolyticus. Microb. Biotechnol. 2020, 13, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Hsu, P.C.; Jen, C.F.; Chen, I.H.; Wang, C.H.; Chan, H.C.; Tsai, P.W.; Tung, K.C.; Wang, C.H.; Lan, C.Y.; et al. Zebrafish as a model host for Candida albicans infection. Infect Immun 2010, 78, 2512–2521. [Google Scholar] [CrossRef] [Green Version]
- Philip, A.M.; Wang, Y.; Mauro, A.; El-Rass, S.; Marshall, J.C.; Lee, W.L.; Slutsky, A.S.; dosSantos, C.C.; Wen, X.Y. Development of a zebrafish sepsis model for high-throughput drug discovery. Mol. Med. 2017, 23, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.N.; Zhang, J.L.; Huang, Y.; Ren, H.T.; Guan, S.H.; Zeng, Q.H. Dibutyltin depressed immune functions via NF-κB, and JAK/STAT signaling pathways in zebrafish (Danio rerio). Environ. Toxicol. 2018, 33, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Ba, Q.; Wang, Z.; Hao, M.; Li, X.; Hu, P.; Zhang, D.; Zhang, R.; Wang, H. Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. Int. J. Biochem. Cell Biol. 2011, 43, 1218–1227. [Google Scholar] [CrossRef]
- Chybowska, A.D.; Childers, D.S.; Farrer, R.A. Nine Things Genomics Can Tell Us About Candida auris. Front. Genet. 2020, 11, 351. [Google Scholar] [CrossRef]

| Strain | Minimum Inhibitory Concentration (MIC, ug/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AND | MF | CAS | AB | 5FC | PZ | VOR | IZ | FZ | |
| C. albicans ATCC24433 | ≤0.015 | ≤0.008 | 0.015 | 0.12 | ≤0.06 | 0.03 | 0.015 | ≤0.015 | 0.5 |
| C. auris SI-18-CAU-HEM | 0.12 | 0.12 | 0.25 | 4 | 0.25 | ≥8 | ≥8 | ≥16 | ≥256 |
| C. haemulonii SI-21-CH-PLF | 0.12 | 0.25 | 0.12 | 2 | 0.5 | 0.25 | 0.25 | 0.5 | 32 |
| Genes | Early Timepoint (8 hpi) | Late Timepoint (96 hpi) | ||
|---|---|---|---|---|
| C. auris | C. haemulonii | C. auris | C. haemulonii | |
| Proinflammatory and Inflammatory Cytokines | ||||
| tnfa | 3.74 **** | 2.60 * | 2.13 * | 2.06 * |
| ifng | 1.22 NS | 1.27 NS | 0.92 NS | 1.76 NS |
| il1b | 2.43 *** | 1.22 NS | 1.12 NS | 1.96 ** |
| il6 | 0.97 NS | 1.25 NS | 0.63 ** | 1.05 NS |
| il8 | 4.72 **** | 2.37 *** | 2.05 NS | 2.51 ** |
| il10 | 2.47 ** | 1.35 NS | 1.33 NS | 2.00 ** |
| il17a | 1.18 NS | 2.31 *** | 1.25 NS | 0.78 NS |
| Leukocyte Activities | ||||
| inos | 1.07 NS | 0.93 NS | 0.96 NS | 0.85 NS |
| mpx | 0.98 NS | 0.83 NS | 0.42**** | 1.04 NS |
| Matrix Metalloproteinases | ||||
| mmp9 | 0.76 NS | 3.82 **** | 2.09 **** | 2.41 **** |
| mmp13 | 1.62 * | 1.72 * | 1.74 * | 1.88 ** |
| Inflammatory Regulators | ||||
| myd88 | 1.54 NS | 1.06 NS | 0.92 NS | 1.05 NS |
| nfkb | 1.02 NS | 1.56 ** | 1.17 NS | 2.11 **** |
| jak2 | 3.04 **** | 0.46 * | 0.93 NS | 0.52 NS |
| stat3 | 0.87 NS | 1.17 NS | 1.23 NS | 1.53 * |
| Regulatory T-Cells | ||||
| foxp3a | 3.16 **** | 1.12 NS | 0.33 **** | 0.92 NS |
| foxp3b | 1.49 ** | 0.96 NS | 0.56 **** | 0.97 NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pharkjaksu, S.; Boonmee, N.; Mitrpant, C.; Ngamskulrungroj, P. Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains. J. Fungi 2021, 7, 725. https://doi.org/10.3390/jof7090725
Pharkjaksu S, Boonmee N, Mitrpant C, Ngamskulrungroj P. Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains. Journal of Fungi. 2021; 7(9):725. https://doi.org/10.3390/jof7090725
Chicago/Turabian StylePharkjaksu, Sujiraphong, Nawarat Boonmee, Chalermchai Mitrpant, and Popchai Ngamskulrungroj. 2021. "Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains" Journal of Fungi 7, no. 9: 725. https://doi.org/10.3390/jof7090725
APA StylePharkjaksu, S., Boonmee, N., Mitrpant, C., & Ngamskulrungroj, P. (2021). Immunopathogenesis of Emerging Candida auris and Candida haemulonii Strains. Journal of Fungi, 7(9), 725. https://doi.org/10.3390/jof7090725






