Isolation and Characterization of Endomycorrhizal Fungi Associated with Growth Promotion of Blueberry Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Bacterial Strain and Plasmid Vector
2.3. Isolation and Identification of Blueberry Endomycorrhizal Fungi
2.4. Screening of Beneficial Endomycorrhizal Fungi
2.5. Determination of Related Physiological Indexes
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.7. Cloning and Identification of VcPHT1s
2.8. Plant Transformation and Function Analysis
2.9. Statistical Analysis
3. Results
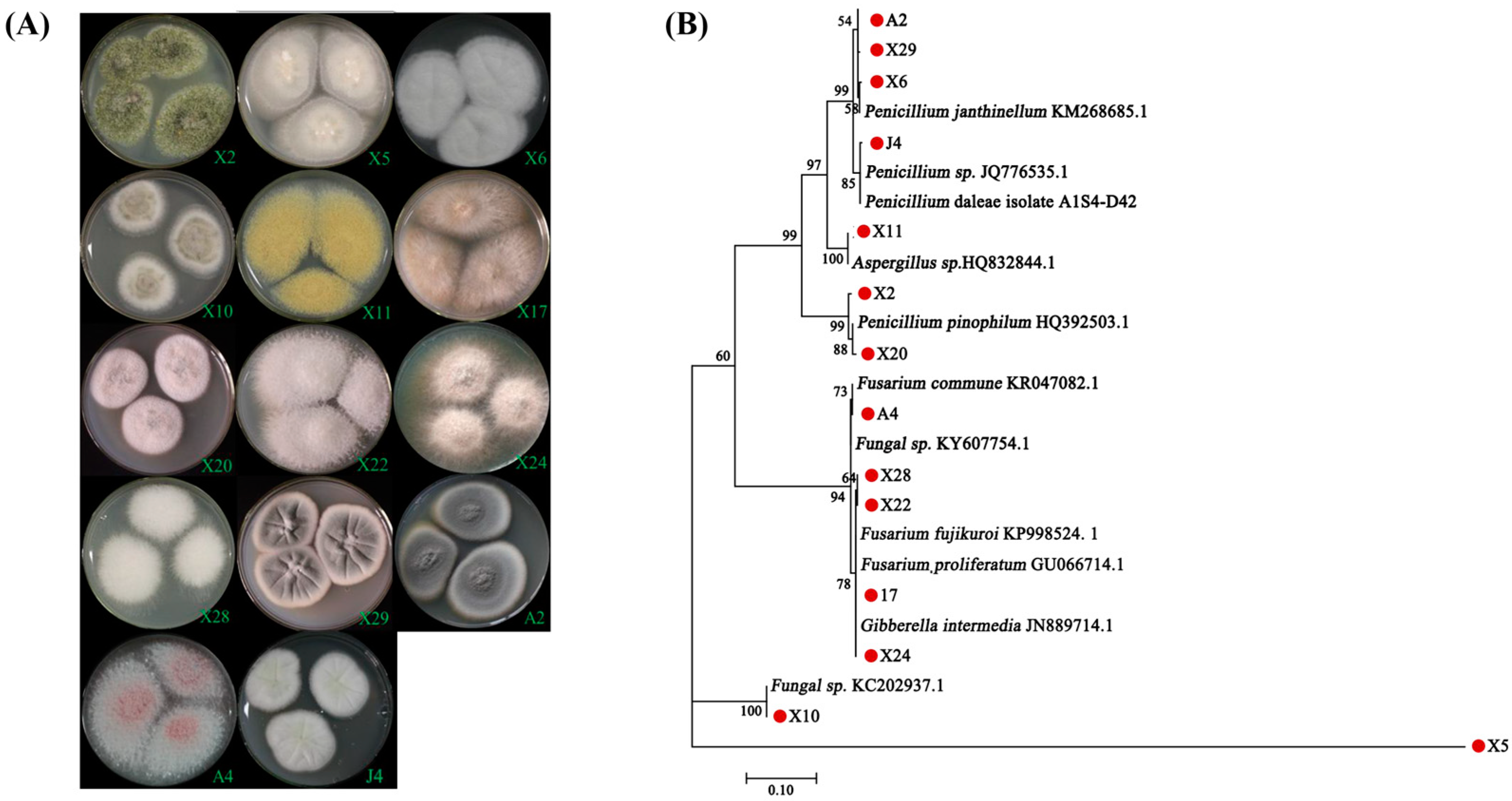
3.1. Isolation and Identification of Endomycorrhizal Fungi from Blueberry Plants
3.2. Inoculation with Beneficial Endomycorrhizal Fungi Significantly Increased the Growth of Blueberry Seedlings
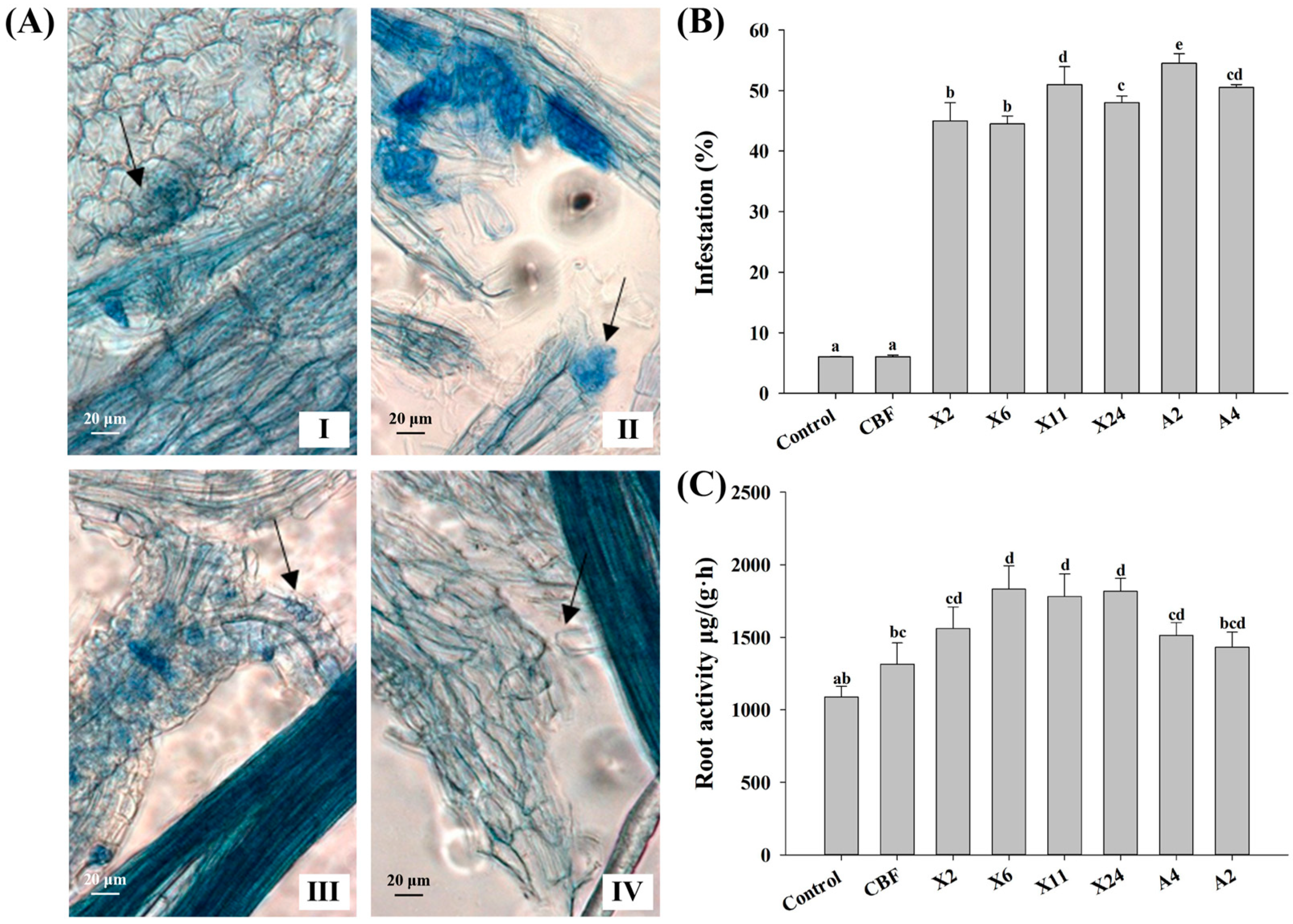
3.3. Infection Rates and Effect on Root Activities of Beneficial Endomycorrhizal Fungi in Blueberries
3.4. Phosphorus and Nitrogen Content of Blueberries after Inoculation with Beneficial Endomycorrhizal Fungi
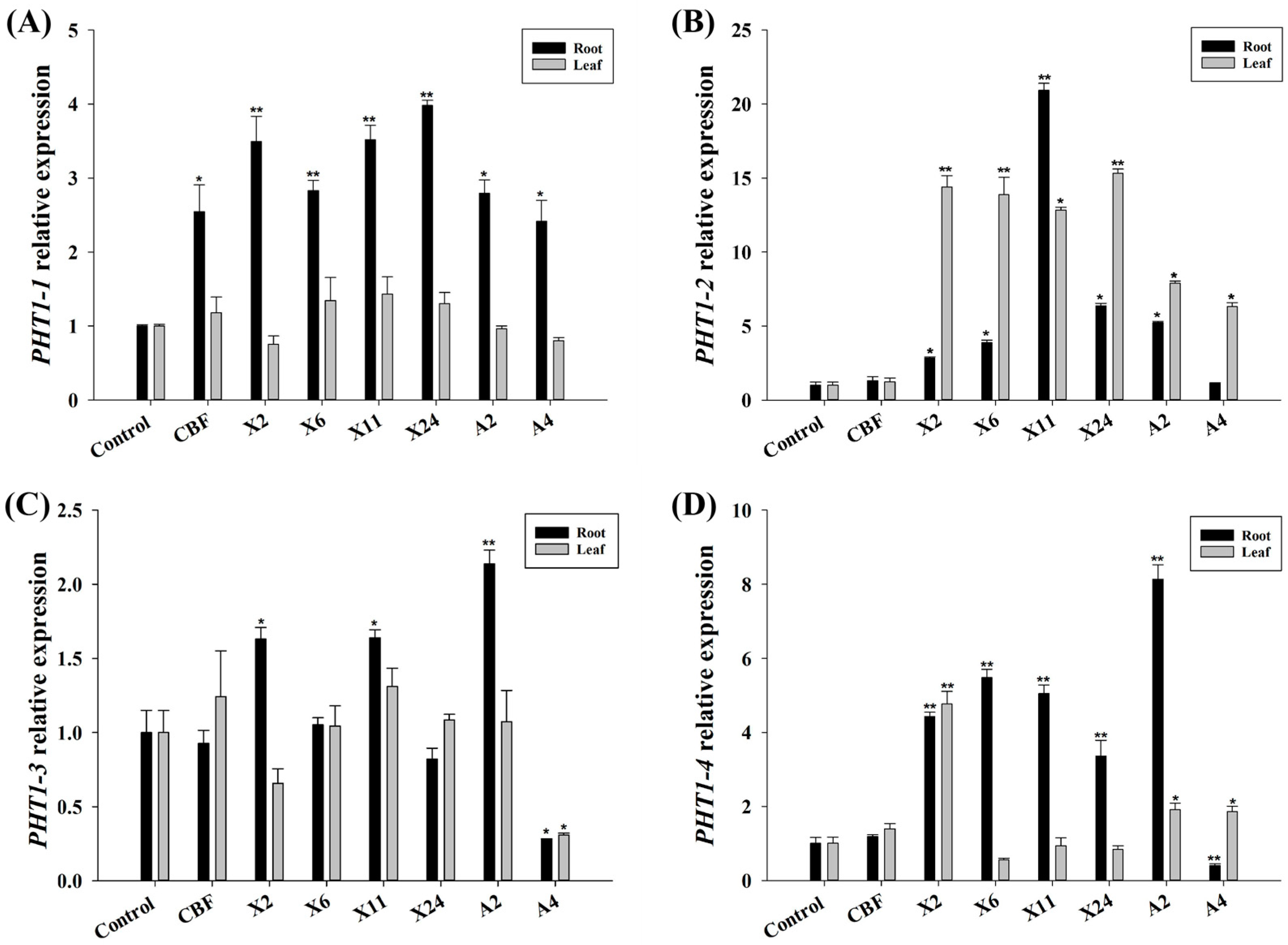
3.5. Impact of Beneficial Endomycorrhizal Fungi on VcPHT1s Expression
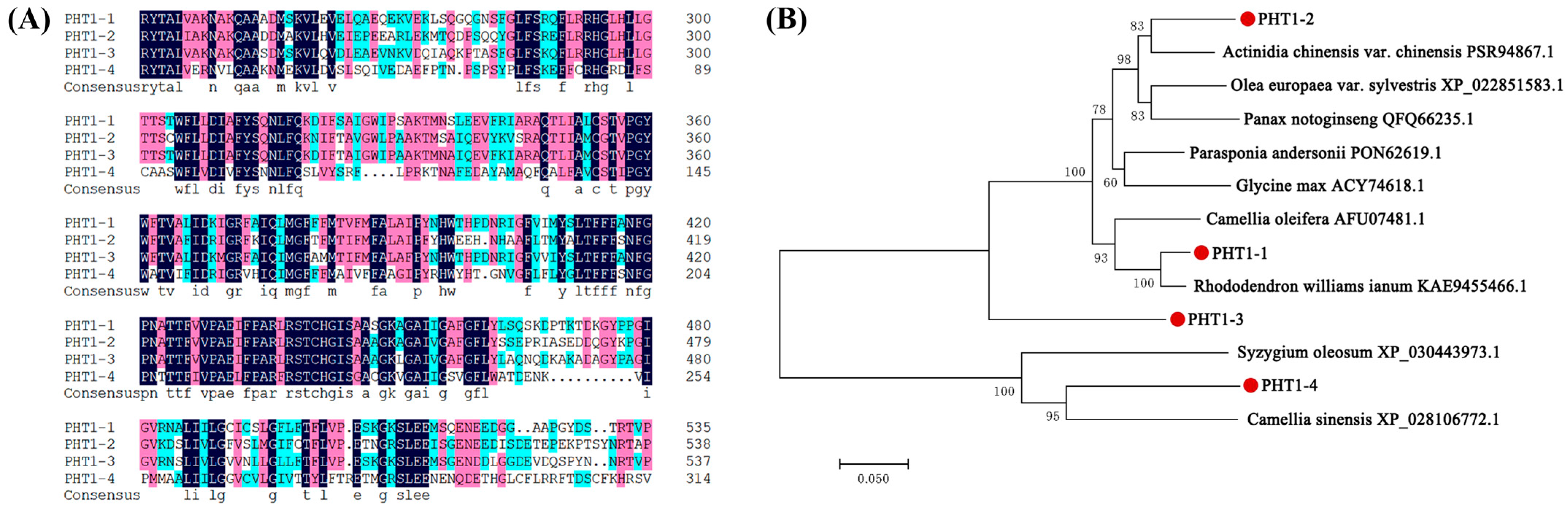
3.6. Isolation and Bioinformatics Analysis of VcPHT1s
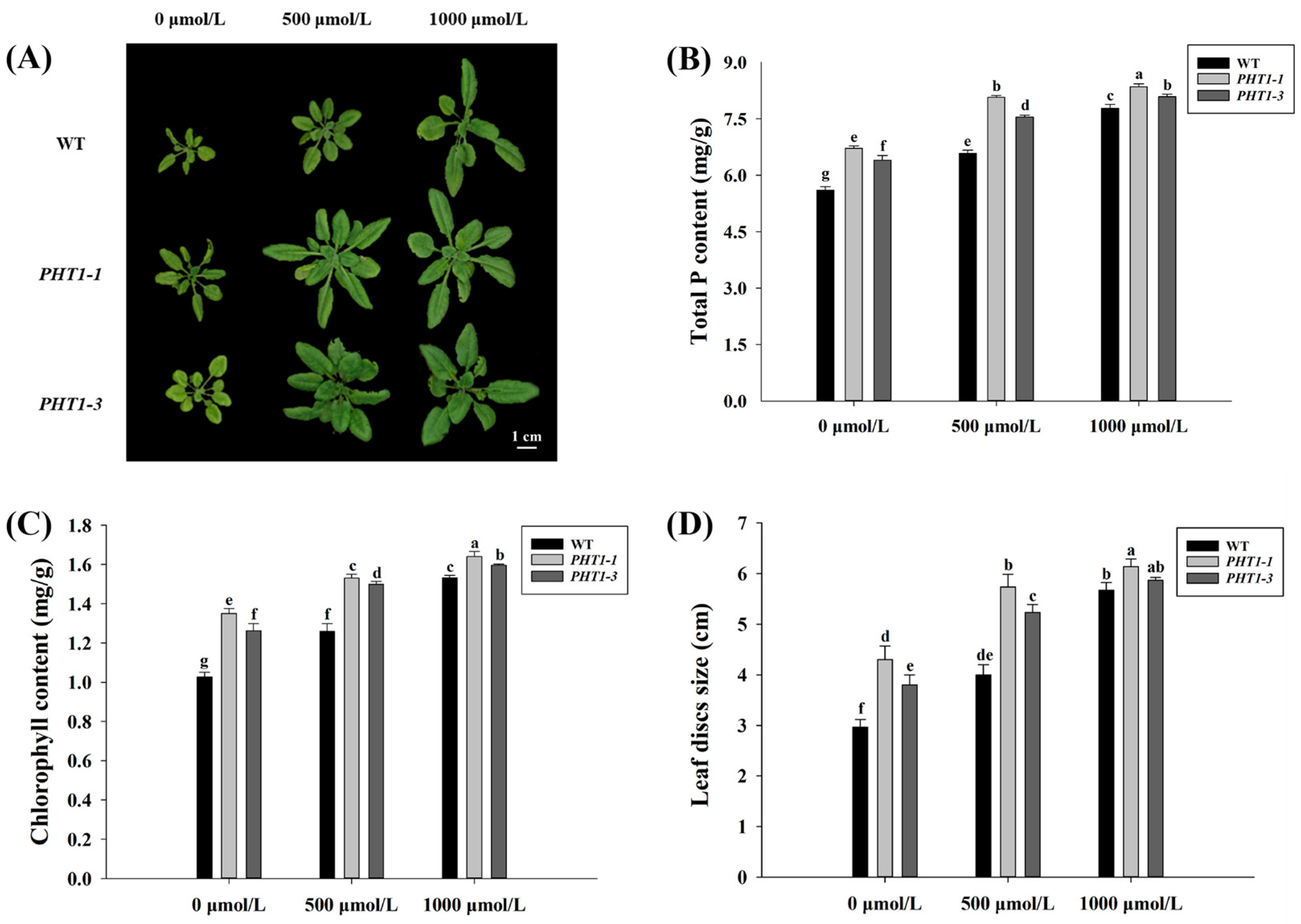
3.7. Expression of VcPHT1s in Transgenic A. thaliana Plants under Different P Concentrations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caspersen, S.; Svensson, B.; Håkansson, T.; Winter, C.; Khalil, S.; Asp, H. Blueberry—Soil interactions from an organic perspective. Sci. Hortic. 2016, 208, 78–91. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Jiang, S.; Meng, J.J.; An, H.S.; Zhang, X.Y. First report of leaf spot caused by Nigrospora oryzae on blueberry in Shanghai, China. Plant Dis. 2019, 103, 2473. [Google Scholar] [CrossRef]
