Assessing Genotypic and Environmental Effects on Endophyte Communities of Fraxinus (Ash) Using Culture Dependent and Independent DNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Fungal and Plant DNA Extraction and Sequencing
2.2.1. DNA Sequencing and Identification of Pure Culture Fungal DNA
2.2.2. High-Throughput Sequencing of nrITS-1 Directly from Leaf and Seed
2.3. OTU Cluster, Species Determination, and Diversity Analyses of HTS Data
3. Results
3.1. Identification of Cultured Fungal Isolates from Fraxinus Excelsior and Other Fraxinus Species
3.1.1. Endophytes in Other Fraxinus Species
3.1.2. OTU Retrieval from Different Leaf Tissue Types and Roots in Fraxinus
3.2. Community Analysis of Samples Using Direct Amplicon High Throughput Sequencing (HTS)
3.2.1. Diversity of the Ash Microbiome by HTS
3.2.2. Taxonomic Composition of HTS Data
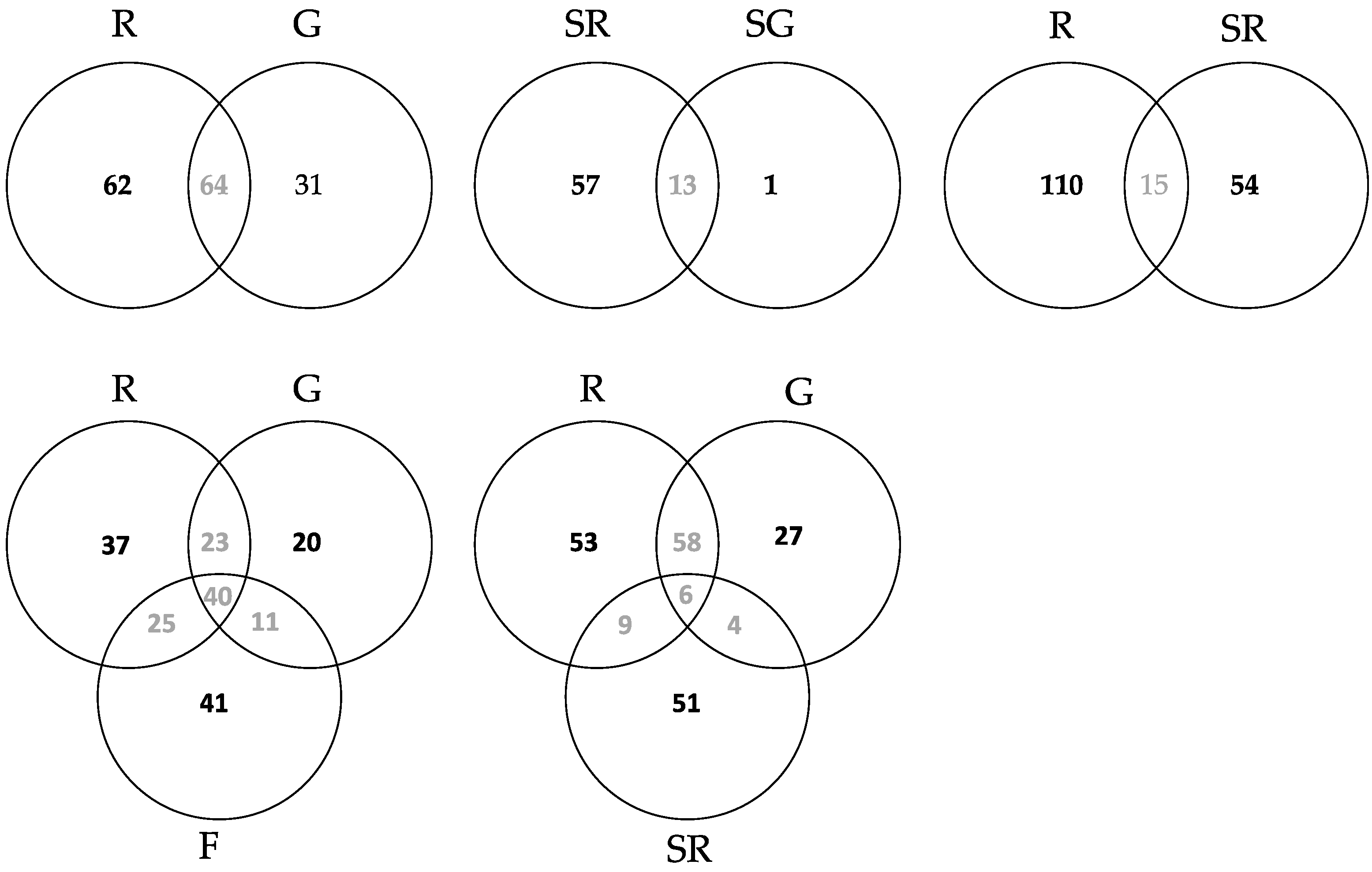
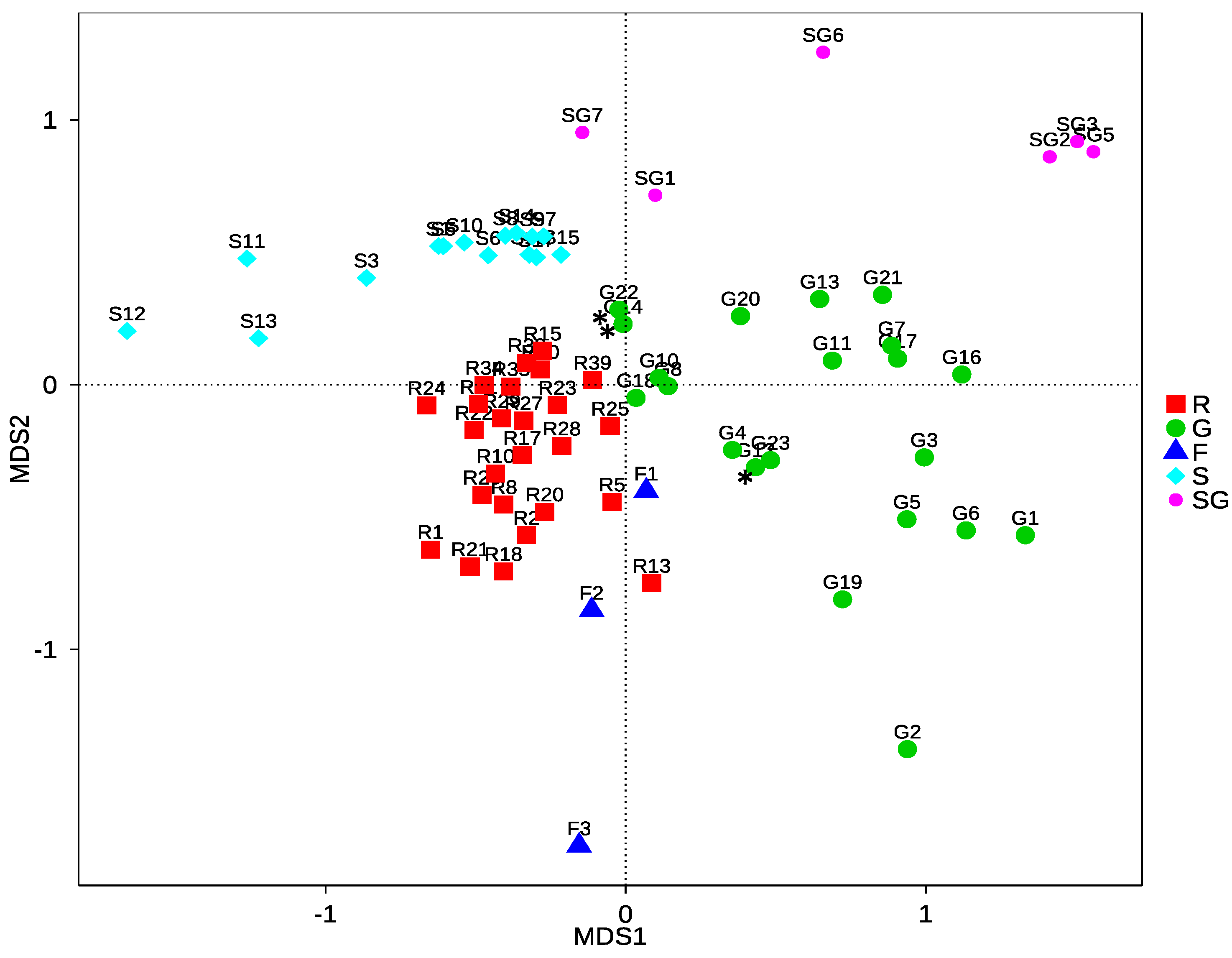
3.2.3. Community Structure and Differences of HTS Data
4. Discussion
4.1. Culture Dependent Fungal Diversity and Community Composition
4.1.1. Diversity and Taxonomy of Endophytes from the Culture Dependent Sample
4.1.2. Community Differences Detected from the Culture Dependent Approach
4.2. Culture Independent Fungal Diversity and Community Composition
4.2.1. Diversity in the Culture Independent Sample
4.2.2. Community Differences
4.3. Comparison of OTUs Obtained from High Throughput Amplicon Sequencing (HTS) and Sanger Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Newton, A.C.; Fitt, B.D.L.; Atkins, S.D.; Walters, D.R.; Daniell, T.J. Pathogenesis, parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 2010, 18, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef] [Green Version]
