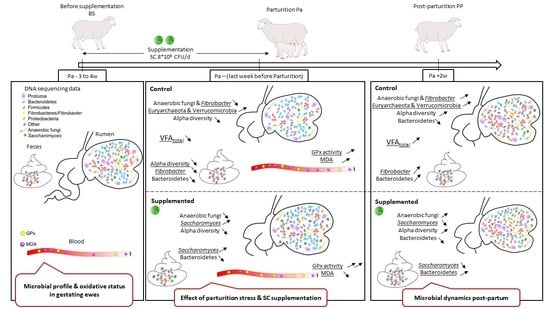
Changes in Digestive Microbiota, Rumen Fermentations and Oxidative Stress around Parturition Are Alleviated by Live Yeast Feed Supplementation to Gestating Ewes
Abstract
1. Introduction
2. Materials and Methods
2.1. Diets and Animals
2.2. Sample Collection
2.3. Measured Parameters
2.4. Statistical Analyses
3. Results
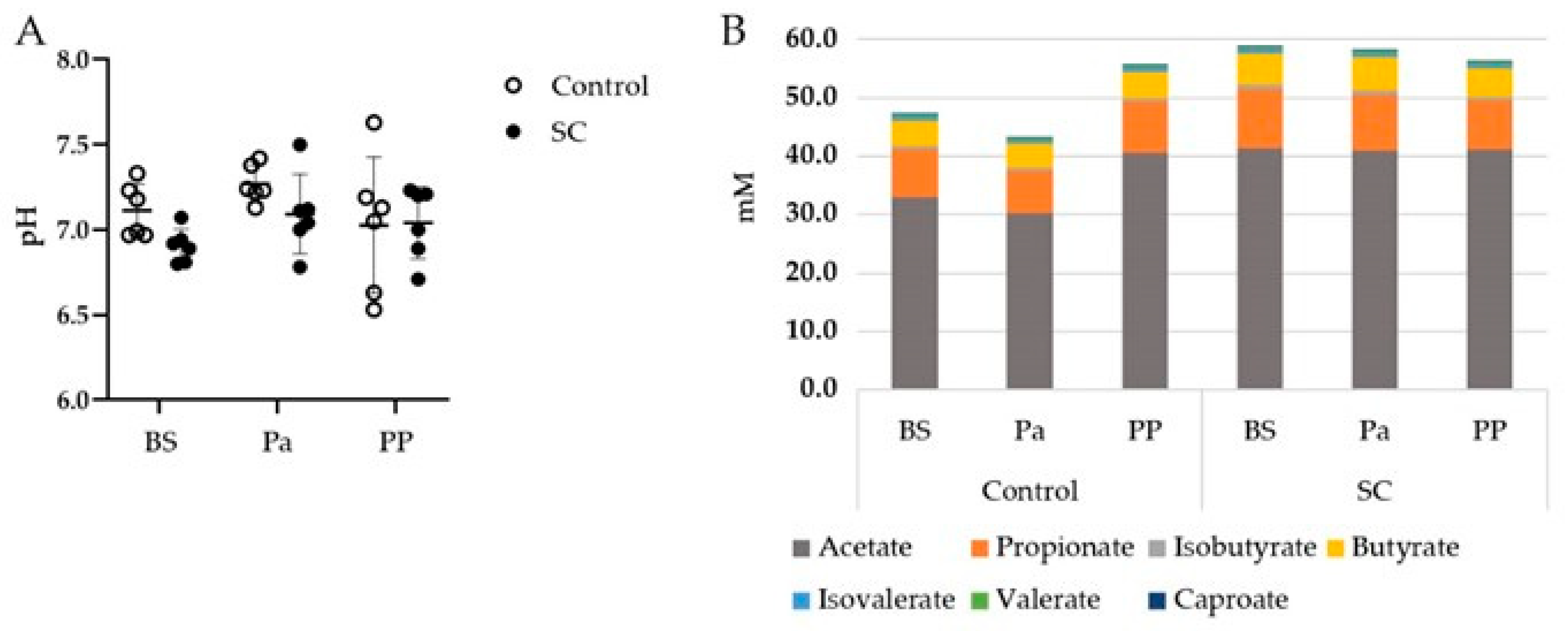
3.1. Rumen pH and VFAs
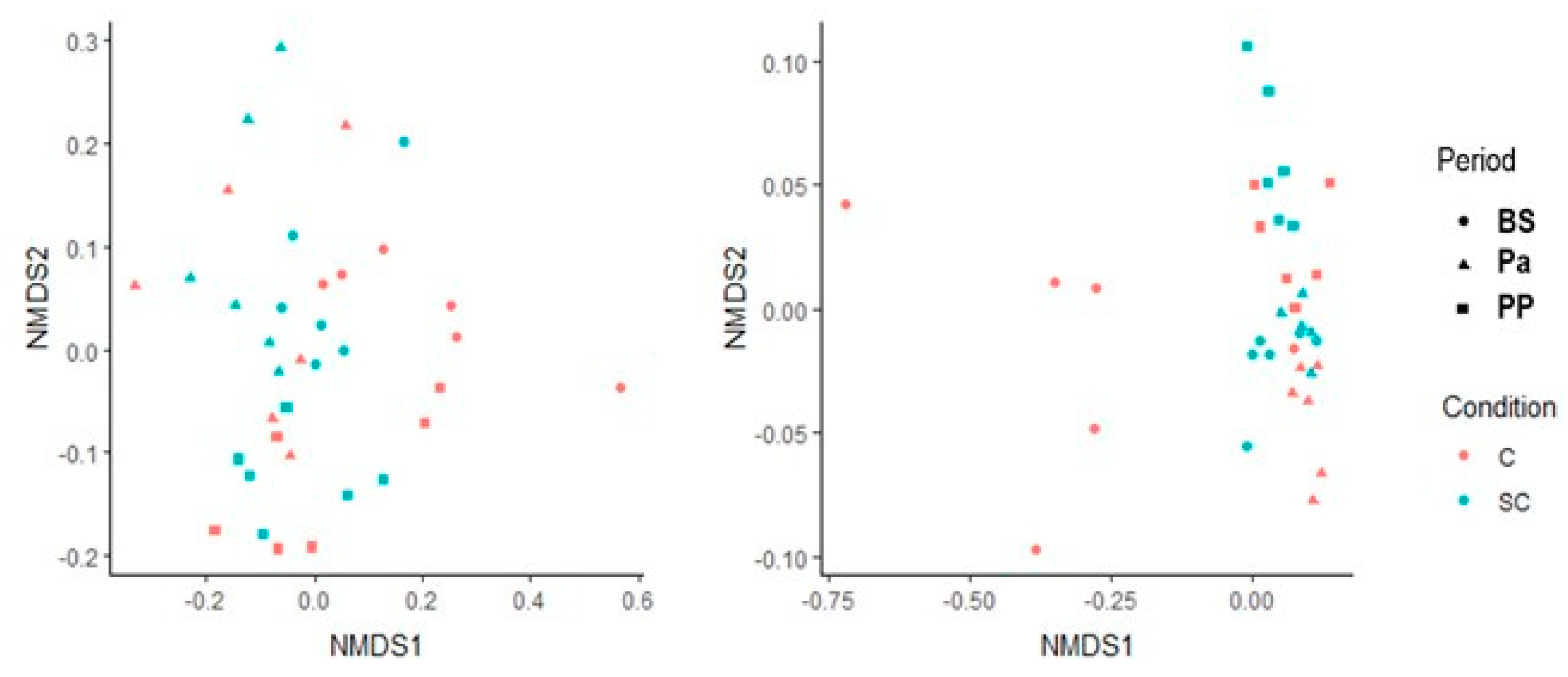
3.2. Rumen and Feces Microbiota
3.2.1. qPCR Results
3.2.2. 16S-DNA Sequencing Results
- Differential analysis of OTUs
3.2.3. 18S-DNA Sequencing Results
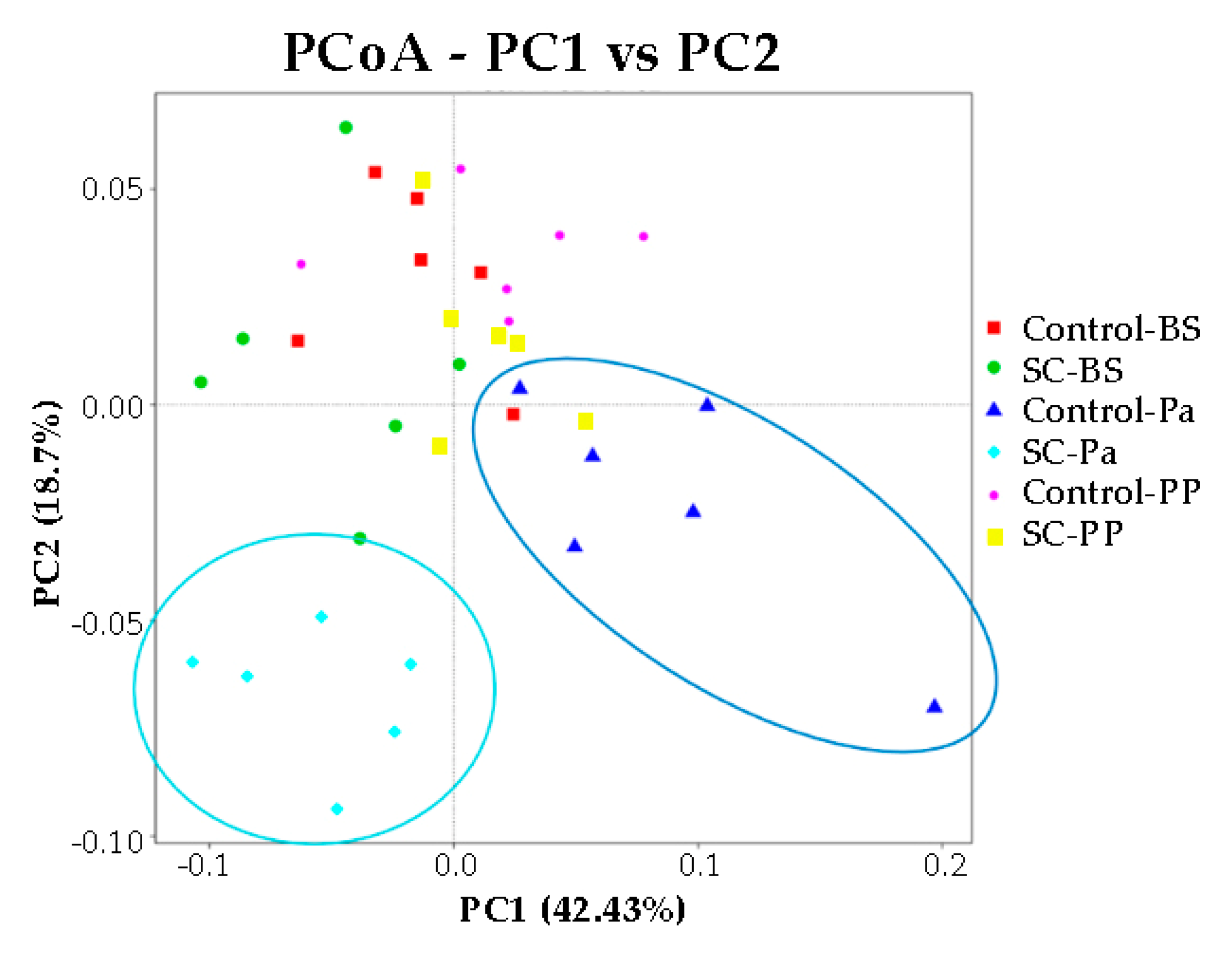
- Alpha and beta diversity measures
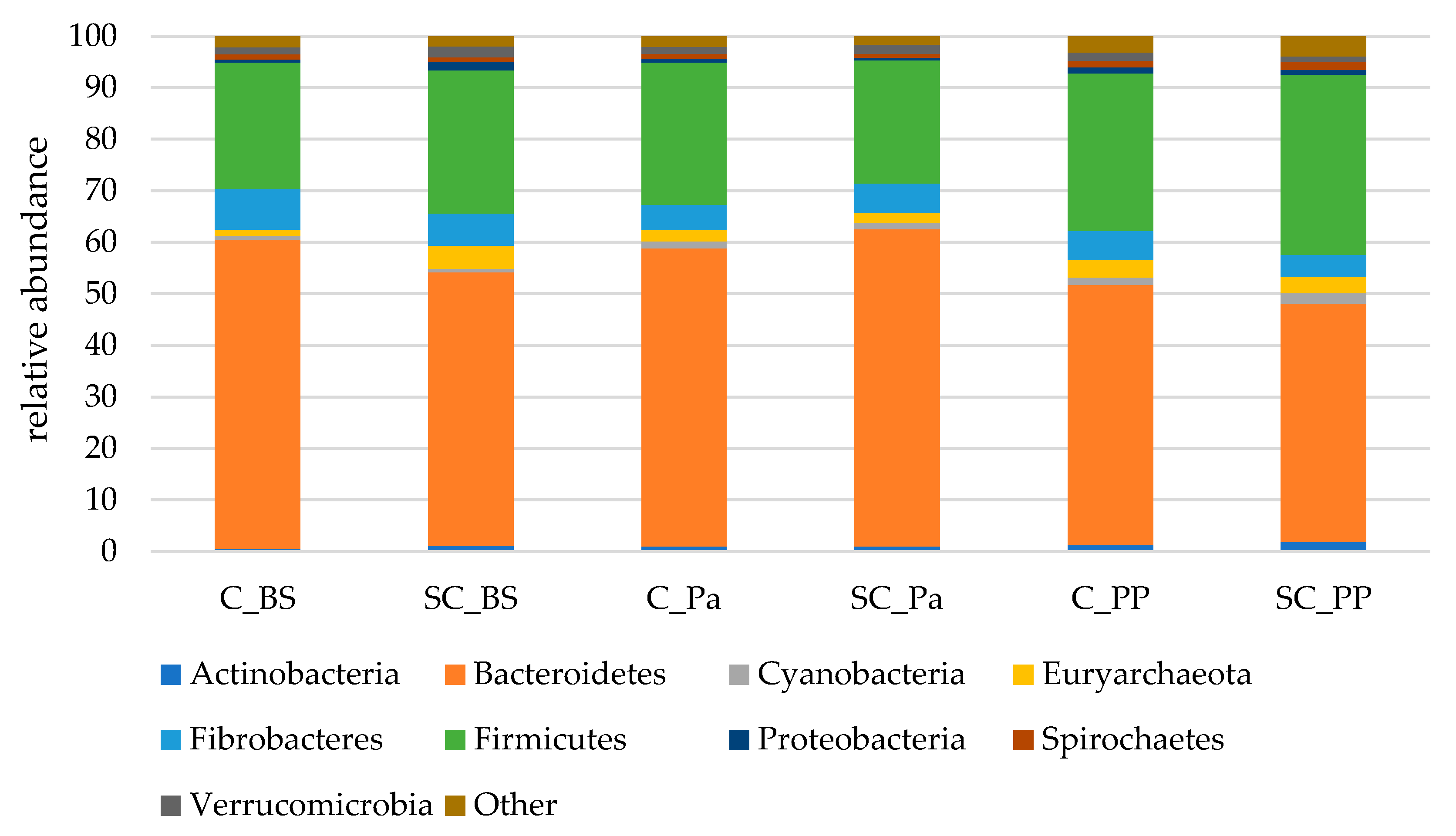
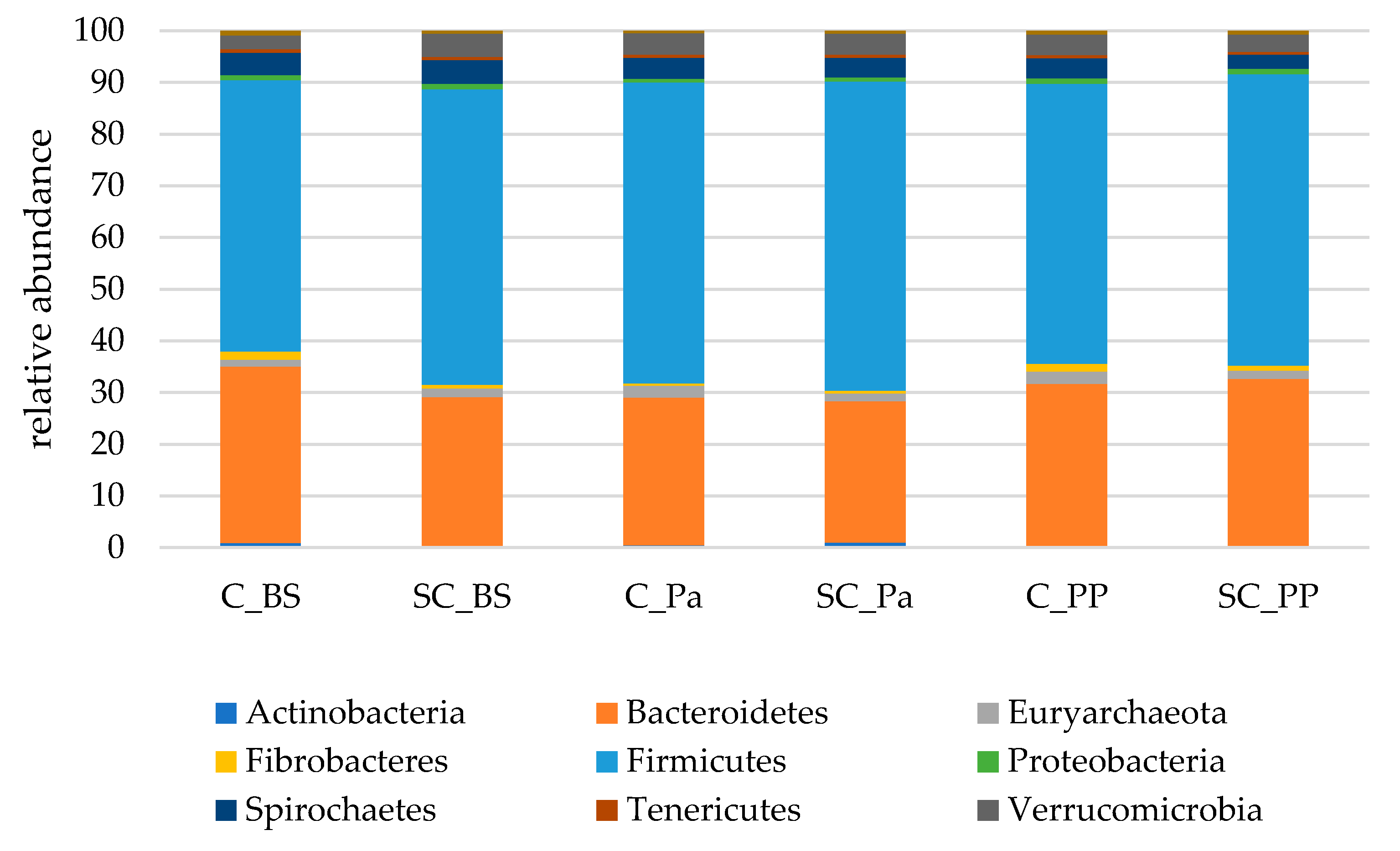
- Relative abundance of the main taxonomic groups
- Differential analysis of OTUs
3.3. Blood Biomarkers
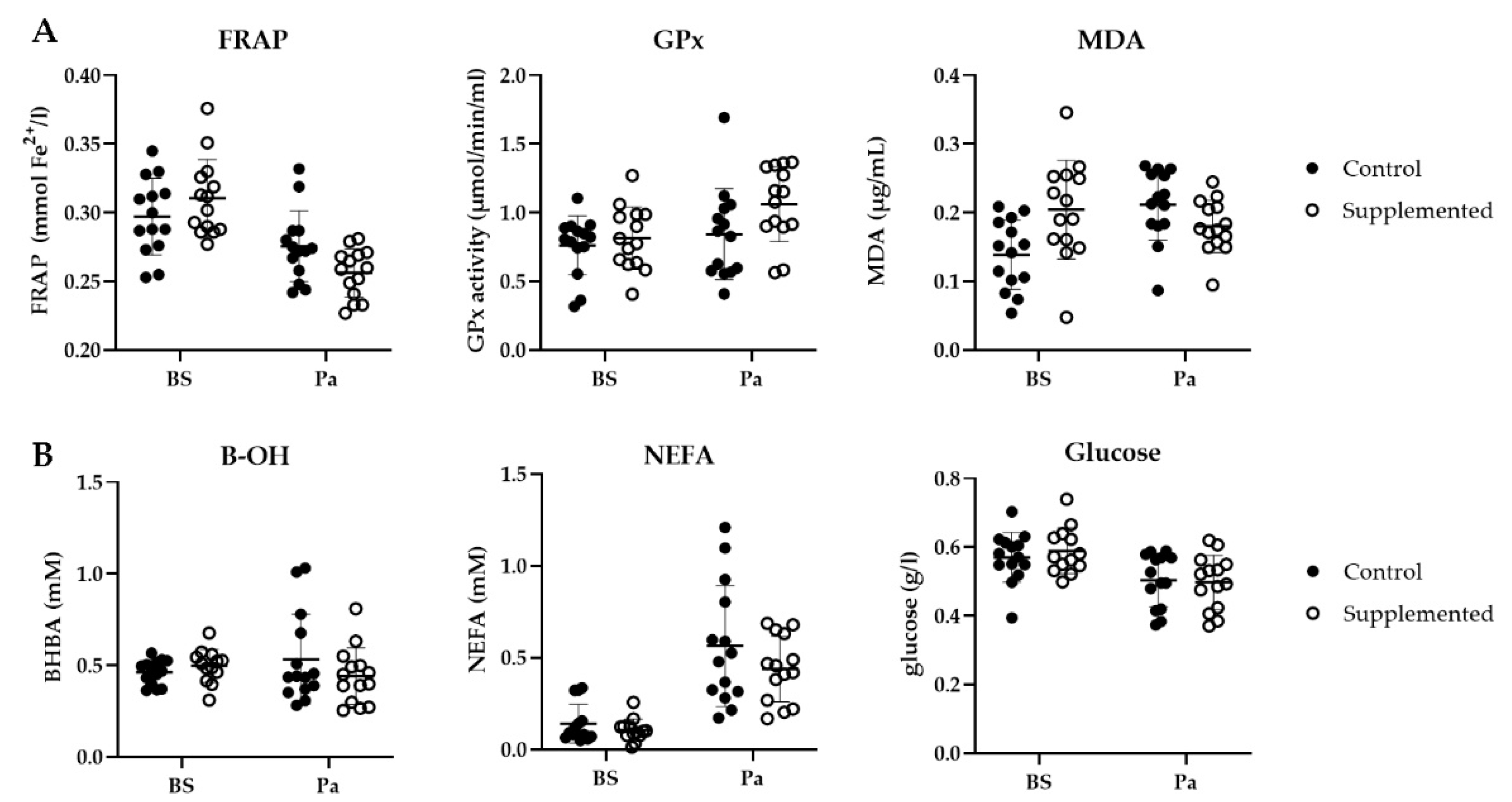
3.3.1. Oxidative Stress Blood Markers
3.3.2. Metabolic Status Blood Markers
3.4. Lambs Birth Weight
4. Discussion
4.1. What Was the Microbial Profile in Gestating Ewes?
4.2. What Happened to the Digestive Microbiota and Activity of Control Ewes Around Parturition?
4.3. What Was the Impact of Parturition on Oxidative and Metabolic Status of the Control Ewes?
4.4. What Were the Effects of S. Cerevisiae I-1077 Supplementation around Parturition?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abuelo, A.; Hernández, J.; Benedito, J.L.; Castillo, C. Redox Biology in Transition Periods of Dairy Cattle: Role in the Health of Periparturient and Neonatal Animals. Antioxidants 2019, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Castellano, L.; Almeida, A.; Castro, N.; Arguello, A. The Colostrum Proteome, Ruminant Nutrition and Immunity: A Review. Curr. Protein Pept. Sci. 2014, 15, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Yeoman, C.J.; Ishaq, S.L.; Bichi, E.; Olivo, S.K.; Lowe, J.; Aldridge, B.M. Biogeographical Differences in the Influence of Maternal Microbial Sources on the Early Successional Development of the Bovine Neonatal Gastrointestinal Tract. Sci. Rep. 2018, 8, 3197. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.; López-García, A.; González-Recio, O.; Elcoso, G.; Fàbregas, F.; Chaucheyras-Durand, F.; Castex, M. Changes in the Rumen and Colon Microbiota and Effects of Live Yeast Dietary Supplementation during the Transition from the Dry Period to Lactation of Dairy Cows. J. Dairy Sci. 2019, 102, 6180–6198. [Google Scholar] [CrossRef] [PubMed]
