The Glycosylphosphatidylinositol-Anchored Superoxide Dismutase of Scedosporium apiospermum Protects the Conidia from Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Genomic DNA Extraction
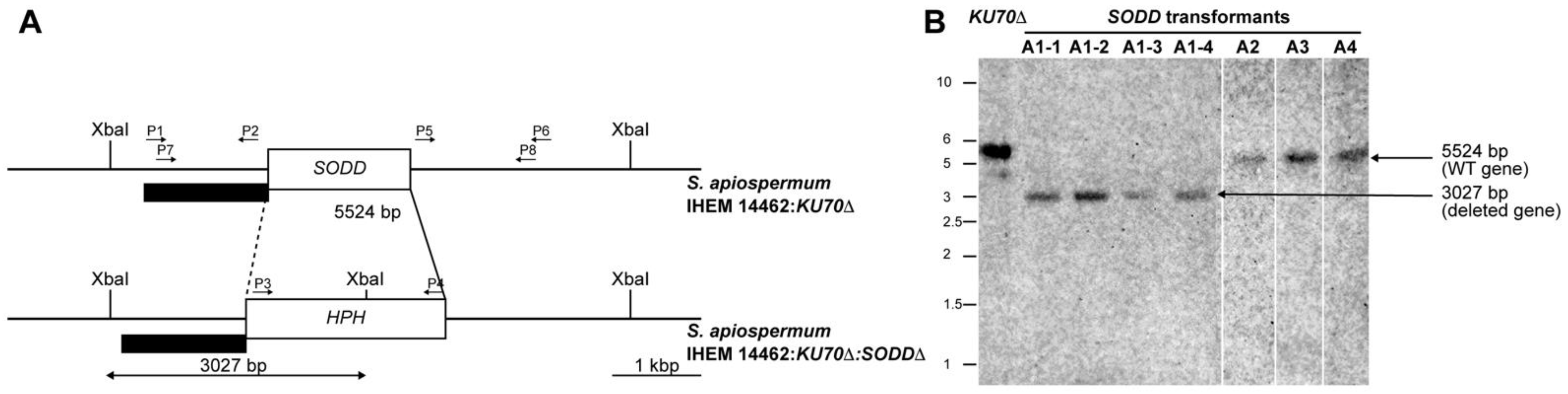
2.3. Disruption of the SODD Gene
2.4. Southern Blot Analysis
2.5. Susceptibility Studies
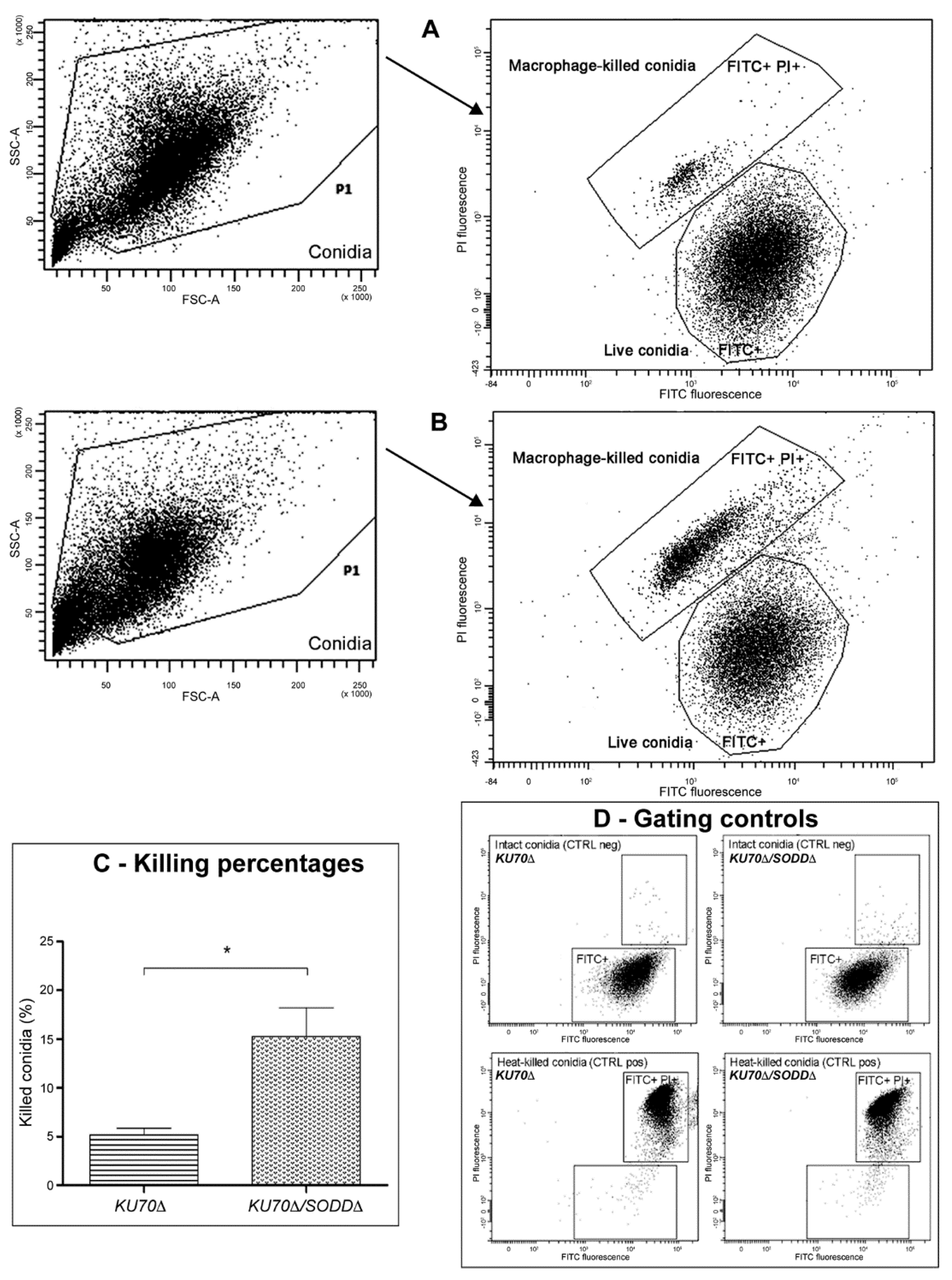
2.6. Phagocytosis Assays
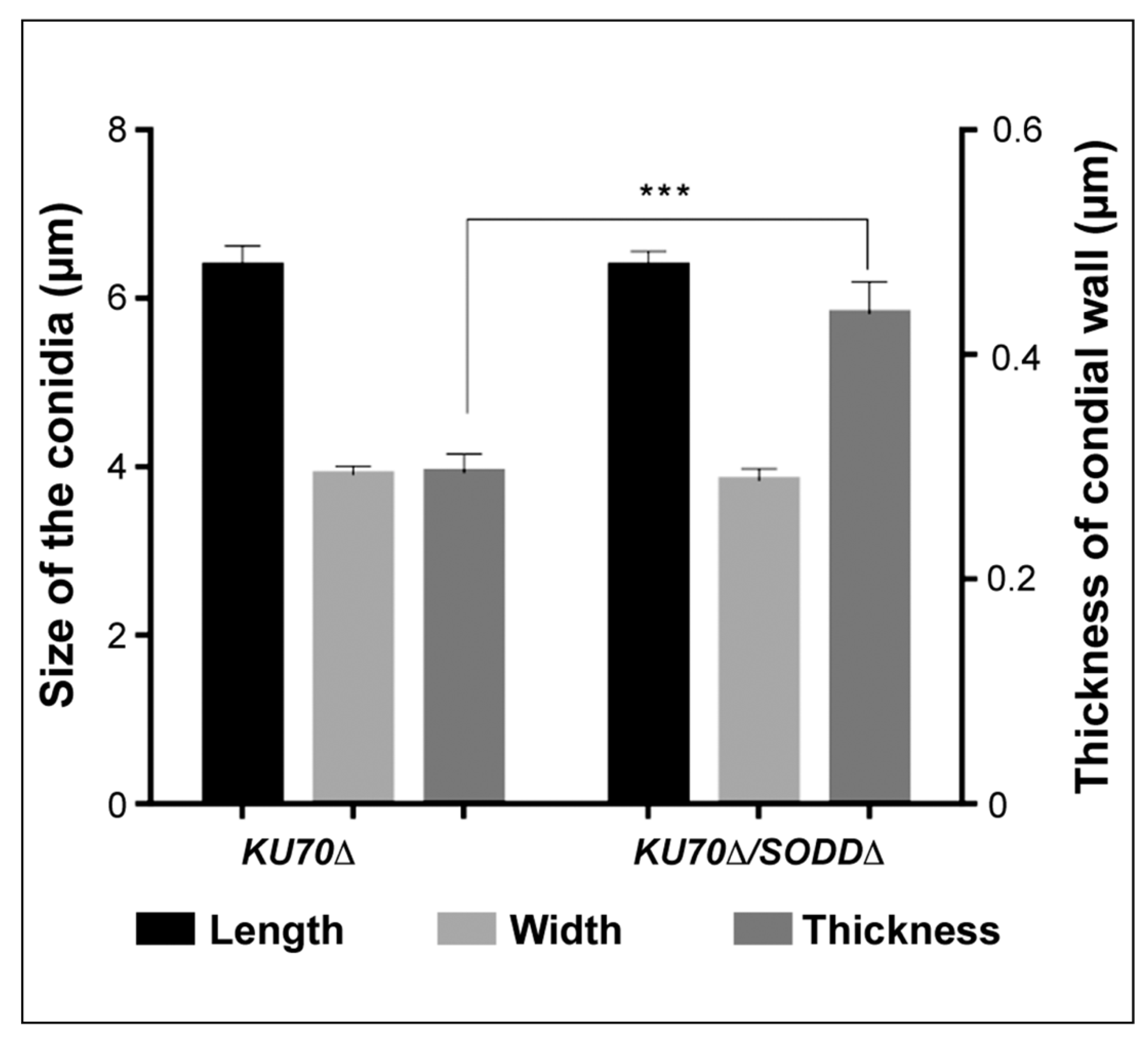
2.7. Transmission Electron Microscopy
2.8. Statistical Analysis
3. Results
3.1. Generation of a SODD Deficient Mutant
3.2. Sensitivity to Temperature and Cell Wall Stressing Chemicals
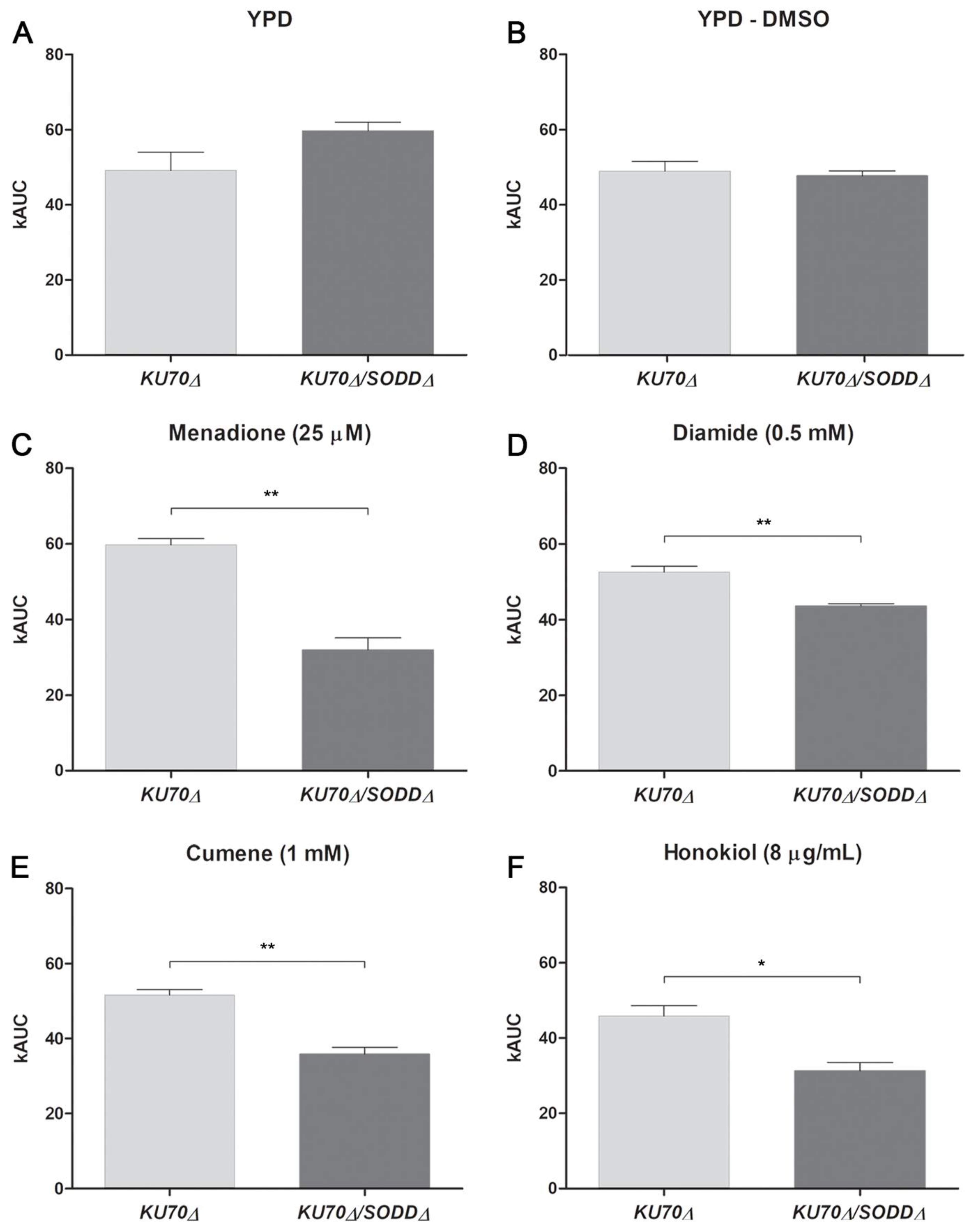
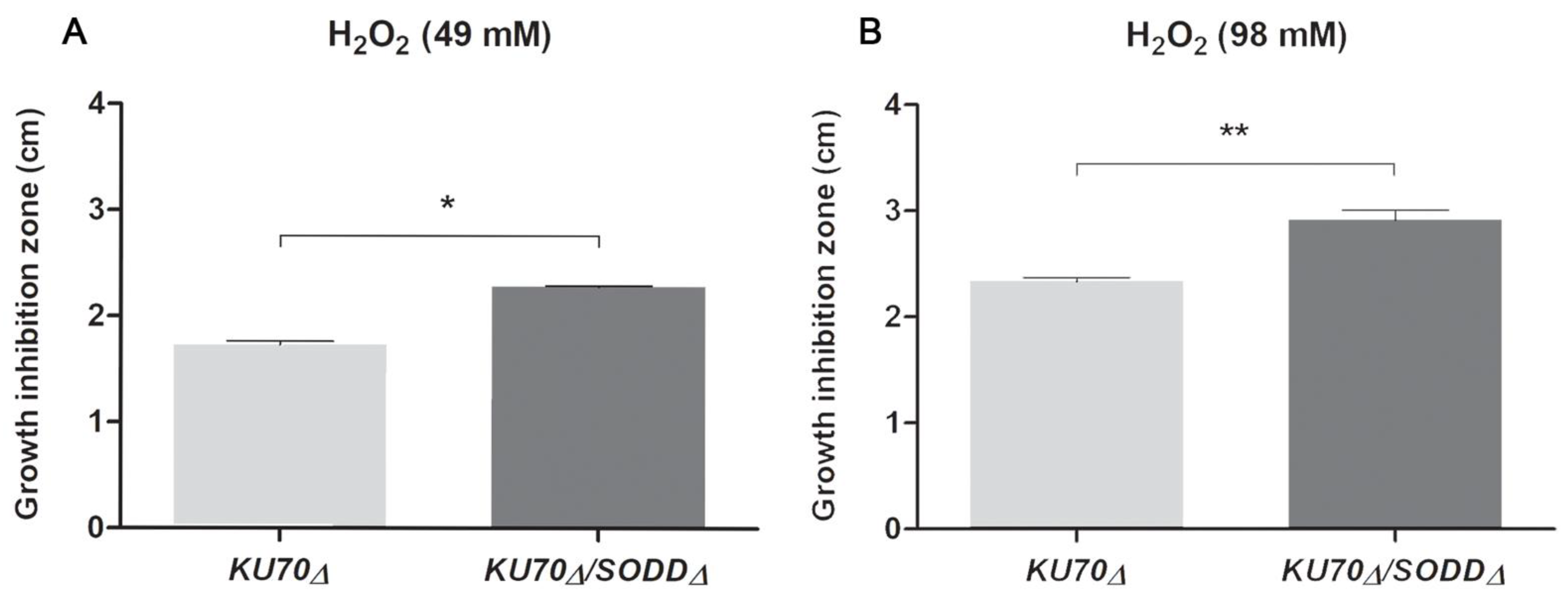
3.3. Growth under Chemically-Induced Oxidative Stress
3.4. Susceptibility to Antifungals
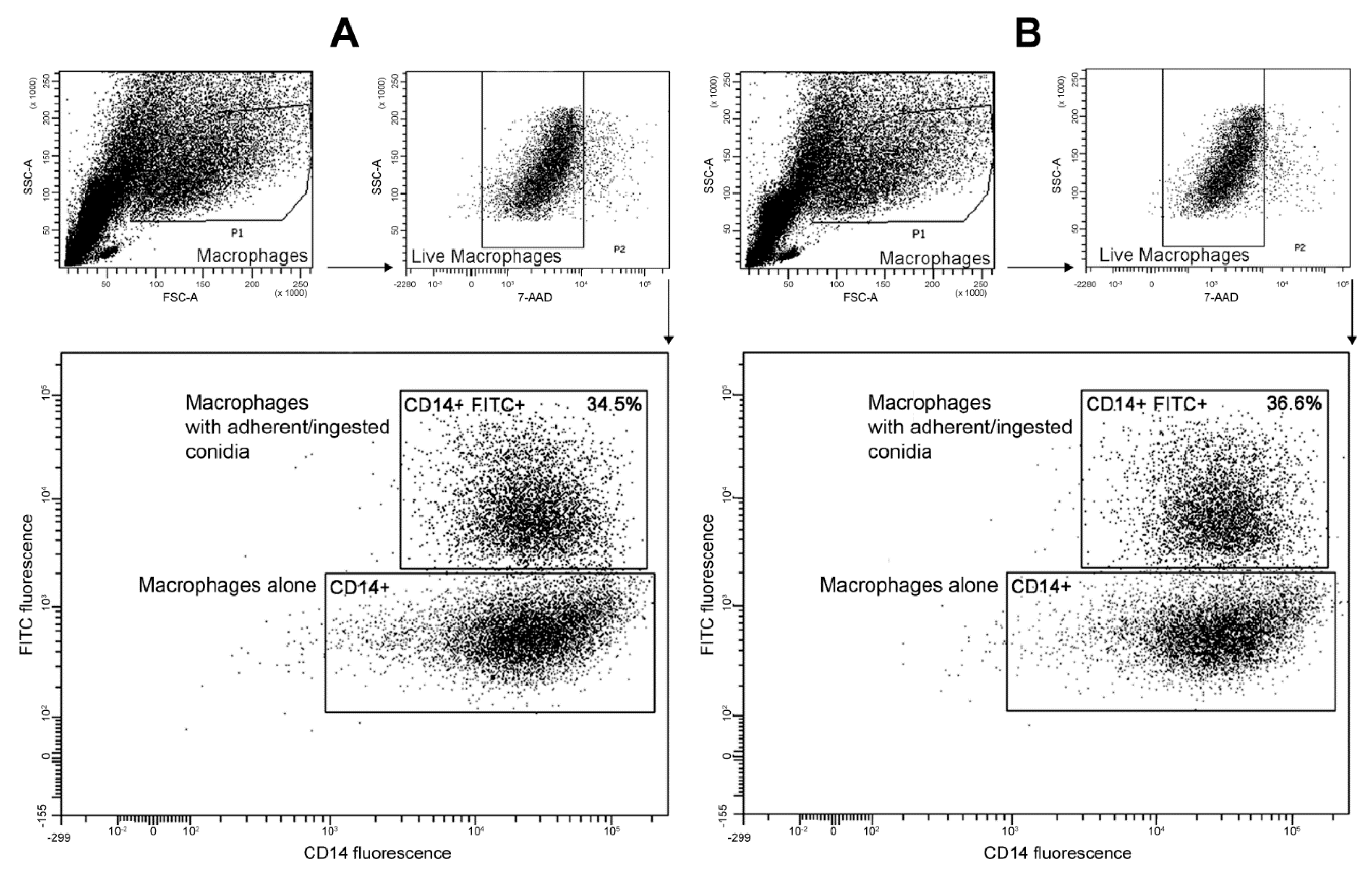
3.5. Interactions with Phagocytes
3.6. Morphological Features of the Conidia and Ultrastructure of Their Cell Wall
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rougeron, A.; Giraud, S.; Alastruey-Izquierdo, A.; Cano-Lira, J.; Rainer, J.; Mouhajir, A.; Le Gal, S.; Nevez, G.; Meyer, W.; Bouchara, J.P. Ecology of Scedosporium Species: Present Knowledge and Future Research. Mycopathologia 2018, 183, 185–200. [Google Scholar] [CrossRef]
- Ramirez-Garcia, A.; Pellon, A.; Rementeria, A.; Buldain, I.; Barreto-Bergter, E.; Rollin-Pinheiro, R.; De Meirelles, J.V.; Xisto, M.I.D.S.; Ranque, S.; Havlicek, V.; et al. Scedosporium and Lomentospora: An updated overview of underrated opportunists. Med. Mycol. 2018, 56, S102–S125. [Google Scholar] [CrossRef] [PubMed]
- Cimon, B.; Carrère, J.; Vinatier, J.F.; Chazalette, J.P.; Chabasse, D.; Bouchara, J.P. Clinical Significance of Scedosporium apiospermum in Patients with Cystic Fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Horré, R.; Marklein, G.; Siekmeier, R.; Nidermajer, S.; Reiffert, S. Selective Isolation of Pseudallescheria and Scedosporium Species from Respiratory Tract Specimens of Cystic Fibrosis Patients. Respiration 2008, 77, 320–324. [Google Scholar] [CrossRef]
- Blyth, C.; Middleton, P.; Harun, A.; Sorrell, T.C.; Meyer, W.; Chen, S.C.A. Clinical associations and prevalence of Scedosporium spp. in Australian cystic fibrosis patients: Identification of novel risk factors? Med. Mycol. 2010, 48 (Suppl. 1), S37–S44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masoud-Landgraf, L.; Badura, A.; Feierl, G.; Marth, E.; Eber, E.; Buzina, W. Modified culture method detects a high diversity of fungal species in cystic fibrosis patients. Med Mycol. 2013, 52, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Sedlacek, L.; Graf, B.; Schwarz, C.; Albert, F.; Peter, S.; Würstl, B.; Wagner, S.; Klotz, M.; Becker, A.; Haase, G.; et al. Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J. Cyst. Fibros. 2015, 14, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Ziesing, S.; Suerbaum, S.; Sedlacek, L. Fungal epidemiology and diversity in cystic fibrosis patients over a 5-year period in a national reference center. Med. Mycol. 