- McEvoy, A.; O’Regan, F.; Fleming, C.C.; Moreland, B.P.; Pollock, J.A.; McGuinness, B.W.; Hodkinson, T.R. Bleeding canker of horse chestnut (Aesculus hippocastanum) in Ireland: Incidence, severity and characterization using DNA sequences and real-time PCR. Plant Pathol. 2016, 65, 1419–1429. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Doohan, F.M.; Saunders, M.J.; Murphy, B.R. Endophytes for a Growing World; Cambridge University Press: Cambridge, UK, 2019; p. 420. [Google Scholar]
- Lahiri, A.; Douglas, G.C.; Murphy, B.R.; Hodkinson, T.R. In vitro methods for plant-microbe interaction and biocontrol studies in European ash (Fraxinus excelsior L.). In Endophytes for a Growing World; Hodkinson, T.R., Doohan, F.M., Saunders, M.J., Murphy, B.R., Eds.; Cambridge University Press: Cambridge, UK, 2019; pp. 328–340. [Google Scholar]
- Strobel, G.A.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef]
- Sudhakar, T.; Dash, S.; Rao, R.; Srinivasan, R.; Zacharia, S.; Atmanand, M.; Subramaniam, B.; Nayak, S. Do endophytic fungi possess pathway genes for plant secondary metabolites? Curr. Sci. 2013, 104, 178–182. [Google Scholar]
- Aly, A.H.; Debbab, A.; Proksch, P. Fifty years of drug discovery from fungi. Fungal Divers. 2011, 50, 3–19. [Google Scholar] [CrossRef]
- Crous, P.W.; Braun, U.; Schubert, K.; Groenewald, J.Z. Delimiting Cladosporium from morphologically similar genera. Stud. Mycol. 2007, 58, 33–56. [Google Scholar] [CrossRef]
- KoKo, T.W.; Stephenson, S.L.; Bahkali, A.H.; Hyde, K.D. From morphology to molecular biology: Can we use sequence data to identify fungal endophytes? Fungal Divers. 2011, 50, 113–120. [Google Scholar] [CrossRef]