- Garren, R. Riego y Poda en Arandanos. Ser. Carillanca. 1988. Available online: https://biblioteca.inia.cl/handle/123456789/38977 (accessed on 3 July 2020).
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Pumplin, N.; Harrison, M.J. Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 2007, 30, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A.; Jakobsen, I. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 2003, 133, 16–20. [Google Scholar] [CrossRef]
- Okumura, S.; Mitsukawa, N.; Shirano, Y.; Shibata, D. Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 1998, 5, 261–269. [Google Scholar] [CrossRef]
- Poirier, Y.; Bucher, M. Phosphate transport and homeostasis in Arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2002, 1. [Google Scholar] [CrossRef]
- Chiou, T.J.; Liu, H.; Harrison, M.J. The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 2001, 25, 281–293. [Google Scholar] [CrossRef]
- Liu, J.; Versaw, W.K.; Pumplin, N.; Gomez, S.K.; Blaylock, L.A.; Harrison, M.J. Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 2008, 283, 24673–24681. [Google Scholar] [CrossRef]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Nagy, R.; Karandashov, V.; Chague, V.; Kalinkevich, K.; Tamasloukht, M.B.; Xu, G.H.; Jakobsen, I.; Levy, A.A.; Amrhein, N.; Bucher, M. The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 2005, 42, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed]
- Read, D.J. The biology of mycorrhiza in the Ericales. Can. J. Bot. 1983, 61, 985–1004. [Google Scholar] [CrossRef]