- Pitta, D.W.; Indugu, N.; Baker, L.; Vecchiarelli, B.; Attwood, G. Symposium Review: Understanding Diet-Microbe Interactions to Enhance Productivity of Dairy Cows. J. Dairy Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Kristensen, L.; Difford, G.F.; Poulsen, M.; Noel, S.J.; Abu Al-Soud, W.; Sørensen, S.J.; Lassen, J.; Løvendahl, P.; Højberg, O. Changes in Rumen Bacterial and Archaeal Communities over the Transition Period in Primiparous Holstein Dairy Cows. J. Dairy Sci. 2018, 101, 9847–9862. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.; Guasch, I.; Elcoso, G.; Chaucheyras-Durand, F.; Castex, M.; Fàbregas, F.; Garcia-Fruitos, E.; Aris, A. Changes in Gene Expression in the Rumen and Colon Epithelia during the Dry Period through Lactation of Dairy Cows and Effects of Live Yeast Supplementation. J. Dairy Sci. 2018, 101, 2631–2640. [Google Scholar] [CrossRef] [PubMed]
- Abaker, J.A.; Xu, T.L.; Jin, D.; Chang, G.J.; Zhang, K.; Shen, X.Z. Lipopolysaccharide Derived from the Digestive Tract Provokes Oxidative Stress in the Liver of Dairy Cows Fed a High-Grain Diet. J. Dairy Sci. 2017, 100, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Golder, H.M.; Lean, I.J.; Rabiee, A.R.; King, R.; Celi, P. Effects of Grain, Fructose, and Histidine Feeding on Endotoxin and Oxidative Stress Measures in Dairy Heifers. J. Dairy Sci. 2013, 96, 7881–7891. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Xue, Y.; Guo, C.; Liu, J.; Mao, S. The Response of Ruminal Fermentation, Epithelium-Associated Microbiota, and Epithelial Barrier Function to Severe Feed Restriction in Pregnant Ewes. J. Anim. Sci. 2018, 96, 4293–4305. [Google Scholar] [CrossRef]
- Abuelo, A.; Hernández, J.; Benedito, J.L.; Castillo, C. The Importance of the Oxidative Status of Dairy Cattle in the Periparturient Period: Revisiting Antioxidant Supplementation. J. Anim. Physiol. Anim. Nutr. 2015, 99, 1003–1016. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Walker, N.D.; Bach, A. Effects of Active Dry Yeasts on the Rumen Microbial Ecosystem: Past, Present and Future. Anim. Feed Sci. Technol. 2008, 145, 5–26. [Google Scholar] [CrossRef]
- Jiang, Y.; Ogunade, I.M.; Qi, S.; Hackmann, T.J.; Staples, C.R.; Adesogan, A.T. Effects of the Dose and Viability of Saccharomyces Cerevisiae. 1. Diversity of Ruminal Microbes as Analyzed by Illumina MiSeq Sequencing and Quantitative PCR. J. Dairy Sci. 2017, 100, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Terré, M.; Maynou, G.; Bach, A.; Gauthier, M. Effect of Saccharomyces Cerevisiae CNCM I-1077 Supplementation on Performance and Rumen Microbiota of Dairy Calves. Prof. Anim. Sci. 2015, 31, 153–158. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, X.; Zou, Y.; Yang, Z.; Li, S.; Cao, Z. Changes in Feed Intake, Nutrient Digestion, Plasma Metabolites, and Oxidative Stress Parameters in Dairy Cows with Subacute Ruminal Acidosis and Its Regulation with Pelleted Beet Pulp. J. Anim. Sci. Biotechnol. 2013, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Villot, C.; Meunier, B.; Bodin, J.; Martin, C.; Silberberg, M. Relative Reticulo-Rumen PH Indicators for Subacute Ruminal Acidosis Detection in Dairy Cows. Anim. Int. J. Anim. Biosci. 