2016, 54, 781–786. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, C.; Brandt, C.; Antweiler, E.; Krannich, A.; Staab, D.; Schmitt-Grohé, S.; Fischer, R.; Hartl, M.; Thronicke, A.; Tintelnot, K. Prospective multicenter German study on pulmonary colonization with Scedosporium /Lomentospora species in cystic fibrosis: Epidemiology and new association factors. PLoS ONE 2017, 12, e0171485. [Google Scholar] [CrossRef] [PubMed]
- Coron, N.; Pihet, M.; Fréalle, E.; Lemeille, Y.; Pinel, C.; Pelloux, H.; Gargala, G.; Favennec, L.; Accoceberry, I.; Durand-Joly, I.; et al. Toward the Standardization of Mycological Examination of Sputum Samples in Cystic Fibrosis: Results from a French Multicenter Prospective Study. Mycopathologia 2017, 183, 101–117. [Google Scholar] [CrossRef]
- Defontaine, A.; Zouhair, R.; Cimon, B.; Carrère, J.; Bailly, E.; Symoens, F.; Diouri, M.; Hallet, J.-N.; Bouchara, J.-P. Genotyping Study of Scedosporium apiospermum Isolates from Patients with Cystic Fibrosis. J. Clin. Microbiol. 2002, 40, 2108–2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zouhair, R.; Rougeron, A.; Razafimandimby, B.; Kobi, A.; Bouchara, J.P.; Giraud, S. Distribution of the different species of the Pseudallescheria boydii/Scedosporium apiospermum complex in French patients with cystic fibrosis. Med. Mycol. 2013, 51, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Symoens, F.; Knoop, C.; Schrooyen, M.; Denis, O.; Estenne, M.; Nolard, N.; Jacobs, F. Disseminated Scedosporium apiospermum Infection in a Cystic Fibrosis Patient After Double-lung Transplantation. J. Hear. Lung Transplant. 2006, 25, 603–607. [Google Scholar] [CrossRef]
- Staerck, C.; Gastebois, A.; Vandeputte, P.; Calenda, A.; Larcher, G.; Gillmann, L.; Papon, N.; Bouchara, J.-P.; Fleury, M.J. Microbial antioxidant defense enzymes. Microb. Pathog. 2017, 110, 56–65. [Google Scholar] [CrossRef]
- Marciano, B.E.; Spalding, C.; Fitzgerald, A.; Mann, D.; Brown, T.; Osgood, S.; Yockey, L.; Darnell, D.N.; Barnhart, L.; Daub, J.; et al. Common Severe Infections in Chronic Granulomatous Disease. Clin. Infect. Dis. 2015, 60, 1176–1183. [Google Scholar] [CrossRef]
- Belozerskaya, T.A.; Gessler, N.N. Reactive oxygen species and the strategy of antioxidant defense in fungi: A review. Appl. Biochem. Microbiol. 2007, 43, 506–515. [Google Scholar] [CrossRef]
- Forrester, M.T.; Foster, M.W. Protection from nitrosative stress: A central role for microbial flavohemoglobin. Free. Radic. Biol. Med. 2012, 52, 1620–1633. [Google Scholar] [CrossRef]
- Galli, F.; Battistoni, A.; Gambari, R.; Pompella, A.; Bragonzi, A.; Pilolli, F.; Iuliano, L.; Piroddi, M.; Dechecchi, M.C.; Cabrini, G. Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim. Et Biophys. Acta BBA Mol. Basis Dis. 2012, 1822, 690–713. [Google Scholar] [CrossRef] [Green Version]