- Yuan, J. The Separation, Identification and Inoculation Effect of Mycorrhizal Fungi Isolated from Wild Blueberries. Master’s Thesis, Beijing Forestry University, Beijing, China, 2013. [Google Scholar]
- Yang, X.; Yan, W. Mycorrhizal fungi diversity of Vaccinium uliginosum L. Acta Microbiol. Sin. 2015, 55, 214–219. [Google Scholar] [CrossRef]
- Bizabani, C.; Fontenla, S.; Dames, J.F. Ericoid fungal inoculation of blueberry under commercial production in South Africa. Sci. Hortic. 2016, 209, 173–177. [Google Scholar] [CrossRef]
- Bizabani, C.; Dames, J. Effects of inoculating Lachnum and Cadophora isolates on the growth of Vaccinium corymbosum. Microbiol. Res. 2015, 181, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Scagel, C.F. Inoculation with ericoid mycorrhizal fungi alters fertilizer use of highbush blueberry cultivars. HortScience 2005, 40, 786–794. [Google Scholar] [CrossRef]
- Read, D.J.; Stribley, D.P. Effect of mycorrhizal infection on nitrogen and phosphorus nutrition of ericaceous plants. Nat. New Biol. 1973, 244, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.T.; Gibson, B.R. Ericoid mycorrhizal association: Ability to adapt to a broad range of habitats. Mycologist 2006, 20, 2–9. [Google Scholar] [CrossRef]
- Lang, G.A.; Tao, J. Postharvest performance of Southern highbush blueberry fruit. HortTechnology 1992, 2, 366–370. [Google Scholar] [CrossRef]
- Ballington, J.R.; Mainland, C.M.; Duke, S.D.; Draper, A.D.; Galletta, G.J. ‘O’Neal’southern highbush blueberry. HortScience 1990, 25, 711–712. [Google Scholar] [CrossRef]
- Williamson, J.G.; Phillips, D.A.; Lyrene, P.; Munoz, P.R. Southern Highbush Blueberry Cultivars from the University of Florida. Edis. 2019. Available online: https://edis.ifas.ufl.edu/publication/HS1245 (accessed on 11 July 2021).