2018, 12, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Mosoni, P.; Chaucheyras-Durand, F.; Béra-Maillet, C.; Forano, E. Quantification by Real-Time PCR of Cellulolytic Bacteria in the Rumen of Sheep after Supplementation of a Forage Diet with Readily Fermentable Carbohydrates: Effect of a Yeast Additive. J. Appl. Microbiol. 2007, 103, 2676–2685. [Google Scholar] [CrossRef]
- Bayat, A.R.; Kairenius, P.; Stefański, T.; Leskinen, H.; Comtet-Marre, S.; Forano, E.; Chaucheyras-Durand, F.; Shingfield, K.J. Effect of Camelina Oil or Live Yeasts (Saccharomyces Cerevisiae) on Ruminal Methane Production, Rumen Fermentation, and Milk Fatty Acid Composition in Lactating Cows Fed Grass Silage Diets. J. Dairy Sci. 2015, 98, 3166–3181. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinforma. Oxf. Engl. 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S RRNA Sequence Formation and Detection in Sanger and 454-Pyrosequenced PCR Amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Utilities for Microbiome Analysis; Release 3.12; Microbiome R Package; Bioconductor. 2012. Available online: https://bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 6 March 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.; Strain, J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996. [Google Scholar] [CrossRef]
- Agergaard, N.; Jensen, P.T. Procedure for Blood Glutathione Peroxidase Determination in Cattle and Swine. Acta Vet. Scand. 1982, 23, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Delosière, M.; Thomas, A.; Terlouw, C.; Durand, D. Plasma Indicators of Bovine Health: Impacts of Diet Supplementations and Pre-Slaughter Stress. Data Brief 2018, 21, 2496–2503. [Google Scholar] [CrossRef]
- Agarwal, R.; Chase, S.D. Rapid, Fluorimetric–Liquid Chromatographic Determination of Malondialdehyde in Biological Samples. J. Chromatogr. B 2002, 775, 121–126. [Google Scholar] [CrossRef]
- Van Breukelen, G.J.P. ANCOVA versus Change from Baseline: More Power in Randomized Studies, More Bias in Nonrandomized Studies [Corrected]. J. Clin. Epidemiol. 2006, 59, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; Tun, H.M.; Cardoso, F.C.; Plaizier, J.C.; Khafipour, E.; Loor, J.J. Linking Peripartal Dynamics of Ruminal Microbiota to Dietary Changes and Production Parameters. Front. Microbiol. 2016, 7, 2143. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Danesh Mesgaran, M.; Derakhshani, H.; Golder, H.; Khafipour, E.; Kleen, J.L.; Lean, I.; Loor, J.; Penner, G.; Zebeli, Q. Review: Enhancing Gastrointestinal Health in Dairy Cows. Anim. Int. J. Anim. Biosci. 2018, 12, s399–s418. [Google Scholar] [CrossRef]
