Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms
2.2. Identification
2.3. Isolates Growth in Solid Culture Media
2.4. Cellulase Activity
2.5. Xylanase Activity
2.6. Manganese Peroxidase Activity
2.7. Lignin Peroxidase Activity
2.8. Experimental Design
2.9. Statistical Analysis
3. Results
3.1. Organisms
3.2. Identification
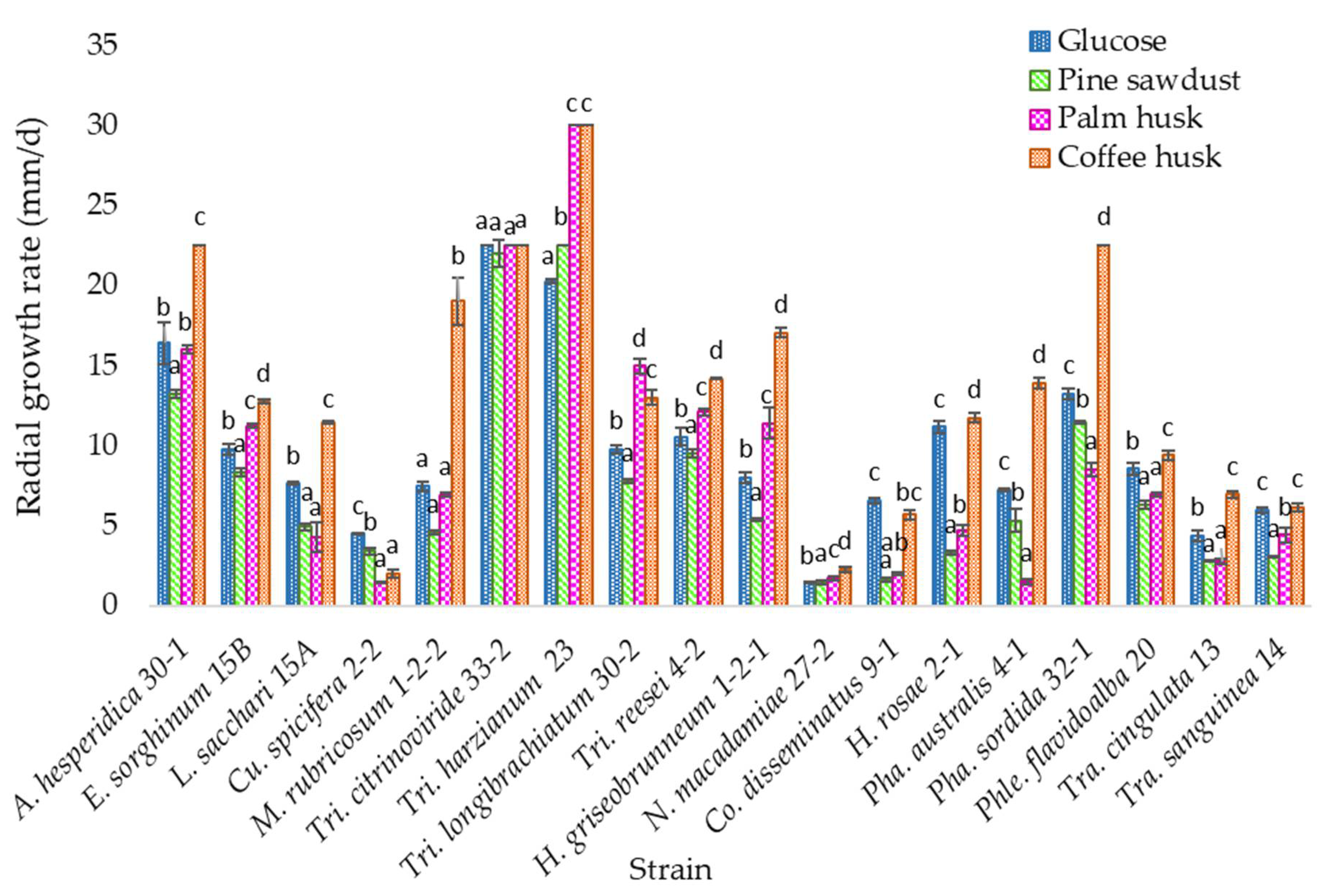
3.3. Solid Media Growth
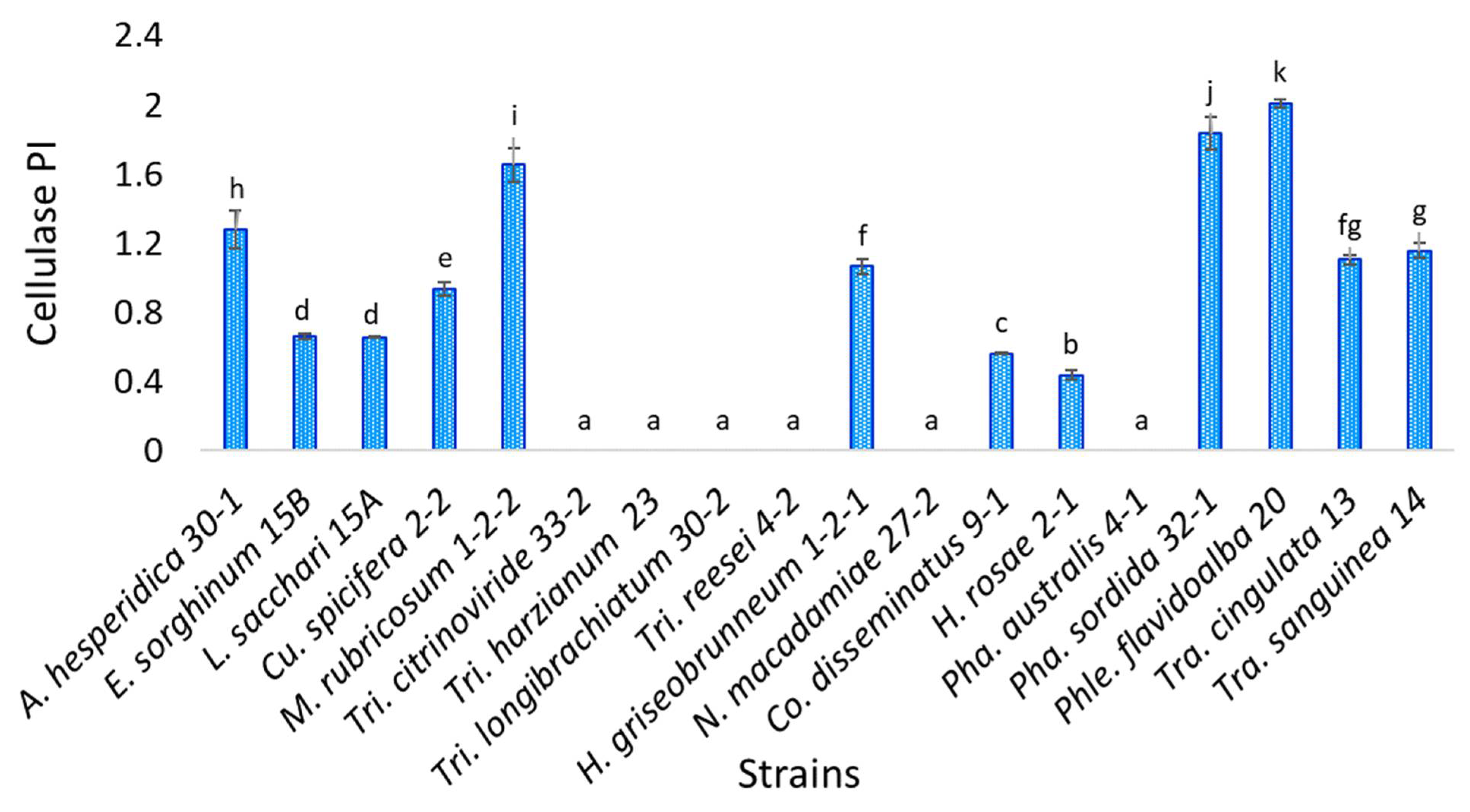
3.4. Cellulase Activity
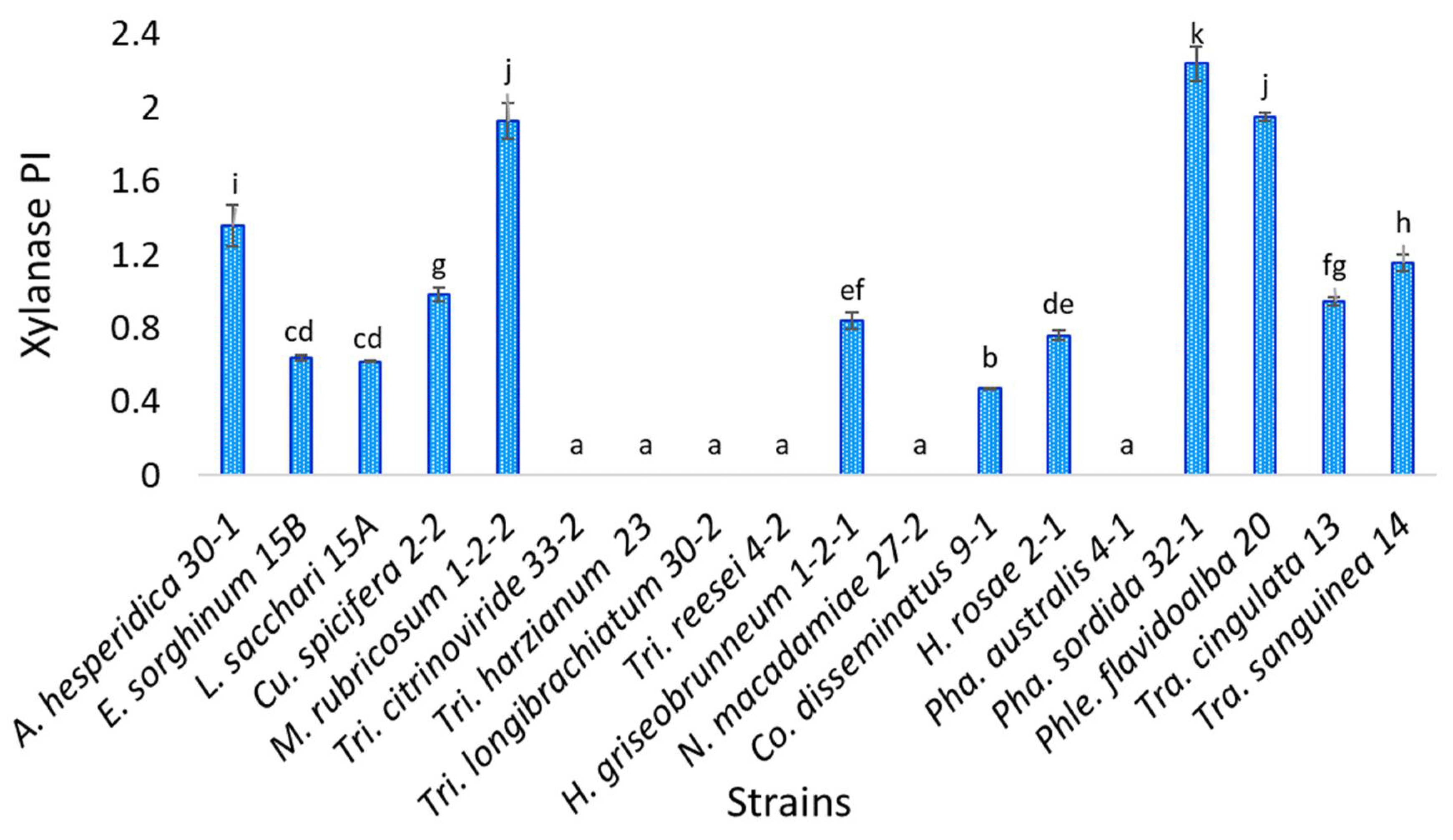
3.5. Xylanase Enzyme Activity
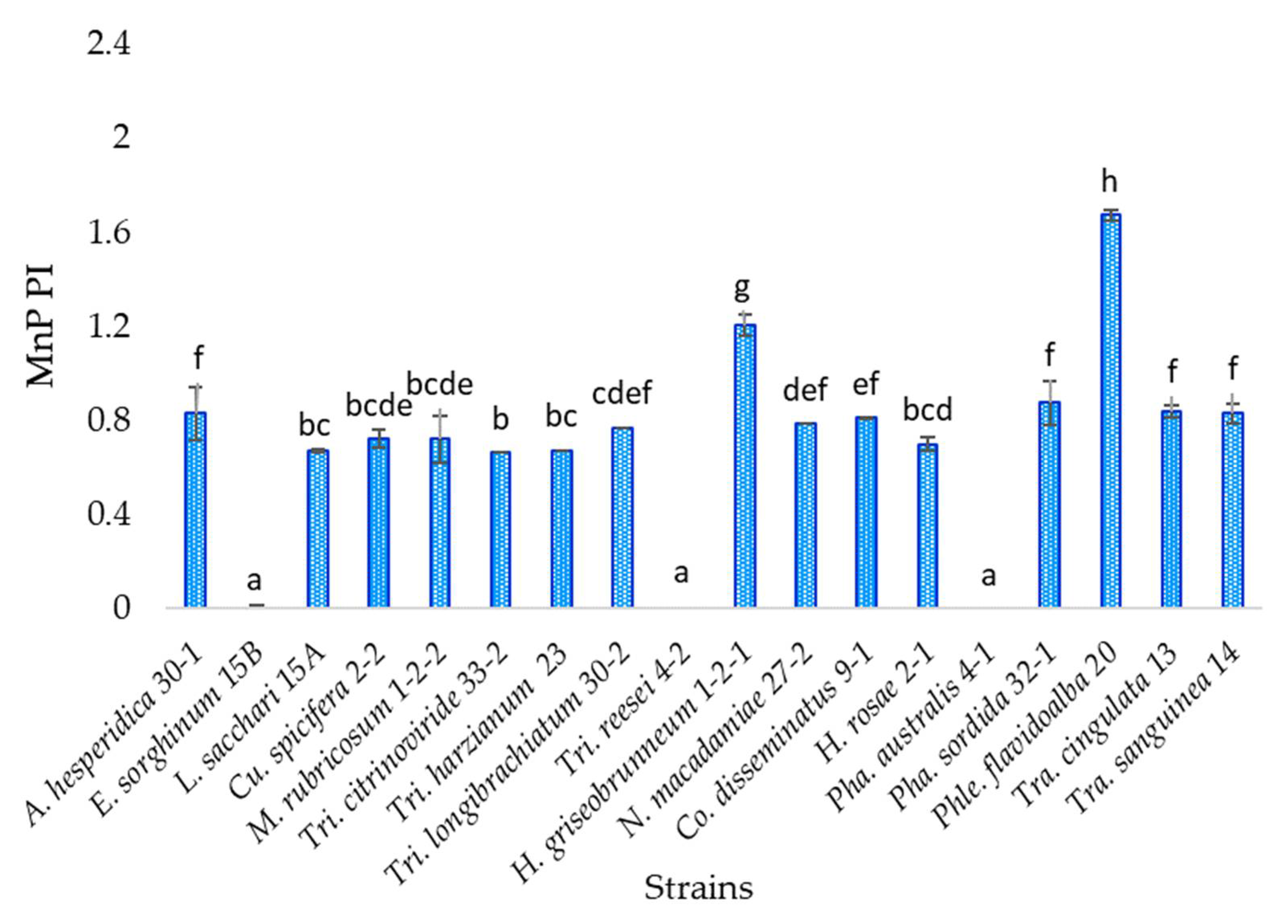
3.6. Manganese Peroxidase Activity