- Smith, N.R.; Dawson, V.T. The bacteriostatic action of rose bengal in media used for plate counts of soil fungi. Soil Sci. 1944, 58, 467–472. [Google Scholar] [CrossRef]
- Stewart, C.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 1993, 14, 748–751. [Google Scholar] [PubMed]
- Ward, A.C. Rapid analysis of yeast transformants using colony-PCR. Biotechniques 1992, 13, 350. [Google Scholar] [PubMed]
- Schmidt, C.S.; Lovecká, P.; Mrnka, L.; Vychodilová, A.; Strejček, M.; Fenclová, M.; Demnerová, K. Distinct communities of poplar endophytes on an unpolluted and a risk element-polluted site and their plant growth-promoting potential in vitro. Microb. Ecol. 2018, 75, 955–969. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Gu, J.; Xu, J.; Guo, Y.; Zong, Y.; Chen, W.; Guo, W. Cloning and functional analysis of ZIP transporters in blueberry. Sci. Hortic. 2021, 278, 109871. [Google Scholar] [CrossRef]
- Kladnik, A. Maize Kernels–Fixation in FAA, Embedding, Sectioning and Feulgen Staining. Bio-Protocol 2013, 3, e835. [Google Scholar] [CrossRef]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Biermann, B.; Linderman, R.G. Quantifying vesicular-arbuscular mycorrhizae: A proposed method towards standardization. New Phytol. 1981, 87, 63–67. [Google Scholar] [CrossRef]
- Islam, E.; Yang, X.; Li, T.; Liu, D.; Jin, X.; Meng, F. Effect of Pb toxicity on root morphology, physiology and ultrastructure in the two ecotypes of Elsholtzia argyi. J. Hazard. Mater. 2007, 147, 806–816. [Google Scholar] [CrossRef]