- Sylvester, J.T.; Karnati, S.K.R.; Dehority, B.A.; Morrison, M.; Smith, G.L.; St-Pierre, N.R.; Firkins, J.L. Rumen Ciliated Protozoa Decrease Generation Time and Adjust 18S Ribosomal DNA Copies to Adapt to Decreased Transfer Interval, Starvation, and Monensin. J. Dairy Sci. 2009, 92, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Firkins, J.L.; Yu, Z.; Park, T.; Plank, J.E. Extending Burk Dehority’s Perspectives on the Role of Ciliate Protozoa in the Rumen. Front. Microbiol. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Silberberg, M.; Chaucheyras-Durand, F.; Commun, L.; Mialon, M.M.; Monteils, V.; Mosoni, P.; Morgavi, D.P.; Martin, C. Repeated Acidosis Challenges and Live Yeast Supplementation Shape Rumen Microbiota and Fermentations and Modulate Inflammatory Status in Sheep. Anim. Int. J. Anim. Biosci. 2013, 7, 1910–1920. [Google Scholar] [CrossRef]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef] [PubMed]
- Dieho, K.; van den Bogert, B.; Henderson, G.; Bannink, A.; Ramiro-Garcia, J.; Smidt, H.; Dijkstra, J. Changes in Rumen Microbiota Composition and in Situ Degradation Kinetics during the Dry Period and Early Lactation as Affected by Rate of Increase of Concentrate Allowance. J. Dairy Sci. 2017, 100, 2695–2710. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. Islands in the Stream: From Individual to Communal Fiber Degradation in the Rumen Ecosystem. FEMS Microbiol. Rev. 2019, 43, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Raut, M.P.; Couto, N.; Karunakaran, E.; Biggs, C.A.; Wright, P.C. Deciphering the Unique Cellulose Degradation Mechanism of the Ruminal Bacterium Fibrobacter Succinogenes S85. Sci. Rep. 2019, 9, 16542. [Google Scholar] [CrossRef]
- Comtet-Marre, S.; Parisot, N.; Lepercq, P.; Chaucheyras-Durand, F.; Mosoni, P.; Peyretaillade, E.; Bayat, A.R.; Shingfield, K.J.; Peyret, P.; Forano, E. Metatranscriptomics Reveals the Active Bacterial and Eukaryotic Fibrolytic Communities in the Rumen of Dairy Cow Fed a Mixed Diet. Front. Microbiol. 2017, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Chaucheyras-Durand, F.; Ameilbonne, A.; Auffret, P.; Bernard, M.; Mialon, M.-M.; Dunière, L.; Forano, E. Supplementation of Live Yeast Based Feed Additive in Early Life Promotes Rumen Microbial Colonization and Fibrolytic Potential in Lambs. Sci. Rep. 2019, 9, 19216. [Google Scholar] [CrossRef] [PubMed]
- Mosoni, P.; Fonty, G.; Gouet, P. Competition between Ruminal Cellulolytic Bacteria for Adhesion to Cellulose. Curr. Microbiol. 1997, 35, 44–47. [Google Scholar] [CrossRef][Green Version]
- Shinkai, T.; Ohji, R.; Matsumoto, N.; Kobayashi, Y. Fibrolytic Capabilities of Ruminal Bacterium Fibrobacter Succinogenes in Relation to Its Phylogenetic Grouping. FEMS Microbiol. Lett. 2009, 294, 183–190. [Google Scholar] [CrossRef]