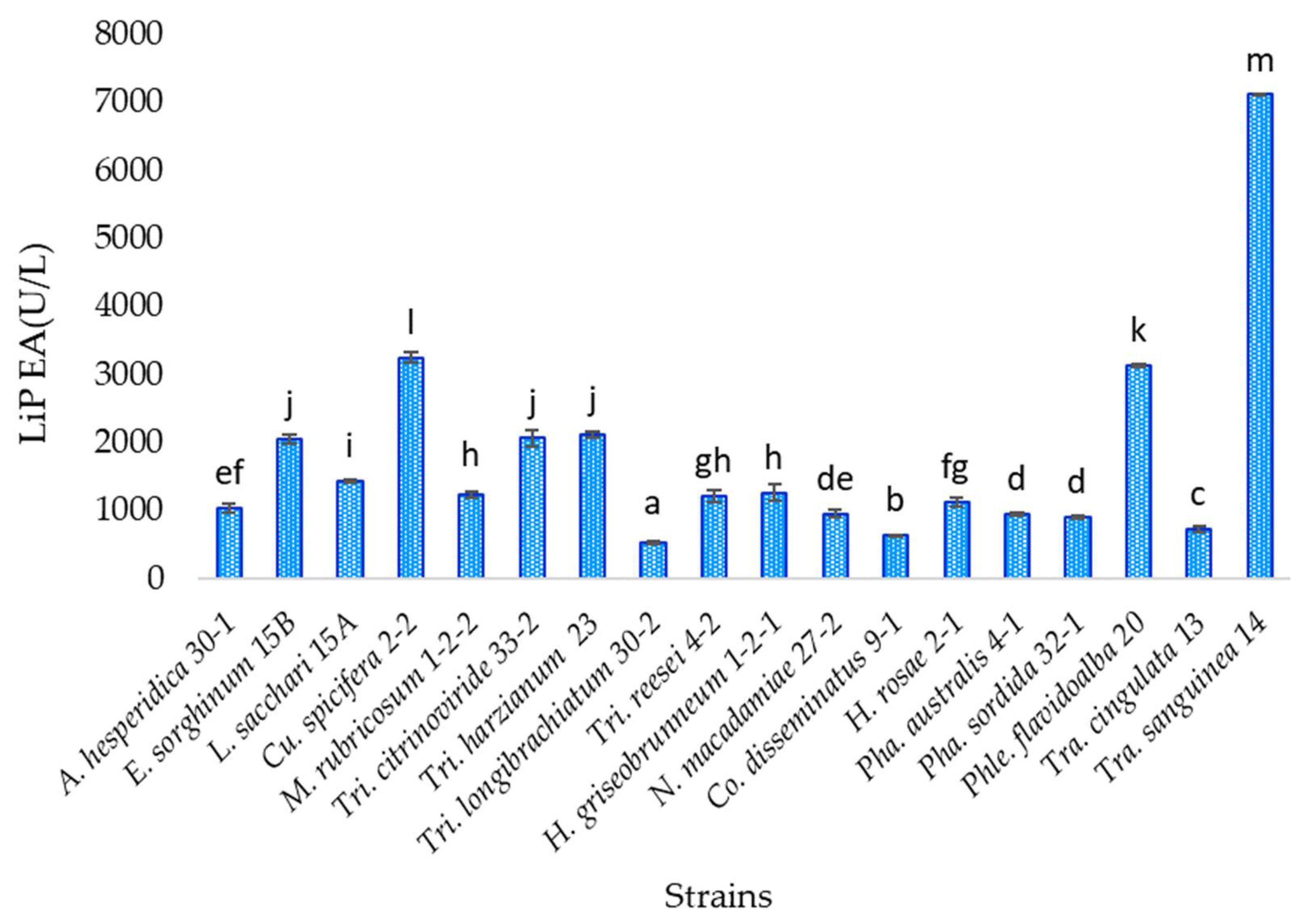
3.7. Lignin Peroxidase Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iram, A.; Cekmecelioglu, D.; Demirci, A. Ideal Feedstock and Fermentation Process Improvements for the Production of Lignocellulolytic Enzymes. Processes 2021, 9, 38. [Google Scholar] [CrossRef]
- Montoya, S.; Patiño, A.; Sánchez, Ó.J. Production of Lignocellulolytic Enzymes and Biomass of Trametes versicolor from Agro-Industrial Residues in a Novel Fixed-Bed Bioreactor with Natural Convection and Forced Aeration at Pilot Scale. Processes 2021, 9, 397. [Google Scholar] [CrossRef]