- Makino, A.; Nakano, H.; Mae, T. Responses of ribulose-1, 5-bisphosphate carboxylase, cytochrome f, and sucrose synthesis enzymes in rice leaves to leaf nitrogen and their relationships to photosynthesis. Plant Physiol. 1994, 105, 173–179. [Google Scholar] [CrossRef]
- Chen, P.S.; Toribara, T.T.; Warner, H. Microdetermination of phosphorus. Anal. Chem. 1956, 28, 1756–1758. [Google Scholar] [CrossRef]
- Yu, K.; Ye, M.; Chen, W.; Zhu, K.; Zhang, C.; Guo, W. Methods for RNA isolation from blueberry tissues. J. Zhejiang Norm. Univ. (Nat. Sci.) 2016, 1. [Google Scholar] [CrossRef]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Soza, V.L.; Lindsley, D.; Waalkes, A.; Ramage, E.; Patwardhan, R.P.; Burton, J.N.; Adey, A.; Kumar, A.; Qiu, R.; Shendure, J.; et al. The Rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae). Genome Biol. Evol. 2019, 11, 3353–3371. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 1–19. [Google Scholar] [CrossRef]
- Guo, X.; Yuan, L.; Shakeel, M.; Wan, Y.; Song, Z.; Wang, D. Isolation and identification of endophytic fungi with screening of promotion growth on mycorrhizal fungi in blueberry. Rhizosphere 2021, 100389. [Google Scholar] [CrossRef]
- Monreal, M.; Berch, S.M.; Berbee, M. Molecular diversity of ericoid mycorrhizal fungi. Can. J. Bot. 2000, 77, 1580–1594. [Google Scholar] [CrossRef]
- Scagel, C.F.; Wagner, A.; Winiarski, P. Inoculation with ericoid mycorrhizal fungi alters root colonization and growth in nursery production of blueberry plants from tissue culture and cuttings. Small Fruits Rev. 2005, 4, 113–135. [Google Scholar] [CrossRef]
- Scagel, C.F.; Wagner, A.; Winiarski, P. Frequency and intensity of root colonization by ericoid mycorrhizal fungi in nursery production of blueberry plants. Small Fruits Rev. 2005, 4, 95–112. [Google Scholar] [CrossRef]