- Larcher, G.; Cimon, B.; Symoens, F.; Tronchin, G.; Chabasse, D.; Bouchara, J.P. A 33 kDa serine proteinase from Scedosporium apiospermum. Biochem. J. 1996, 315, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Lima, O.C.; Larcher, G.; Vandeputte, P.; Lebouil, A.; Chabasse, D.; Simoneau, P.; Bouchara, J.-P. Molecular cloning and biochemical characterization of a Cu,Zn-superoxide dismutase from Scedosporium apiospermum. Microbes Infect. 2007, 9, 558–565. [Google Scholar] [CrossRef] [Green Version]
- Mina, S.; Marot-Leblond, A.; Cimon, B.; Fleury, M.J.J.; Larcher, G.; Bouchara, J.-P.; Robert, R. Purification and Characterization of a Mycelial Catalase from Scedosporium boydii, a Useful Tool for Specific Antibody Detection in Patients with Cystic Fibrosis. Clin. Vaccine Immunol. 2014, 22, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, S.; Larcher, G.; Landreau, A.; Richomme, P.; Duval, O.; Bouchara, J.-P. Hydroxamate siderophores of Scedosporium apiospermum. BioMetals 2009, 22, 1019–1029. [Google Scholar] [CrossRef] [Green Version]
- Le Govic, Y.; Havlíček, V.; Capilla, J.; Luptáková, D.; Dumas, D.; Papon, N.; Le Gal, S.; Bouchara, J.P.; Vandeputte, P. Synthesis of the hydroxamate siderophore Na-methylcoprogen B in Scedosporium apiospermum is mediated by SIDD ortholog and is required for virulence. Front Cell. Infect. Microbiol. 2020, 10, 587909. [Google Scholar] [CrossRef]
- Mina, S.; Staerck, C.; D’Almeida, S.M.; Marot, A.; Delneste, Y.; Calenda, A.; Tabiasco, J.; Bouchara, J.-P.; Fleury, M.J. Identification of Scedosporium boydii catalase A1 gene, a reactive oxygen species detoxification factor highly expressed in response to oxidative stress and phagocytic cells. Fungal Biol. 2015, 119, 1322–1333. [Google Scholar] [CrossRef]
- Vandeputte, P.; Ghamrawi, S.; Rechenmann, M.; Iltis, A.; Giraud, S.; Fleury, M.; Thornton, C.; Delhaès, L.; Meyer, W.; Papon, N.; et al. Draft genome sequence of the pathogenic fungus Scedosporium apiospermum. Genome Announc. 2014, 2, e00988-14. [Google Scholar] [CrossRef] [Green Version]
- Staerck, C.; Vandeputte, P.; Gastebois, A.; Calenda, A.; Giraud, S.; Papon, N.; Bouchara, J.P.; Fleury, M.J.J. Enzymatic Mechanisms Involved in Evasion of Fungi to the Oxidative Stress: Focus on Scedosporium apiospermum. Mycopathologia 2017, 183, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Staerck, C.; Tabiasco, J.; Godon, C.; Delneste, Y.; Bouchara, J.P.; Fleury, M.J.J. Transcriptional profiling of Scedosporium apiospermum enzymatic antioxidant gene battery unravels the involvement of thioredoxin reductases against chemical and phagocytic cells oxidative stress. Med. Mycol. 2019, 57, 363–373. [Google Scholar] [CrossRef]
- Ghamrawi, S.; Gastebois, A.; Zykwinska, A.; Vandeputte, P.; Marot, A.; Mabilleau, G.; Cuenot, S.; Bouchara, J.-P. A Multifaceted Study of Scedosporium boydii Cell Wall Changes during Germination and Identification of GPI-Anchored Proteins. PLoS ONE 2015, 10, e0128680. [Google Scholar] [CrossRef] [Green Version]
- Pateau, V.; Razafimandimby, B.; Vandeputte, P.; Thornton, C.R.; Guillemette, T.; Bouchara, J.-P.; Giraud, S. Gene Disruption in Scedosporium aurantiacum: Proof of Concept with the Disruption of SODC Gene Encoding a Cytosolic Cu,Zn-Superoxide Dismutase. Mycopathologia 2018, 183, 241–249. [Google Scholar] [CrossRef]