- Olson, Å.; Stenlid, J. Pathogenic fungal species hybrids infecting plants. Microbes Infect. 2002, 4, 1353–1359. [Google Scholar] [CrossRef]
- Kohn, L.M. Mechanism of fungal speciation. Ann. Rev. Phytopathol. 2005, 43, 279–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foltz, M.J.; Perez, K.E.; Volk, T.J. Molecular phylogeny and morphology reveal three new species of Cantharellus within 20 m of one another in western Wisconsin, USA. Mycologia 2013, 105, 447–461. [Google Scholar] [CrossRef] [Green Version]
- Giraud, T.; Refrégier, G.; Le Gac, M.; de Vienne, D.M.; Hood, M.E. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef]
- Moore, D.; Robson, G.D.; Trinci, A.P.J. 21st Century Guidebook to Fungi; Cambridge University Press: Cambridge, UK, 2011; pp. 41–81. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomial RNA genes for phyologenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity of basidiomycetes: Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorfc, S.; Robertd, V.; Spougea, J.L.; Levesqueb, C.A.; Chen, W. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begerow, D.; Nilsson, H.; Unterseher, M.; Maier, W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 2010, 87, 99–108. [Google Scholar] [CrossRef]
- Seifert, K.A. Progress towards DNA barcoding of fungi. Mol. Ecol. Res. 2009, 9 (Suppl. S1), 83–89. [Google Scholar] [CrossRef]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the Pezizomycotina (Euasco-mycetes, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef]
- Rehner, S. Primers for Elongation Factor 1-a (EF1-a). 2001. Available online: http://ocid.nacse.org/research/deephyphae/EF1primer.pdf (accessed on 1 October 2017).