- Suen, G.; Stevenson, D.M.; Bruce, D.C.; Chertkov, O.; Copeland, A.; Cheng, J.-F.; Detter, C.; Detter, J.C.; Goodwin, L.A.; Han, C.S.; et al. Complete Genome of the Cellulolytic Ruminal Bacterium Ruminococcus Albus 7. J. Bacteriol. 2011, 193, 5574–5575. [Google Scholar] [CrossRef] [PubMed]
- Mousaie, A.; Valizadeh, R.; Chamsaz, M. Selenium-Methionine and Chromium-Methionine Supplementation of Sheep around Parturition: Impacts on Dam and Offspring Performance. Arch. Anim. Nutr. 2017, 71, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Castillo, C.; Hernández, J.; Bravo, A.; Lopez-Alonso, L.; Pereira, V.; Benedito, J. Oxidative Status during Late Pregnancy and Early Lactation in Dairy Cows. Vet. J. Lond. Engl. 1997 2005, 2, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Aydin, I.; Bulbul, T.; Polat, E.S.; Yazar, E. Serum Antioxidant Status and Adenosine Deaminase Activity during the Gestational Period of Sheep. Rev. Méd Vét 2010, 161, 479–484. [Google Scholar]
- Mohebbi-Fani, M.; Mirzaei, A.; Nazifi, S.; Shabbooie, Z. Changes of Vitamins A, E, and C and Lipid Peroxidation Status of Breeding and Pregnant Sheep during Dry Seasons on Medium-to-Low Quality Forages. Trop. Anim. Health Prod. 2012, 44, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.W. Regulation of Organic Nutrient Metabolism during Transition from Late Pregnancy to Early Lactation. J. Anim. Sci. 1995, 73, 2804–2819. [Google Scholar] [CrossRef] [PubMed]
- Durand-Chaucheyras, F.; Fonty, G.; Bertin, G.; Théveniot, M.; Gouet, P. Fate of Levucell SC I-1077 Yeast Additive during Digestive Transit in Lambs. Reprod. Nutr. Dev. 1998, 38, 275–280. [Google Scholar] [CrossRef]
- Chaucheyras Durand, F.; Duniere, L. The use of probiotics as supplements for ruminants. In Improving Rumen Function; Burleigh Dodds Science Publishing: Cambridge, UK, 2020; pp. 775–818. ISBN 978-1-78676-332-7. [Google Scholar]
- Chaucheyras-Durand, F.; Ameilbonne, A.; Bichat, A.; Mosoni, P.; Ossa, F.; Forano, E. Live Yeasts Enhance Fibre Degradation in the Cow Rumen through an Increase in Plant Substrate Colonization by Fibrolytic Bacteria and Fungi. J. Appl. Microbiol. 2016, 120, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.; Brazel, D.M.; Swan, B.K.; Arnosti, C.; Chain, P.S.G.; Reitenga, K.G.; Xie, G.; Poulton, N.J.; Lluesma Gomez, M.; Masland, D.E.D.; et al. Capturing Single Cell Genomes of Active Polysaccharide Degraders: An Unexpected Contribution of Verrucomicrobia. PLoS ONE 2012, 7, e35314. [Google Scholar] [CrossRef] [PubMed]