- Dao, A.T.N.; Smits, M.; Dang, H.T.C.; Brouwer, A.; de Boer, T.E. Elucidating fungal Rigidoporus species FMD21 lignin-modifying enzyme genes and 2,3,7,8-tetrachlorodibenzo-p-dioxin degradation by laccase isozymes. Enzym. Microb. Technol. 2021, 147, 109800. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 23, 110–119. [Google Scholar] [CrossRef]
- Schmidt, O. Wood and Tree Fungi. Biology, Damage, Protection, and Use, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 134–146. [Google Scholar]
- Papinutti, L. Hongos causantes de pudrición blanca: La utilización de sus enzimas ligninolíticas para el desarrollo de tecnologías de biorremediación. Cienc. Nat. 2011, 24, 40–42. [Google Scholar]
- Alshammari, N.; Ameen, F.; AlKahtani, M.D.F.; Stephenson, S. Characterizing the Assemblage of Wood-Decay Fungi in the Forests of Northwest Arkansas. J. Fungi 2021, 7, 309. [Google Scholar] [CrossRef] [PubMed]
- Tanruean, K.; Penkhrue, W.; Kumla, J.; Suwannarach, N.; Lumyong, S. Valorization of LignocellulosicWastes to Produce Phytase and Cellulolytic Enzymes from a Thermophilic Fungus, Thermoascus aurantiacus SL16W, under Semi-Solid State Fermentation. J. Fungi 2021, 7, 286. [Google Scholar] [CrossRef]
- Secretaría del Medio Ambiente e Historia Natural (SEMAHN). Available online: http://www.semahn.chiapas.gob.mx/portal/san_jose/conocenos (accessed on 21 January 2019).
- Chanona-Gómez, F.; Alvarez-Gutiérrez, P.E.; Pérez-Luna, Y.C. Macromycetes of the San José educational park, municipality of Zinacantan, Chiapas, Mexico. Acta Univ. 2019, 29, 1–13. [Google Scholar] [CrossRef]
- Instituto Nacional para el Federalismo y el Desarrollo Municipal. Enciclopedia de los Municipios y Delegaciones de México. Available online: http://www.inafed.gob.mx/work/enciclopedia/EMM07chiapas/municipios/07018a.html (accessed on 4 April 2019).