- Turgeon, B.; Condon, B.; Liu, J.; Zhang, N. Protoplast Transformation of Filamentous Fungi. Methods Mol. Biol. 2010, 638, 3–19. [Google Scholar] [CrossRef]
- Liu, Z.; Friesen, T.L. Polyethylene Glycol (PEG)-Mediated Transformation in Filamentous Fungal Pathogens. Methods Mol. Biol. 2012, 835, 365–375. [Google Scholar] [CrossRef]
- Joubert, A.; Calmes, B.; Berruyer, R.; Pihet, M.; Bouchara, J.-P.; Simoneau, P.; Guillemette, T. Laser nephelometry applied in an automated microplate system to study filamentous fungus growth. Biotechniques 2010, 48, 399–404. [Google Scholar] [CrossRef] [Green Version]
- Martin-Vicente, A.; Guarro, J.; González, G.M.; Lass-Flörl, C.; Lackner, M.; Capilla, J. Voriconazole MICs are predictive for the outcome of experimental disseminated scedosporiosis. J. Antimicrob. Chemother. 2017, 72, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Paolini, L.; Adam, C.; Beauvillain, C.; Preisser, L.; Blanchard, S.; Pignon, P.; Seegers, V.; Chevalier, L.-M.; Campone, M.; Wernert, R.; et al. Lactic Acidosis Together with GM-CSF and M-CSF Induces Human Macrophages toward an Inflammatory Protumor Phenotype. Cancer Immunol. Res. 2020, 8, 383–395. [Google Scholar] [CrossRef] [Green Version]
- Marr, K.A.; Koudadoust, M.; Black, M.; Balajee, S.A. Early Events in Macrophage Killing of Aspergillus fumigatus Conidia: New Flow Cytometric Viability Assay. Clin. Diagn. Lab. Immunol. 2001, 8, 1240–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staerck, C.; Godon, C.; Bouchara, J.-P.; Fleury, M.J.J. Varying susceptibility of clinical and environmental Scedosporium isolates to chemical oxidative stress in conidial germination. Arch. Microbiol. 2018, 200, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V. Genetic engineering of filamentous fungi-progress, obstacles and future trends. Biotechnol. Adv. 2008, 26, 177–185. [Google Scholar] [CrossRef]
- Krappmann, S. Gene targeting in filamentous fungi: The benefits of impaired repair. Fungal Biol. Rev. 2007, 21, 25–29. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Holdom, M.D.; Hay, R.J. Specific recognition of purified Cu,Zn superoxide dismutase from Aspergillus fumigatus by immune human sera. J. Clin. Microbiol. 1995, 33, 495–496. [Google Scholar] [CrossRef] [Green Version]
- Desai, G.; Nassaft, F.; Brummer, E.; Stevens, D.A. Killing of Histoplasma capsulatum by macrophage colony stimulating factor-treated human monocyte-derived macrophages: Role for reactive oxygen intermediates. J. Med. Microbiol. 1995, 43, 224–229. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.-S.; Rhie, G.-E.; Oh, J.-H.; Huh, W.-K.; Yim, H.-S.; Kang, S.-O. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 2002, 148, 3705–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narasipura, S.D.; Ault, J.G.; Behr, M.J.; Chaturvedi, V.; Chaturvedi, S. Characterization of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: Role in biology and virulence. Mol. Microbiol. 2003, 47, 1681–1694. [Google Scholar] [CrossRef]