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, I.; Crespo, A.; Divakar, P.K.; Fankhauser, J.D.; Herman-Sackett, E.; Kalb, K.; Nelsen, M.P.; Nelson, N.A.; Rivas-Plata, E.; Shimp, A.D.; et al. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 2009, 23, 35–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.; Abarenkov, K.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Wang, Q.; Crous, P.; Robert, V.; Helgason, T.; et al. Sequence-based classification and identification of fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, T.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [Green Version]
- Maignien, L.; Deforce, E.A.; Chafee, M.E.; Eren, A.M.; Simmons, S.L. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 2014, 5, e00682-13. [Google Scholar] [CrossRef] [Green Version]
- Williams, T.R.; Moyne, A.L.; Harris, L.J.; Marco, M.L. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS ONE 2013, 8, e68642. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Ashton, P.D.; Aziz, N.; Feng, G.; Nelson, M.; Dytham, C.; Fitter, A.H.; Helgason, T. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 2011, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Gottel, N.R.; Castro, H.F.; Kerley, M.; Yang, Z.M.; Pelletier, D.A.; Podar, M.; Karpinets, T.; Uberbacher, E.; Tuskan, G.A.; Vilgalys, R.; et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011, 77, 5934–5944. [Google Scholar] [CrossRef] [Green Version]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [Green Version]
- Scholtysik, A.; Unterseher, M.; Otto, P.; Wirth, C. Spatio-temporal dynamics of endophyte diversity in the canopy of European ash (Fraxinus excelsior). Mycol. Prog. 2012, 12, 291–304. [Google Scholar] [CrossRef]
- Schlegel, M.; Queloz, V.; Sieber, T.N. The endophytic mycobiome of European ash and sycamore maple leaves–geographic patterns, host specificity and influence of ash dieback. Front. Microbiol. 2018, 9, 2345. [Google Scholar] [CrossRef] [PubMed]
- Bialek, R.; González, G.M.; Begerow, D.; Zelck, U.E. Coccidioido mycosis and blastomycosis: Advances in molecular diagnosis. FEMS Immunol. Med. Microbiol. 2005, 45, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Davydenko, K.; Vasaitis, R.; Stenlid, J.; Menkis, A. Fungi in foliage shoots of Fraxinus excelsior in eastern Ukraine: A first report on Hymenoschyphus pseudoalbidus. For. Pathol. 2013, 43, 462–467. [Google Scholar] [CrossRef]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Thomsen, I.M.; Stenlid, J. Occurrence and pathogenicity of fungi in necrotic and nonsymptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur. J. For. Res. 2009, 128, 51–60. [Google Scholar] [CrossRef]
- Kowalski, T.; Kraj, W.; Bednarz, B. Fungi on stems and twigs in initial and advanced stages of dieback of European ash (Fraxinus excelsior) in Poland. Eur. J. For. Res. 2016, 135, 565–579. [Google Scholar] [CrossRef] [Green Version]
- Cross, H.; Sønstebø, J.H.; Nagy, N.E.; Timmermann, V.; Solheim, H.; Børja, I.; Kauserud, H.; Carlsen, T.; Rzepka, B.; Wasak, K.; et al. Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol. 2017, 213, 1405–1417. [Google Scholar] [CrossRef]
- Agostinelli, M. Fungal Assemblages in Forest Trees—Influence of External and Internal Conditions. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2018; pp. 1–68. [Google Scholar]