- Instituto Nacional para el Federalismo y el Desarrollo Municipal. Enciclopedia de los Municipios y Delegaciones de México. Available online: http://www.inafed.gob.mx/work/enciclopedia/EMM07chiapas/municipios/07101a.html (accessed on 4 April 2019).
- Guzmán, G. Identificación de los Hongos Comestibles, Venenosos, Alucinantes y Destructores de la Madera, 1st ed.; Limusa: Monterrey, México, 1977; pp. 435–436. [Google Scholar]
- Ryvarden, L.; Gilbertson, R.L. European Polyporus; Grolands Grafiske Als: Oslo, Norway, 1983; pp. 149–170. [Google Scholar]
- Ryvarden, L.; Gilbertson, R.L. North American Polyporus Vol. I; Grolands Grafiske Als: Oslo, Norway, 1986; pp. 1–437. [Google Scholar]
- Ryvarden, L.; Gilbertson, R.L. North American Polyporus Vol. II; Grolands Grafiske Als: Oslo, Norway, 1987; pp. 438–885. [Google Scholar]
- Ryvarden, L. Chapter 9: Tropical polypores. In Aspects of Tropical Mycology; Simposium of the British Mycological Society, held at the University of Liverpool, Reino Unido; Cambridge University Press: Cambridge, UK, 1991; pp. 149–170. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 1st ed.; Innis, M.A., Gelgard, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database Indexing for Production MegaBLAST Searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Cáceres, M.; Scott, F.; Díaz-Robles, L.; Aroca, G.; Vergara-Fernández, A. Biodegradation of benzo[α]pyrene, toluene, and formaldehyde from the gas phase by a consortium of Rhodococcus erythropolis and Fusarium solani. Appl. Microbiol. Biotechnol. 2017, 101, 6765–6777. [Google Scholar] [CrossRef]
- Gómez, G. Caracterización Bioquímica y molecular de Macromicetos Degradadores de Madera del Rancho El Arco, Cintalapa, Chiapas. Bachelor’s Thesis, Instituto De Ciencias Biológicas, Universidad De Ciencias y Artes De Chiapas, Tuxtla Gutiérrez, Chiapas, México, October 2015. [Google Scholar]
- Rojas-Solís, D.; Santoyo, G. Data on the effect of Pseudomonas stutzeri E25 and Stenotrophomonas maltophilia CR71 culture supernatants on the mycelial growth of Botrytiscinerea. Data Brief 2018, 17, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Amadi, O.C.; Egong, E.J.; Nwagu, T.N.; Okpala, G.; Onwosi, C.O.; Chukwu, G.C.; Okolo, B.N.; Agu, R.C.; Moneke, A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon 2020, 6, e04566. [Google Scholar] [CrossRef] [PubMed]
- Orwa, P.; Mugambi, G.; Wekesa, V.; Mwirichia, R. Isolation of haloalkaliphilic fungi from Lake Magadi in Kenya. Heliyon 2020, 6, e02823. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.S.; Polonio, J.C.; Quecine, M.C.; Almeida, T.T.; Bogas, A.C.; Pamphile, J.A.; Pereira, J.O.; Astolfi-Filho, S.; Azevedo, J.L. Endophytic cultivable bacterial community obtained from the Paullinia cupana seed in Amazonas and Bahia regions and its antagonistic effects against Colletotrichum gloeosporioides. Microb. Pathog. 2016, 98, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Bustamante, S.R.; Mondaca-Fernández, I.; Caro-Reyes, R.B.; Gámez-Gutiérrez, L.A.; De los Santos-Villalobos, S.; Meza-Montenegro, M.M.; Balderas-Cortés, J.J. Selección de cepas productoras de enzimas ligninolíticas nativas del Valle del Yaqui. Nova Sci. 2017, 9, 24–36. [Google Scholar] [CrossRef][Green Version]
- Muñoz, L.D. Evaluación de Enzimas Degradadoras de Lignina Producidas por Aislamientos Fúngicos de Cultivos de Arroz. Bachelor’s Thesis, Facultad de ciencias, Pontificia Universidad Javeriana, Bogotá, Colombia, December 2012. [Google Scholar]
- Páez, M. Determinación de la Actividad Enzimática de Lacasas y Lignina Peroxidasas de Hongos Degradadores de Colorantes Seleccionados para el Tratamiento de Aguas Residuales de la Industria Textil. Bachelor’s Thesis, Departamento de Ciencias de la Vida, Escuela Politécnica del Ejército, Sangolquí, Ecuador, 20 April 2012. [Google Scholar]