- Martchenko, M.; Alarco, A.-M.; Harcus, D.; Whiteway, M. Superoxide Dismutases in Candida albicans: Transcriptional Regulation and Functional Characterization of the Hyphal-inducedSOD5Gene. Mol. Biol. Cell 2004, 15, 456–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirach, S.; Cooper, C.R.; Vanittanakom, P.; Vanittanakom, N. The Copper, Zinc Superoxide Dismutase Gene of Penicillium marneffei: Cloning, Characterization, and Differential Expression During Phase Transition and Macrophage Infection. Med. Mycol. 2007, 45, 409–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frohner, I.E.; Bourgeois, C.; Yatsyk, K.; Majer, O.; Kuchler, K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 2009, 71, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Lambou, K.; Lamarre, C.; Beau, R.; Dufour, N.; Latgé, J.P. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol. Microbiol. 2010, 75, 910–923. [Google Scholar] [CrossRef]
- Miramon, P.; Dunker, C.; Windecker, H.; Bohovych, I.M.; Brown, A.J.; Kurzai, O.; Hube, B. Cellular Responses of Candida albicans to Phagocytosis and the Extracellular Activities of Neutrophils Are Critical to Counteract Carbohydrate Starvation, Oxidative and Nitrosative Stress. PLoS ONE 2012, 7, e52850. [Google Scholar] [CrossRef] [Green Version]
- Youseff, B.H.; Holbrook, E.D.; Smolnycki, K.A.; Rappleye, C.A. Extracellular Superoxide Dismutase Protects Histoplasma Yeast Cells from Host-Derived Oxidative Stress. PLOS Pathog. 2012, 8, e1002713. [Google Scholar] [CrossRef] [Green Version]
- Gleason, J.E.; Galaleldeen, A.; Peterson, R.L.; Taylor, A.B.; Holloway, S.P.; Waninger-Saroni, J.; Cormack, B.P.; Cabelli, D.E.; Hart, P.J.; Culotta, V.C. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. USA 2014, 111, 5866–5871. [Google Scholar] [CrossRef] [Green Version]
- Garfoot, A.L.; Rappleye, C.A. Histoplasma capsulatum surmounts obstacles to intracellular pathogenesis. FEBS J. 2016, 283, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, D.; Muñoz, J.F.; Lopez, Á.; Urán, M.; Herrera, J.; Borges, C.L.; Restrepo, Á.; Soares, C.M.; Taborda, C.P.; Almeida, A.J.; et al. Identification and analysis of the role of superoxide dismutases isoforms in the pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 2016, 10, e0004481. [Google Scholar] [CrossRef]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latgé, J.-P. Glycosylphosphatidylinositol-anchored Glucanosyltransferases Play an Active Role in the Biosynthesis of the Fungal Cell Wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [Green Version]
- Rittenour, W.R.; Harris, S.D. Glycosylphosphatidylinositol-Anchored Proteins in Fusarium graminearum: Inventory, Variability, and Virulence. PLoS ONE 2013, 8, e81603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Chen, S.M.; Liu, W.; Zhu, F.; He, L.J.; Zhang, J.D.; Zhang, S.Q.; Yan, L.; Xu, Z.; Xu, G.T.; et al. Abolishing Cell Wall Glycosylphosphatidylinositol-Anchored Proteins in Candida albicans Enhances Recognition by Host Dectin-1. Infect. Immun. 2015, 83, 2694–2704. [Google Scholar] [CrossRef] [Green Version]