- Kamke, J.; Kittelmann, S.; Soni, P.; Li, Y.; Tavendale, M.; Ganesh, S.; Janssen, P.H.; Shi, W.; Froula, J.; Rubin, E.M.; et al. Rumen Metagenome and Metatranscriptome Analyses of Low Methane Yield Sheep Reveals a Sharpea-Enriched Microbiome Characterised by Lactic Acid Formation and Utilisation. Microbiome 2016, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.C.; Costa, D.F.A.; Palma, A.S.V.; Camargo, K.D.V.; Lima, L.O.; Harper, K.J.; Meale, S.J.; Silva, L.F.P. The Use of Live Yeast to Increase Intake and Performance of Cattle Receiving Low-Quality Tropical Forages. J. Anim. Sci. 2021, 99. [Google Scholar] [CrossRef] [PubMed]
- Lefler, J.; Minini, S.; Morandi, A.; Embree, M. Effect of Direct-Fed Microbial on Commercial Dairy Drylot; Elsevier Science Inc: New York, NY, USA, 2020; p. M132. [Google Scholar]
- Oikonomou, G.; Teixeira, A.G.V.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal Microbial Diversity in Pre-Weaned Dairy Calves as Described by Pyrosequencing of Metagenomic 16S RDNA. Associations of Faecalibacterium Species with Health and Growth. PLoS ONE 2013, 8, e63157. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.M.; Raphael, W. Significance of Metabolic Stress, Lipid Mobilization, and Inflammation on Transition Cow Disorders. Vet. Clin. North Am. Food Anim. Pract. 2013, 29, 267–278. [Google Scholar] [CrossRef]
- Abuelo, Á.; Hernández, J.; Benedito, J.L.; Castillo, C. The Connexion between Serum Redox Balance and Concentration of Lactic Acid Enantiomers in Dairy Cows around the Time of Calving. Comp. Clin. Pathol. 2015, 24, 465–468. [Google Scholar] [CrossRef]
- Cerretani, L.; Bendini, A. Chapter 67–Rapid Assays to Evaluate the Antioxidant Capacity of Phenols in Virgin Olive Oil. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 625–635. ISBN 978-0-12-374420-3. [Google Scholar]
- Mavrommatis, A.; Mitsiopoulou, C.; Christodoulou, C.; Karabinas, D.; Nenov, V.; Zervas, G.; Tsiplakou, E. Dietary Supplementation of a Live Yeast Product on Dairy Sheep Milk Performance, Oxidative and Immune Status in Peripartum Period. J. Fungi. 2020, 6, 334. [Google Scholar] [CrossRef] [PubMed]
- Doucet, M.; Sagot, L. Des Brebis Bien Nourries, Des Agneaux Plus Vigoureux; Institut De L’elevage: Paris, France, 2018; Volume 6. [Google Scholar]
- Roca Fraga, F.J.; Lagisz, M.; Nakagawa, S.; Lopez-Villalobos, N.; Blair, H.T.; Kenyon, P.R. Meta-Analysis of Lamb Birth Weight as Influenced by Pregnancy Nutrition of Multiparous Ewes. J. Anim. Sci. 2018, 96, 1962–1977. [Google Scholar] [CrossRef] [PubMed]
- Sales, F.; Peralta, O.A.; Narbona, E.; McCoard, S.; De Los Reyes, M.; González-Bulnes, A.; Parraguez, V.H. Hypoxia and Oxidative Stress Are Associated with Reduced Fetal Growth in Twin and Undernourished Sheep Pregnancies. Anim. Open Access J. MDPI 2018, 8, 217. [Google Scholar] [CrossRef]
- Chang, H.C.; Leaw, S.N.; Huang, A.H.; Wu, T.L.; Chang, T.C. Rapid Identification of Yeasts in Positive Blood Cultures by a Multiplex PCR Method. J. Clin. Microbiol. 2001, 39, 3466–3471. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and Low Abundance of Classical Ruminal Bacterial Species in the Bovine Rumen Revealed by Relative Quantification Real-Time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef] [PubMed]