- Córdoba, R.; Cultid, G. Estudio Comparativo de la Actividad Enzimática de Lacasa (Lac), Lignina Peroxidasa (LiP) y Manganeso Peroxidasa (MnP) de “Pleurotus ostreatus” Cultivado en Residuos Lignocelulósicos de Raquis de Palma de Aceite, Bagazo de Fique y Pulpa de Café. Bachelor’s Thesis, Facultad de Ciencias exactas y naturales, Universidad de Nariño, San Juan de Pasto, Colombia, January 2015. [Google Scholar]
- Martínez, S.; Nakasone, K.K.; Bettucci, L. Diversity of wood-inhabiting Agaricomycotina on wood of different size classes in riparian forests of Uruguay. Mycoscience 2019, 60, 156–164. [Google Scholar] [CrossRef]
- Arora, A.; Nandal, P.; Singh, J.; Verma, M.L. Nanobiotechnological advancements in lignocellulosic biomass pretreatment. Mater. Sci. Energy Technol. 2020, 3, 308–318. [Google Scholar] [CrossRef]
- Mallerman, J.; Itria, R.; Babay, P.; Saparrat, M.; Levin, L. Biodegradation of nonylphenol polyethoxylates by litter-basidiomycetous fungi. J. Environ. Chem. Eng. 2019, 7, 103316. [Google Scholar] [CrossRef]
- Dao, A.T.N.; Vonck, J.; Janssens, T.K.S.; Dang, H.T.C.; Brouwer, A.; de Boer, T.E. Screening white-rot fungi for bioremediation potential of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Ind. Crop. Prod. 2019, 128, 153–161. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Q.; Sun, X.; Chen, D.; Insam, H.; Koide, R.T.; Zhang, S. Effects of mixed-species litter on bacterial and fungal lignocellulose degradation functions during litter decomposition. Soil Biol. Biochem. 2020, 141, 107690. [Google Scholar] [CrossRef]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.G.; Chu, H. Microbial resistance promotes plant production in a four-decade nutrient fertilization experiment. Soil Biol. Biochem. 2020, 141, 107679. [Google Scholar] [CrossRef]
- Segaran, G.; Sathiavelu, M. Fungal endophytes: A potent biocontrol agent and a bioactive metabolites reservoir. Biocatal. Agric. Biotechnol. 2019, 21, 101284. [Google Scholar] [CrossRef]
- Haridas, S.; Albert, R.; Binder, M.; Bloem, J.; LaButti, K.; Salamov, A.; Andreopoulos, B.; Baker, S.E.; Barry, K.; Bills, G.; et al. 101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens. Stud. Mycol. 2020, 96, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Heleria, C.G. Macromicetos de “Laguna Verde”, Municipio de Coapilla, Chiapas. Bachelor’s Thesis, Instituto de Ciencias Biológicas, Universidad de Ciencias y Artes de Chiapas, Tuxtla Gutiérrez, Chiapas, México, 2 February 2018. [Google Scholar]
- García, A. Evaluación de la Cascarilla de Café para Utilizarse Como Sustrato en Cultivo Sin Suelo de Hortalizas. Master’s Thesis, Instituto Politécnico Nacional, Oaxaca, México, January 2008. Available online: https://tesis.ipn.mx/jspui/handle/123456789/152 (accessed on 19 February 2021).
- Servicio de Información Agroalimentaria y Pesquera (SIAP). Café: Datos Preliminares a 2017 Indican una Producción Nacional de 839 Mil Toneladas. Available online: https://www.gob.mx/siap/articulos/cafe-datos-preliminares-a-2017-indican-una-produccion-nacional-de-839-mil-toneladas (accessed on 19 February 2021).
- Secretaría de Agricultura y Desarrollo Rural. El Café Una Producción en Manos Sabias. Available online: https://www.gob.mx/agricultura/articulos/el-cafe-una-produccion-en-manos-sabias?idiom=es (accessed on 19 February 2021).
- Servicio de Información Agroalimentaria y Pesquera (SIAP). Palma Africana o de Aceite en México: Cultivo Tropical Aceitero. Available online: https://www.gob.mx/siap/articulos/palma-africana-o-de-aceite-en-mexico-cultivo-tropical-aceitero?idiom=es (accessed on 19 February 2021).