- Matsushika, A.; Negi, K.; Suzuki, T.; Goshima, T.; Hoshino, T. Identification and characterization of a novel issatchenkia orientalis GPI-anchored protein, IoGas1, required for resistance to low pH and salt stress. PLoS ONE 2016, 11, e0161888. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.M.; Harrison, T.S.; McDade, H.C.; Taborda, C.; Heinrich, G.; Casadevall, A.; Perfect, J.R. Superoxide Dismutase Influences the Virulence of Cryptococcus neoformans by Affecting Growth within Macrophages. Infect. Immun. 2003, 71, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Narasipura, S.D.; Chaturvedi, V.; Chaturvedi, S. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 2005, 55, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Holdom, M.D.; Lechenne, B.; Hay, R.J.; Hamilton, A.J.; Monod, M. Production and Characterization of Recombinant Aspergillus fumigatus Cu,Zn Superoxide Dismutase and Its Recognition by Immune Human Sera. J. Clin. Microbiol. 2000, 38, 558–562. [Google Scholar] [CrossRef] [Green Version]
- Leal, S.; Vareechon, C.; Cowden, S.; Cobb, B.A.; Latgé, J.-P.; Momany, M.; Pearlman, E. Fungal antioxidant pathways promote survival against neutrophils during infection. J. Clin. Investig. 2012, 122, 2482–2498. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Chen, L.; Zhong, S.; Wei, X.; Zhao, Q.; Pan, Q.; Kang, Z.; Liu, J. A Cu-only superoxide dismutase from stripe rust fungi functions as a virulence factor deployed for counter defense against host-derived oxidative stress. Environ. Microbiol. 2020, 22, 5309–5326. [Google Scholar] [CrossRef]
- Dantas, A.D.S.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative Stress Responses in the Human Fungal Pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef] [Green Version]
- Chaput, M.; Brygier, J.; Lion, Y.; Sels, A. Potentiation of oxygen toxicity by menadione in Saccharomyces cerevisiae. Biochimie 1983, 65, 501–512. [Google Scholar] [CrossRef]
- Sun, L.; Liao, K.; Hang, C.; Wang, D. Honokiol induces reactive oxygen species-mediated apoptosis in Candida albicans through mitochondrial dysfunction. PLoS ONE 2017, 12, e0172228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Pokhrel, A.; Coleman, J.J. The Extracellular Superoxide Dismutase Sod5 From Fusarium oxysporum Is Localized in Response to External Stimuli and Contributes to Fungal Pathogenicity. Front. Plant Sci. 2021, 12, 608861. [Google Scholar] [CrossRef] [PubMed]
- Hink, H.U.; Santanam, N.; Dikalov, S.; McCann, L.; Nguyen, A.D.; Parthasarathy, S.; Harrison, D.G.; Fukai, T. Peroxidase properties of extracellular superoxide dismutase: Role of uric acid in modulating in vivo activity. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1402–1408. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, D.; Kondo, K.; Uehara, N.; Otokozawa, S.; Tsuji, N.; Yagihashi, A.; Watanabe, N. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob. Agents Chemother. 2002, 46, 3113–3117. [Google Scholar] [CrossRef] [Green Version]