- Golldack, J.; Schubert, P.; Tauschke, M.; Schwarzel, H.; Hofflich, G.; Lentzsch, P.; Munzenberger, B. Mycorrhization and plant growth of highbush blueberry (Vaccinium corymbosum L.) on arable land in Germany. In Proceedings of the 3rd International Conference on Mycorrhiza, Adelaide, Australia, 11 July 2001. [Google Scholar]
- Zhang, H.; Xu, N.; Li, X.; Long, J.; Sui, X.; Wu, Y.; Li, J.; Wang, J.; Zhong, H.; Sun, G.Y. Arbuscular mycorrhizal fungi (Glomus mosseae) improves growth, photosynthesis and protects photosystem II in leaves of Lolium perenne L. in cadmium contaminated soil. Front. Plant Sci. 2018, 9, 1156. [Google Scholar] [CrossRef]
- Resendes, M.L.; Bryla, D.R.; Eissenstat, D.M. Early events in the life of apple roots: Variation in root growth rate is linked to mycorrhizal and nonmycorrhizal fungal colonization. Plant Soil 2008, 313, 175–186. [Google Scholar] [CrossRef]
- Li, W.C.; Zhou, J.; Guo, S.Y.; Guo, L.D. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers 2007, 25, 69–80. [Google Scholar]
- Walder, F.; Brulé, D.; Koegel, S.; Wiemken, A.; Boller, T.; Courty, P.E. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef]
- Xie, X.; Huang, W.; Liu, F.; Tang, N.; Liu, Y.; Lin, H.; Zhao, B. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef]
- Grunwald, U.; Guo, W.; Fischer, K.; Isayenkov, S.; Ludwig-Müller, J.; Hause, B.; Küster, H.; Yan, X.; Franken, P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 2009, 229, 1023–1034. [Google Scholar] [CrossRef]
- Tamura, Y.; Kobae, Y.; Mizuno, T.; Hata, S. Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Biosci. Biotechnol. Biochem. 2012, 76, 309–313. [Google Scholar] [CrossRef][Green Version]