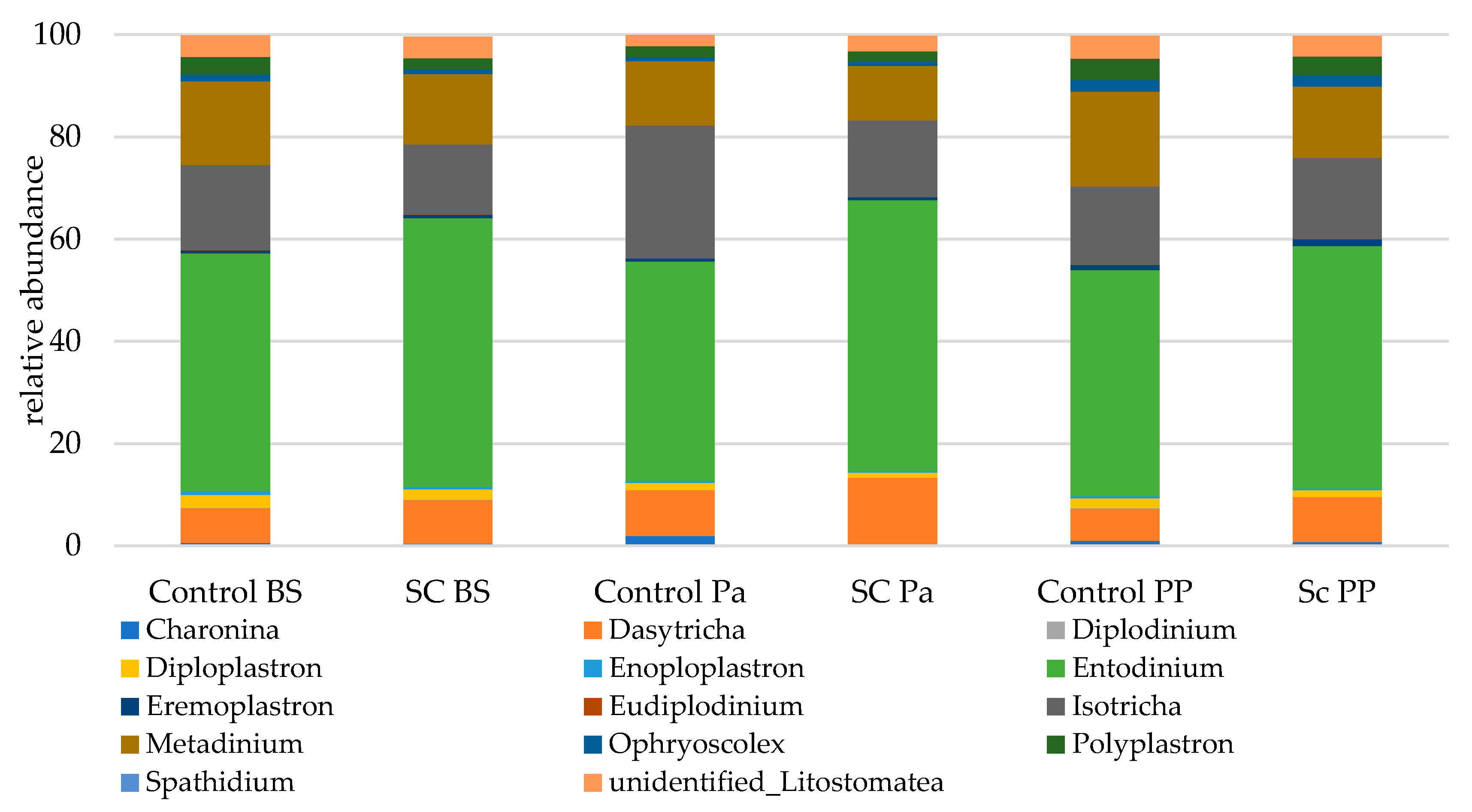
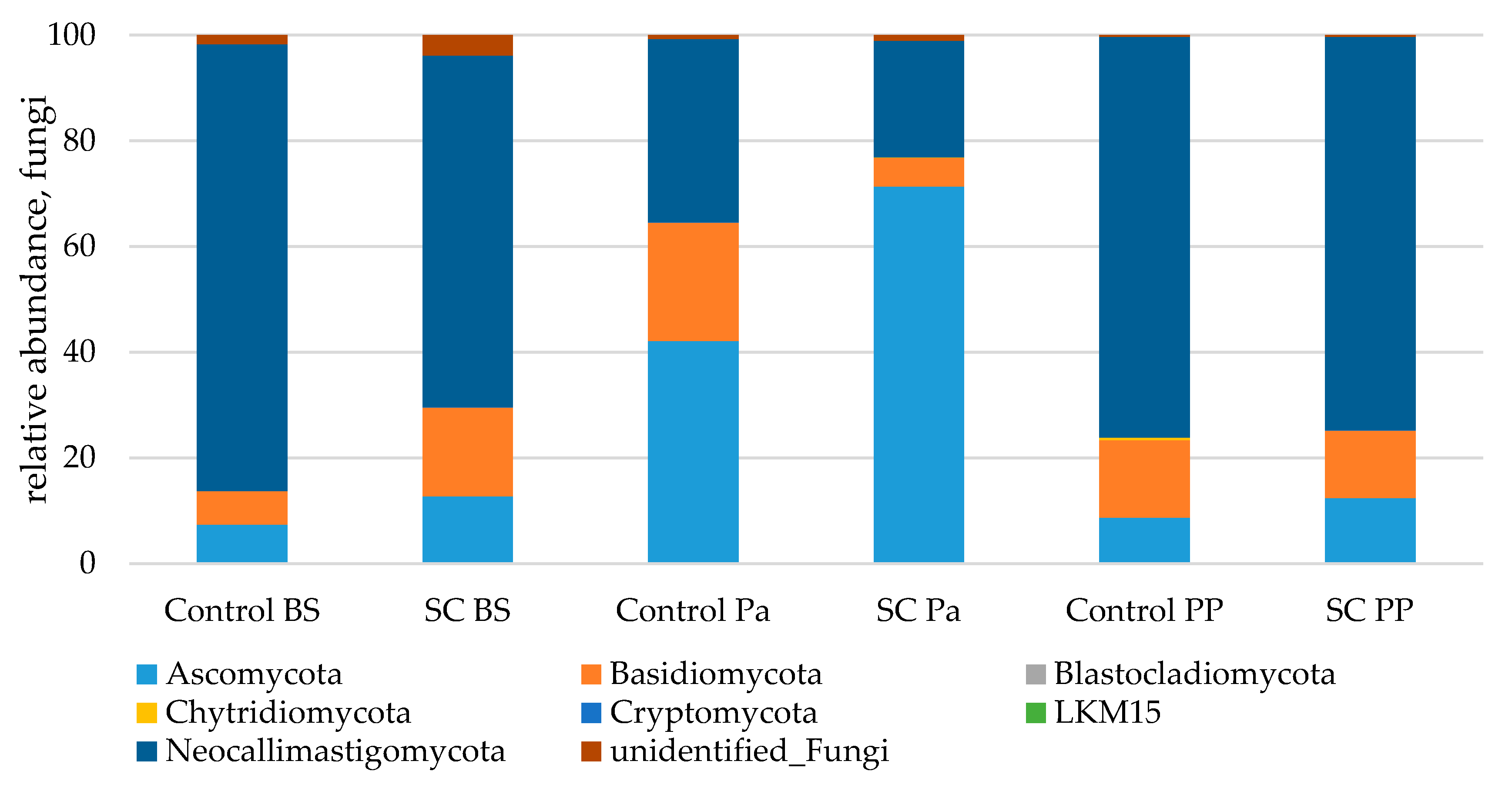
| Measured Parameter | Group | Time | Interaction G × T |
|---|---|---|---|
| Total VFA | 0.309 | 0.164 | 0.060 |
| Acetate | 0.357 | 0.080 | 0.086 |
| Propionate | 0.131 | 0.722 | 0.047 |
| Butyrate | 0.569 | 0.647 | 0.055 |
| Isobutyrate | 0.891 | 0.168 | 0.136 |
| Isovalerate | 0.870 | 0.607 | 0.131 |
| Valerate | 0.535 | 0.002 | 0.085 |
| Caproate | 0.142 | 0.002 | 0.180 |
| Log10 of Copy Numbers/g Pelleted Rumen Content | BS | Pa | PP | p-Value from Linear Mixed Model on Ydiff | |||||
|---|---|---|---|---|---|---|---|---|---|
| Target | Control | SC | Control | SC | Control | SC | Group | Time | Interaction G X T |
| Total bacteria | 10.86 ± 0.09 | 10.83 ± 0.18 | 10.91 ± 0.11 | 10.91 ± 0.24 | 10.78 ± 0.07 | 10.78 ± 0.17 | 0.648 | 0.019 | 0.941 |
| Ruminococcus flavefaciens | 7.72 ± 0.32 | 8.09 ± 0.42 | 8.03 ± 0.16 | 7.79 ± 0.29 | 7.8 ± 0.12 | 7.8 ± 0.23 | 0.013 | 0.181 | 0.152 |
| Ruminococcus albus | 6.64 ± 0.16 | 6.9 ± 0.24 | 6.85 ± 0.35 | 6.99 ± 0.47 | 7.16 ± 0.25 | 7.19 ± 0.22 | 0.157 | 0.003 | 0.403 |
| Fibrobacter succinogenes | 9.61 ± 0.17 | 9.37 ± 0.38 | 9.28 ± 0.10 | 9.32 ± 0.19 | 9.32 ± 0.38 | 9.24 ± 0.16 | NA | NA | NA |
| Prevotella | 8.93 ± 0.12 | 8.94 ± 0.19 | 8.96 ± 0.11 | 9.01 ± 0.16 | 8.84 ± 0.17 | 8.76 ± 0.18 | 0.831 | 0.013 | 0.315 |
| Megasphaera elsdenii | 2.48 ± 0.32 | 2.42 ± 0.17 | 2.41 ± 0.5 | 2.68 ± 0.17 | 2.55 ± 0.32 | 2.70 ± 0.31 | 0.438 | 0.234 | 0.825 |
| Methanogenic Archaea | 7.66 ± 0.23 | 7.87 ± 0.23 | 7.95 ± 0.29 | 8.07 ± 0.19 | 7.85 ± 0.41 | 8.02 ± 0.21 | 0.726 | 0.129 | 0.923 |
| Protozoa | 9.84 ± 0.14 | 9.52 ± 0.66 | 9.92 ± 0.34 | 9.33 ± 0.52 | 9.51 ± 0.15 | 9.28 ± 0.31 | 0.786 | 0.106 | 0.200 |
| Anaerobic fungi | 6.27 ± 0.37 | 6.19 ± 0.64 | 6.36 ± 0.71 | 6.13 ± 0.49 | 5.94 ± 0.72 | 6.16 ± 0.39 | 0.819 | 0.486 | 0.426 |
| S. cerevisiae | 5.89 ± 0.49 | 5.65 ± 0.38 | 6.12 ± 0.6 | 7.49 ± 0.34 | 5.6 ± 0.18 | 5.91 ± 0.54 | 0.027 | 0.001 | 0.003 |
| Alpha-Div Indexes | Group | Time | G × T |
|---|---|---|---|
| Observed OTUs | 0.589 | 0.001 | 0.626 |
| Shannon | 0.851 | 0.003 | 0.178 |
| Alpha-Div Indexes | Group | Time | G × T |
|---|---|---|---|
| Observed OTUs | 0.017 | 0.010 | 0.335 |
| Shannon | 0.013 | 0.264 | 0.229 |
| Phylum | Group | Time | G × T |
|---|---|---|---|
| Actinobacteria | 0.589 | 0.039 | 0.503 |
| Bacteroidetes | 0.225 | 0.044 | 0.445 |
| Cyanobacteria | 0.254 | 0.138 | 0.312 |
| Euryarchaeota | 0.051 | 0.260 | 0.952 |
| Fibrobacteres | 0.565 | 0.747 | 0.241 |
| Firmicutes | 0.562 | 0.138 | 0.371 |
| Proteobacteria | 0.259 | 0.006 | 0.776 |
| Spirochaetes | 0.672 | 0.013 | 0.219 |