- Bink, A.; VandenBosch, D.; Coenye, T.; Nelis, H.; Cammue, B.P.A.; Thevissen, K. Superoxide Dismutases Are Involved in Candida albicans Biofilm Persistence against Miconazole. Antimicrob. Agents Chemother. 2011, 55, 4033–4037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shekhova, E.; Kniemeyer, O.; Brakhage, A.A. Induction of Mitochondrial Reactive Oxygen Species Production by Itraconazole, Terbinafine, and Amphotericin B as a Mode of Action against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2017, 61, 00978-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, C.A.; Gaertner, A.A.E.; Henriquez, S.A.; Fang, D.; Colon-Reyes, R.J.; Brumaghim, J.; Kozubowski, L. Fluconazole induces ROS in Cryptococcus neoformans and contributes to DNA damage in vitro. PLoS ONE 2018, 13, e0208471. [Google Scholar] [CrossRef] [PubMed]
- Dbouk, N.H.; Covington, M.B.; Nguyen, K.; Chandrasekaran, S. Increase of reactive oxygen species contributes to growth inhibition by fluconazole in Cryptococcus neoformans. BMC Microbiol. 2019, 19, 243. [Google Scholar] [CrossRef]
- Linares, C.E.B.; Giacomelli, S.R.; Altenhofen, D.; Alves, S.H.; Morsch, V.M.; Schetinger, M.R.C. Fluconazole and amphotericin-B resistance are associated with increased catalase and superoxide dismutase activity in Candida albicans and Candida dubliniensis. Rev. Soc. Bras. Med. Trop. 2013, 46, 752–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schruefer, S.; Böhmer, I.; Dichtl, K.; Spadinger, A.; Kleinemeier, C.; Ebel, F. The response regulator Skn7 of Aspergillus fumigatus is essential for the antifungal effect of fludioxonil. Sci. Rep. 2021, 11, 5317. [Google Scholar] [CrossRef] [PubMed]
- Kopecká, M.; Gabriel, M. The influence of Congo red on the cell wall and (1→3)-β-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 1992, 158, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Rollin-Pinheiro, R.; Xisto, M.I.D.D.S.; Rochetti, V.P.; Barreto-Bergter, E. Scedosporium cell wall: From carbohydrate-containing structures to host-pathogen interactions. Mycopathologia 2020, 185, 931–946. [Google Scholar] [CrossRef] [PubMed]
- Benachour, A.; Sipos, G.; Flury, I.; Reggiori, F.; Canivenc-Gansel, E.; Vionnet, C.; Conzelmann, A.; Benghezal, M. Deletion of GPI7, a Yeast Gene Required for Addition of a Side Chain to the Glycosylphosphatidylinositol (GPI) Core Structure, Affects GPI Protein Transport, Remodeling, and Cell Wall Integrity. J. Biol. Chem. 1999, 274, 15251–15261. [Google Scholar] [CrossRef] [Green Version]
- Bowman, S.M.; Piwowar, A.; Al Dabbous, M.; Vierula, J.; Free, S.J. Mutational Analysis of the Glycosylphosphatidylinositol (GPI) Anchor Pathway Demonstrates that GPI-Anchored Proteins Are Required for Cell Wall Biogenesis and Normal Hyphal Growth in Neurospora crassa. Eukaryot. Cell 2006, 5, 587–600. [Google Scholar] [CrossRef] [Green Version]