- Saia, S.; Rappa, V.; Ruisi, P.; Abenavoli, M.R.; Sunseri, F.; Giambalvo, D.; Frenda, A.S.; Martinelli, F. Soil inoculation with symbiotic microorganisms promotes plant growth and nutrient transporter genes expression in durum wheat. Front. Plant Sci. 2015, 6, 815. [Google Scholar] [CrossRef] [PubMed]
- Bayle, V.; Arrighi, J.F.; Creff, A.; Nespoulous, C.; Vialaret, J.; Rossignol, M.; Gonzalez, E.; Paz-Ares, J.; Nussaume, L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 2011, 23, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Kobae, Y.; Omoto, E.; Tanaka, A.; Banba, M.; Takai, S.; Tamura, Y.; Hirose, A.; Komatsu, K.; Otagaki, S.; et al. The soybean mycorrhiza-inducible phosphate transporter gene, GmPT7, also shows localized expression at the tips of vein endings of senescent leaves. Plant Cell Physiol. 2014, 55, 2102–2111. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Miao, J.; Guo, J.; Shi, Z.; He, M.; Chen, Y.; Zhao, X.; Li, B.; Han, F.; et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann. Bot. 2013, 111, 1139–1153. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Genetic Transformation Analyses and Functional Identification of Tomato Phosphate Transporter SlMPT3;1 Gene in Rice. Ph.D. Thesis, China Agricultural University, Beijing, China, 2019. [Google Scholar]


| Reagent | Concentration (g/L) | Reagent | Concentration (g/L) |
|---|---|---|---|
| K2HPO4 | 0.140 | H3BO3 | 2.86 |
| MgSO4 | 0.490 | MnSO4 | 1.81 |
| Ca(NO3)2·4H2O | 1.180 | ZnSO4 | 0.22 |
| KNO3 | 0.150 | Na2MoO4·2H2O | 0.03 |
| FeSO4·7H2O | 0.011 | CuSO4·5H2O | 0.08 |
| EDTA·2Na | 0.015 |
| Gene Name | Sequence (5′→3′)-Forward | Sequence (5′→3′)-Reverse |
|---|---|---|