- Cleary, M.; Nguyen, D.; Marčiulynienė, D.; Berlin, A.; Vasaitis, R.; Stenlid, J. Friend or foe? Biological and ecological traits of the European ash dieback pathogen Hymenoscyphus fraxineus in its native environment. Sci. Rep. 2016, 6, 21895. [Google Scholar] [CrossRef] [Green Version]
- Kosawang, C.; Amby, D.B.; Bussaban, B.; Mckinney, L.V.; Xu, J.; Kjær, E.D.; Collinge, D.B.; Nielsen, L.R. Fungal communities associated with species of Fraxinus tolerant to ash dieback and their potential for biological control. Fungal Biol. 2018, 122, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.R.; Batke, S.P.; Doohan, F.M.; Hodkinson, T.R. Media manipulations and the culture of beneficial fungal root endophytes. Int. J. Biol. 2015, 7, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Hodkinson, T.R.; Waldren, S.; Parnell, J.A.N.; Kelleher, C.T.; Salamin, K.; Salamin, N. DNA banking for plant breeding, biotechnology and biodiversity evaluation. J. Plant Res. 2007, 120, 17–29. [Google Scholar] [CrossRef]
- Perdereau, A.C.; Kelleher, C.T.; Douglas, G.C.; Hodkinson, T.R. High levels of gene flow and genetic diversity in Irish populations of Salix caprea L. inferred from chloroplast and nuclear SSR markers. BMC Plant Biol. 2014, 14, 202. [Google Scholar] [CrossRef] [Green Version]
- Hodkinson, T.R.; de Cesare, M.; Barth, S. Nuclear SSR markers for Miscanthus, Saccharum, and related grasses (Saccharinae, Poaceae). Appl. Plant Sci. 2013, 1, 1300042. [Google Scholar] [CrossRef]
- Lahiri, A. Endophytic Diversity of Fraxinus excelsior L. (European Ash) and Its Interaction with the Dieback Pathogen Hymenoscyphus fraxineus. Ph.D. Thesis, University of Dublin, Dublin, Ireland, 2020. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetic analysis using maximum likeli-hood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. Microb. Ecol. 2011, 5, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Murphy, B.R.; Nieto, L.M.; Doohan, F.M.; Hodkinson, T.R. Profundae diversitas: The uncharted genetic diversity in a newly studied group of fungal root endophytes. Mycology 2015, 6, 139–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Ihrmark, K.; Stenlid, J. Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol. 2009, 58, 284–292. [Google Scholar] [CrossRef]
- Kowalski, T.; Łukomska, A. Studies on Fraxinus excelsior L. dieback in Włoszczowa Forest Unit stands. Acta Agrobot. 2005, 59, 429–440. [Google Scholar]
- Parnell, J.; Curtis, T. Webb’s an Irish Flora, 8th ed.; Cork University Press: Cork, Ireland, 2012; pp. 1–560. ISBN 978-185918-478-3. [Google Scholar]
- Trapiello, E.; Schoebel, C.N.; Rigling, D. Fungal community in symptomatic ash leaves in Spain. Balt. For. 2017, 23, 68–73. [Google Scholar]
- González, V.; Tello, M.L. The endophytic mycota associated with Vitis vinifera in central Spain. Fungal Divers. 2011, 47, 29–42. [Google Scholar] [CrossRef]
- Quilluam, R.S.; Jones, D.L. Evidence for host-specificity of culturable fungal root endophytes from the carnivorous plant Pinguicula vulgaris (Common Butterwort). Mycol. Prog. 2012, 11, 583–585. [Google Scholar] [CrossRef]