| Primer Name and Use | Sequence 5′→3′ | Tm (°C) | Size of the Amplicon (bp) |
|---|---|---|---|
| PCR amplification of the 5′ flanking region of SODD | |||
| P1-SODD | ATTCATAGACTCAATAATTAGAACTCGACT | 64 | 925 |
| P2-SODD | TCGTGAATCTTTTACCAGATCGGAAGCAATAAATGTAATTTATCTCTTTCAATCCCAAGC | 64 | |
| PCR amplification of the hygromycin B resistance gene | |||
| P3-SODD | GCTTGGGATTGAAAGAGATAAATTACATTTATTGCTTCCGATCTGGTAAAAGATTCACGA | 68 | 2633 |
| P4-SODD | AATTGATTCTTGTCGATCATTAATTTGGTCATCAGAGCAGATTGTACTGAGAGTGCACCA | 68 | |
| PCR amplification of the 5′ flanking region of SODD | |||
| P5-SODD | TGGTGCACTCTCAGTACAATCTGCTCTGATGACCAAATTAATGATCGACAAGAATCAATT | 64 | 901 |
| P6-SODD | GACGTTGTATATATATCCTGGAAGAATCTT | 64 | |
| Fusion of the amplicons | |||
| P7-SODD | GAAACGCCCGACTAGTTAAATC | 64 | 4448 |
| P8-SODD | CTGCAAATGCCAAATTCCAA | 64 | |
| Antifungal | Parent Strain (KU70Δ Mutant) | KU70Δ/SODDΔ Double Mutant | p-Value | ||
|---|---|---|---|---|---|
| MIC Mean (µg/mL) | SD | MIC Mean (µg/mL) | SD | ||
| Isavuconazole | >32 | ND | 0.142 | 0.093 | <0.05 |
| Itraconazole | 0.552 | 0.187 | 0.064 | 0.000 | <0.05 |
| Posaconazole | 0.476 | 0.053 | 0.131 | 0.035 | <0.05 |
| Voriconazole | 0.251 | 0.146 | 0.018 | 0.004 | <0.05 |
| Fluconazole | >256 | ND | >256 | ND | NS |
| Amphotericin B | 5 | 2 | 3.9 | 1.597 | NS |
| Caspofungin | >32 | ND | >32 | ND | NS |
| Micafungin | 0.082 | 0.016 | 0.113 | 0.048 | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staerck, C.; Yaakoub, H.; Vandeputte, P.; Tabiasco, J.; Godon, C.; Gastebois, A.; Giraud, S.; Guillemette, T.; Calenda, A.; Delneste, Y.; et al. The Glycosylphosphatidylinositol-Anchored Superoxide Dismutase of Scedosporium apiospermum Protects the Conidia from Oxidative Stress. J. Fungi 2021, 7, 575. https://doi.org/10.3390/jof7070575
Staerck C, Yaakoub H, Vandeputte P, Tabiasco J, Godon C, Gastebois A, Giraud S, Guillemette T, Calenda A, Delneste Y, et al. The Glycosylphosphatidylinositol-Anchored Superoxide Dismutase of Scedosporium apiospermum Protects the Conidia from Oxidative Stress. Journal of Fungi. 2021; 7(7):575. https://doi.org/10.3390/jof7070575
Chicago/Turabian StyleStaerck, Cindy, Hajar Yaakoub, Patrick Vandeputte, Julie Tabiasco, Charlotte Godon, Amandine Gastebois, Sandrine Giraud, Thomas Guillemette, Alphonse Calenda, Yves Delneste, and et al. 2021. "The Glycosylphosphatidylinositol-Anchored Superoxide Dismutase of Scedosporium apiospermum Protects the Conidia from Oxidative Stress" Journal of Fungi 7, no. 7: 575. https://doi.org/10.3390/jof7070575
APA StyleStaerck, C., Yaakoub, H., Vandeputte, P., Tabiasco, J., Godon, C., Gastebois, A., Giraud, S., Guillemette, T., Calenda, A., Delneste, Y., Fleury, M., & Bouchara, J.-P. (2021). The Glycosylphosphatidylinositol-Anchored Superoxide Dismutase of Scedosporium apiospermum Protects the Conidia from Oxidative Stress. Journal of Fungi, 7(7), 575. https://doi.org/10.3390/jof7070575