| Phylum | Group | Time | G × T |
|---|---|---|---|
| Bacteroidetes | 0.044 | 0.034 | 0.523 |
| Euryarchaeota | 0.355 | 0.928 | 0.920 |
| Fibrobacteres | 0.066 | 0.0001 | 0.013 |
| Firmicutes | 0.442 | 0.146 | 0.891 |
| Proteobacteria | 0.614 | 0.004 | 0.936 |
| Spirochaetes | 0.241 | 0.414 | 0.495 |
| Tenericutes | 0.914 | 0.357 | 0.953 |
| Verrucomicrobia | 0.183 | 0.453 | 0.712 |
| Protozoa | Group | Time | G × T |
|---|---|---|---|
| Dasytricha | 0.224 | 0.007 | 0.514 |
| Entodinium | 0.843 | 0.291 | 0.085 |
| Eudiplodinium | 0.871 | 0.899 | 0.041 |
| Isotricha | 0.625 | 0.032 | 0.015 |
| Metadinium | 0.799 | 0.0002 | 0.115 |
| Polyplastron | 0.14 | 0.001 | 0.550 |
| Unidentified Litostomatea | 0.811 | 0.013 | 0.284 |
| Total Litostomatea | 0.150 | 0.302 | 0.445 |
| Phylum | Group | Time | G × T |
|---|---|---|---|
| Ascomycota | 0.103 | <0.0001 | 0.065 |
| Basidiomycota | 0.009 | 0.971 | 0.289 |
| Neocallimastigomycota | 0.229 | <0.0001 | 0.528 |
| Unidentified fungi | 0.209 | 0.001 | 0.111 |
| Anaerobic Fungi | Group | Time | G × T |
|---|---|---|---|
| Cyllamyces | 0.500 | 0.007 | 0.179 |
| Orpinomyces | 0.825 | 0.201 | 0.704 |
| Unidentified Neocallimastigomycota | 0.180 | <0.0001 | 0.604 |
| Total | 0.236 | <0.0001 | 0.543 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunière, L.; Esparteiro, D.; Lebbaoui, Y.; Ruiz, P.; Bernard, M.; Thomas, A.; Durand, D.; Forano, E.; Chaucheyras-Durand, F. Changes in Digestive Microbiota, Rumen Fermentations and Oxidative Stress around Parturition Are Alleviated by Live Yeast Feed Supplementation to Gestating Ewes. J. Fungi 2021, 7, 447. https://doi.org/10.3390/jof7060447
Dunière L, Esparteiro D, Lebbaoui Y, Ruiz P, Bernard M, Thomas A, Durand D, Forano E, Chaucheyras-Durand F. Changes in Digestive Microbiota, Rumen Fermentations and Oxidative Stress around Parturition Are Alleviated by Live Yeast Feed Supplementation to Gestating Ewes. Journal of Fungi. 2021; 7(6):447. https://doi.org/10.3390/jof7060447
Chicago/Turabian StyleDunière, Lysiane, Damien Esparteiro, Yacine Lebbaoui, Philippe Ruiz, Mickael Bernard, Agnès Thomas, Denys Durand, Evelyne Forano, and Frédérique Chaucheyras-Durand. 2021. "Changes in Digestive Microbiota, Rumen Fermentations and Oxidative Stress around Parturition Are Alleviated by Live Yeast Feed Supplementation to Gestating Ewes" Journal of Fungi 7, no. 6: 447. https://doi.org/10.3390/jof7060447
APA StyleDunière, L., Esparteiro, D., Lebbaoui, Y., Ruiz, P., Bernard, M., Thomas, A., Durand, D., Forano, E., & Chaucheyras-Durand, F. (2021). Changes in Digestive Microbiota, Rumen Fermentations and Oxidative Stress around Parturition Are Alleviated by Live Yeast Feed Supplementation to Gestating Ewes. Journal of Fungi, 7(6), 447. https://doi.org/10.3390/jof7060447