- Paul, N.C.; Lee, H.B. First record of endophytic Paraconiothyrium brasiliense isolated from Chinese maple leaves in Korea. Korea J. Mycol. 2014, 42, 349–352. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Raja, H.A.; Swenson, D.C.; Agarwal, R.; Deep, G.; Falkinham, J.O., III; Oberlies, N.H. Talarolutins A–D: Meroterpenoids from an endophytic fungal Isolate of Talaromyces minioluteus. Phytochemistry 2016, 126, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Soca-Chafre, G.; Rivera-Orduña, F.N.; Hidalgo-Lara, M.E.; Hernandez-Rodriguez, C.; Marsch, R.; Flores-Cotera, L.B. Molecular phylogeny and paclitaxel screening of fungal endophytes from Taxus globosa. Fungal Biol. 2011, 115, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Hayatgheibi, H. Studies on the Microflora Associated with the SEEDS of European Ash (Fraxinus excelsior) and the Infection Biology of the Pathogen Hymenoscyphus pseudoalbidus Causing Ash Dieback. Master’s Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2013; pp. 1–32. [Google Scholar]
- Cleary, M.R.; Arhipova, N.; Gaitnieks, T.; Stenlid, J.; Vasaitis, R. Natural infection of Fraxinus excelsior seeds by Chalara fraxinea. For. Pathol. 2013, 43, 83–85. [Google Scholar]
- Aveskamp, M.M.; de Gruyter, J.; Woudenberg, J.H.; Verkley, G.J.; Crous, P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 2010, 65, 1–60. [Google Scholar] [CrossRef]


| Sanger Culture Dependent | HTS Culture Independent |
|---|---|
| Acremonium sp. | Alternaria infectoria * |
| Alternaria humuli * | Bacidina flavoleprosa |
| Aureobasidium pullulans | Cadophora orchidicola |
| Aureobasidium sp. | Diplodia mutila |
| Boeremia exigua | Microstroma bacarum |
| Cladosporium sp. | Nematostoma parasiticum |
| Diaporthe rudis | Paraleptosphaeria rumicis |
| Diaporthe viticola | Peniophora cinerea |
| Fusarium lateritium | Phaeosphaeria caricicola * |
| Fusarium oxysporum | Phyllactinia fraxini |
| Fusarium proliferatum | Pseudomicrostroma juglandis |
| Mycosphaerella coacervata | Sclerostagonospora opuntiae |
| Paraloratospora gahniae * | Septoriella phragmitis |
| Parengyodontium album | Septoriella vagans * |
| Phaeosphaeria pontiformis * | Strelitziana eucalypti |
| Phoma sp. | Taphrina antarctica |
| Phomopsis velata | Trimmatostroma cordae |
| Pyronema domesticum | Xanthoria parietina |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lahiri, A.; Murphy, B.R.; Hodkinson, T.R. Assessing Genotypic and Environmental Effects on Endophyte Communities of Fraxinus (Ash) Using Culture Dependent and Independent DNA Sequencing. J. Fungi 2021, 7, 565. https://doi.org/10.3390/jof7070565
Lahiri A, Murphy BR, Hodkinson TR. Assessing Genotypic and Environmental Effects on Endophyte Communities of Fraxinus (Ash) Using Culture Dependent and Independent DNA Sequencing. Journal of Fungi. 2021; 7(7):565. https://doi.org/10.3390/jof7070565
Chicago/Turabian StyleLahiri, Anindita, Brian R. Murphy, and Trevor R. Hodkinson. 2021. "Assessing Genotypic and Environmental Effects on Endophyte Communities of Fraxinus (Ash) Using Culture Dependent and Independent DNA Sequencing" Journal of Fungi 7, no. 7: 565. https://doi.org/10.3390/jof7070565
APA StyleLahiri, A., Murphy, B. R., & Hodkinson, T. R. (2021). Assessing Genotypic and Environmental Effects on Endophyte Communities of Fraxinus (Ash) Using Culture Dependent and Independent DNA Sequencing. Journal of Fungi, 7(7), 565. https://doi.org/10.3390/jof7070565