| PHT1-1 | CTGGTTCACCGTAGCACTTATC | GAGTCCAGTGGTTGTATGGAATAG |
| PHT1-2 | CCGACTACGTTTGGAGGATTAT | AGTGTATCGGGCAGTTTCAG |
| PHT1-3 | CCCGTTACACTGCCCTTATC | GGCCTCTTCGGGTTCTATTT |
| PHT1-4 | CGCTTTACTAGGAGCCGTAATC | ATAGAGAATCCGCAACCGAAC |
| Gene Name | Sequence (5′→3′)-Forward | Sequence (5′→3′)-Reverse |
|---|---|---|
| PHT1-1 | GGAATTCCCAGCTATGGCCAAAGAACAATTACA | GCTCTAGAGCGAAATAGCGTAAGCATAAAAAAGTA |
| PHT1-2 | GGAATTCCAGAAAGAAGTGTTGAGAAAGGGAGA | CGGGATCCGGATTCGAAAAGTACAGGACCTCTA |
| PHT1-3 | GGAATTCCATGCCTAGAGAACAGTT | CGGGATCCGTTAAACCGGAGCGGTC |
| PHT1-4 | CGGGATCCCGTTCACAAAAGGAAA | GCTCTAGAGCTACAAATTAATAGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, B.; Vancov, T.; Si, H.; Yang, W.; Tong, K.; Chen, W.; Fang, Y. Isolation and Characterization of Endomycorrhizal Fungi Associated with Growth Promotion of Blueberry Plants. J. Fungi 2021, 7, 584. https://doi.org/10.3390/jof7080584
Cai B, Vancov T, Si H, Yang W, Tong K, Chen W, Fang Y. Isolation and Characterization of Endomycorrhizal Fungi Associated with Growth Promotion of Blueberry Plants. Journal of Fungi. 2021; 7(8):584. https://doi.org/10.3390/jof7080584
Chicago/Turabian StyleCai, Binbin, Tony Vancov, Hanqi Si, Wenru Yang, Kunning Tong, Wenrong Chen, and Yunying Fang. 2021. "Isolation and Characterization of Endomycorrhizal Fungi Associated with Growth Promotion of Blueberry Plants" Journal of Fungi 7, no. 8: 584. https://doi.org/10.3390/jof7080584
APA StyleCai, B., Vancov, T., Si, H., Yang, W., Tong, K., Chen, W., & Fang, Y. (2021). Isolation and Characterization of Endomycorrhizal Fungi Associated with Growth Promotion of Blueberry Plants. Journal of Fungi, 7(8), 584. https://doi.org/10.3390/jof7080584