- Al-Zaban, M.I.; AlHarbi, M.A.; Mahmoud, M.A. Hydrocarbon biodegradation and transcriptome responses of cellulase, peroxidase, and laccase encoding genes inhabiting rhizospheric fungal isolates. Saudi J. Biol. Sci. 2021, 28, 2083–2090. [Google Scholar] [CrossRef]
| Phylum | Class | Order | Family | Name | Collection Site | Record | Rot Type |
|---|---|---|---|---|---|---|---|
| Ascomycota | Dothideomycetes | Botryosphaeriales | Aplosporellaceae | Aplosporella hesperidica TecNM-ITTG L30-1-19 | LVEP 1 | NR 4 | BR 5 |
| Pleosporales | Didymellaceae | Epicoccum sorghinum TecNM-ITTG L15B-19 | SJEP 2 | NR 4 | ND 6 | ||
| Leptosphaeriaceae | Leptosphaeria spegazzinii TecNM-ITTG L15A-19 | SJEP 2 | NR 4 | ND 6 | |||
| Pleosporaceae | Curvularia spicifera TecNM-ITTG L2-2-19 | SJEP 2 | NR 4 | ND 6 | |||
| Valsariales | Valsariaceae | Myrmaecium rubricosum TecNM-ITTG L1-2-2-19 | SJEP 2 | NR 4 | ND 6 | ||
| Sordariomycetes | Hypocreales | Hypocreaceae | Trichoderma citrinoviride TecNM-ITTG L33-2-19 | LVEP 1 | NR 4 | ND 6 | |
| Trichoderma harzianum TecNM-ITTG L23-19 | LVEP 1 | NR 4 | ND 6 | ||||
| Trichoderma longibrachiatum TecNM-ITTG L30-2-19 | LVEP 1 | NR 4 | ND 6 | ||||
| Trichoderma reesei TecNM-ITTG L4-2-19 | SJEP 2 | NR 4 | ND 6 | ||||
| Xylariales | Hypoxylaceae | Hypoxylon griseobrunneum TecNM-ITTG L1-2-1-19 | SJEP 2 | NR 4 | ND 6 | ||
| Sporocadaceae | Neopestalotiopsis macadamiae TecNM-ITTG L27-2-19 | LVEP 1 | NR 4 | WR 7 | |||
| Basidiomycota | Agaricomycetes | Agaricales | Psathyrellaceae | Coprinellus disseminatus TecNM-ITTG L9-1-19 | SJEP 2 | - | WR 7 |
| Polyporales | Phanerochaetaceae | Hyphodermella rosae TecNM-ITTG L2-1-19 | SJEP 2 | NR 4 | ND 6 | ||
| Phanerochaete australis TecNM-ITTG L4-1-19 | SJEP 2 | NR 4 | WR 7 | ||||
| Phanerochaete sordida TecNM-ITTG L32-1-19 | LVEP 1 | NR 4 | WR 7 | ||||
| Phlebiopsis flavidoalba TecNM-ITTG L20-19 | LVEP 1 | NR 4 | WR 7 | ||||
| Polyporaceae | Trametes cingulata TecNM-ITTG L13-19 | TecNM-ITTG 3 | NR 4 | WR 7 | |||
| Trametes sanguinea TecNM-ITTG L14-19 | TecNM-ITTG 3 | - | WR 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Corzo, L.D.; Álvarez-Gutiérrez, P.E.; Meza-Gordillo, R.; Villalobos-Maldonado, J.J.; Enciso-Pinto, S.; Enciso-Sáenz, S. Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material. J. Fungi 2021, 7, 450. https://doi.org/10.3390/jof7060450
Sánchez-Corzo LD, Álvarez-Gutiérrez PE, Meza-Gordillo R, Villalobos-Maldonado JJ, Enciso-Pinto S, Enciso-Sáenz S. Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material. Journal of Fungi. 2021; 7(6):450. https://doi.org/10.3390/jof7060450
Chicago/Turabian StyleSánchez-Corzo, Lina Dafne, Peggy Elizabeth Álvarez-Gutiérrez, Rocío Meza-Gordillo, Juan José Villalobos-Maldonado, Sofía Enciso-Pinto, and Samuel Enciso-Sáenz. 2021. "Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material" Journal of Fungi 7, no. 6: 450. https://doi.org/10.3390/jof7060450
APA StyleSánchez-Corzo, L. D., Álvarez-Gutiérrez, P. E., Meza-Gordillo, R., Villalobos-Maldonado, J. J., Enciso-Pinto, S., & Enciso-Sáenz, S. (2021). Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material. Journal of Fungi, 7(6), 450. https://doi.org/10.3390/jof7060450







