Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Pythium Insidiosum Strain and Culture Conditions
2.2. Protein Sample Extraction
2.3. Serum Samples from Infected Horses and Humans
2.4. Two-Dimensional Gel Electrophoresis
2.5. 2D Immunoblotting of P. insidiosum Proteins
2.6. Trypsin Digestion and Mass Spectrometry
2.7. Bioinformatic Analysis
3. Results
3.1. Pythium Insidiosum Protein Profile and Immunoreactive Proteins
3.2. Protein Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaastra, W.; Lipman, L.J.; De Cock, A.W.; Exel, T.K.; Pegge, R.B.; Scheurwater, J.; Vilela, R.; Mendoza, L. Pythium insidiosum: An overview. Vet. Microbiol. 2010, 146, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Krajaejun, T.; Sathapatayavongs, B.; Pracharktam, R.; Nitiyanant, P.; Leelachaikul, P.; Wanachiwanawin, W.; Chaiprasert, A.; Assanasen, P.; Saipetch, M.; Mootsikapun, P.; et al. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin. Infect. Dis. 2006, 43, 569–576. [Google Scholar] [CrossRef]
- Mendoza, L.; Ajello, L.; McGinnis, M.R. Infection caused by the Oomycetous pathogen Pythium insidiosum. J. Mycol. Med. 1996, 6, 151–164. [Google Scholar]
- Mendoza, L.; Hernandez, F.; Ajello, L. Life Cycle of the Human and Animal Oomycete Pathogen Pythium insidiosum. J. Clin. Microbiol. 1993, 31, 2967–2973. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.L.; Carvalho, E.C.; Nogueira, R.H.; Lemos, L.S.; Mendoza, L. Disseminated pythiosis in three horses. Vet. Microbiol. 2003, 96, 289–295. [Google Scholar] [CrossRef]
- Brown, C.C.; Roberts, E.D. Intestinal pythiosis in a horse. Aust. Vet. J. 1988, 65, 88–89. [Google Scholar] [CrossRef]
- Krajaejun, T.; Pracharktam, R.; Wongwaisayawan, S.; Rochanawutinon, M.; Kunakorn, M.; Kunavisarut, S. Ocular pythiosis: Is it under-diagnosed? Am. J. Ophthalmol. 2004, 137, 370–372. [Google Scholar] [CrossRef]
- Triscott, J.A.; Weedon, D.; Cabana, E. Human subcutaneous pythiosis. J. Cutan. Pathol. 1993, 20, 267–271. [Google Scholar] [CrossRef]
- Bosco, S.M.G.; Bagagli, E.; Araújo, J.P., Jr.; Candeias, J.M.G.; Franco, M.F.; Marques, M.E.A.; Mendoza, L.; Camargo, R.P.; Alencar Marques, A.S. Human pythiosis Brazil. Emerg. Infect. Dis. 2005, 11, 715–717. [Google Scholar] [CrossRef]
- Marques, A.S.; Bagagli, E.; Bosco, S.M.G.; Camargo, R.P.; Marques, M.E.A. Pythium insidiosum: Relato do primeiro caso de infecção humana no Brasil. An. Bras. Dermatol. 2006, 81, 483–485. [Google Scholar] [CrossRef]
- Chareonsirisuthigul, T.; Khositnithikul, R.; Intaramat, A.; Inkomlue, R.; Sriwanichrak, K.; Piromsontikorn, S.; Kitiwanwanich, S.; Lowhnoo, T.; Yingyong, W.; Chaiprasert, A.; et al. Performance comparison of immunodiffusion, enzyme-linked immunosorbent assay, immunochromatography and hemagglutination for serodiagnosis of human pythiosis. Diagn. Microbiol. Infect. Dis. 2013, 76, 42–45. [Google Scholar] [CrossRef]
- Intaramat, A.; Sornprachum, T.; Chantrathonkul, B.; Chaisuriya, P.; Lohnoo, T.; Yingyong, W.; Jongruja, N.; Kumsang, Y.; Sandee, A.; Chaiprasert, A.; et al. Protein A/G-based immunochromatographic test for serodiagnosis of pythiosis in human and animal subjects from Asia and Americas. Med. Mycol. 2016, 54, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Keeratijarut, A.; Lohnoo, T.; Yingyong, W.; Rujirawat, T.; Srichunrusami, C.; Onpeaw, P.; Chongtrakool, P.; Brandhorst, T.T.; Krajaejun, T. Detection of the oomycete Pythium insidiosum by real-time PCR targeting the gene coding for exo-1,3-β-glucanase. J. Med. Microbiol. 2015, 64, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Inkomlue, R.; Larbcharoensub, N.; Karnsombut, P.; Lerksuthirat, T.; Aroonroch, R.; Lohnoo, T.; Yingyong, W.; Santanirand, P.; Sansopha, L.; Krajaejun, T. Development of an anti-elicitin antibody-based immunohistochemical assay for diagnosis of pythiosis. J. Clin. Microbiol. 2016, 54, 43–48. [Google Scholar] [CrossRef]
- Krajaejun, T.; Imkhieo, S.; Intaramat, A.; Ratanabanangkoon, K. Development of an immunochromatographic test for rapid serodiagnosis of human pythiosis. Clin. Vaccine Immunol. 2009, 16, 506–509. [Google Scholar] [CrossRef]
- Chaiprasert, A.; Samerpitak, K.; Wanachiwanawin, W.; Thasnakorn, P. Induction of zoospore formation in Thai isolates of Pythium insidiosum. Mycoses 1990, 33, 317–323. [Google Scholar] [CrossRef]
- Mendoza, L.; Prendas, J. A method to obtain rapid zoosporogenesis of Pythium insidiosum. Mycopathologia 1988, 104, 59–62. [Google Scholar] [CrossRef]
- Grooters, A.M.; Whittington, A.; Lopez, M.K.; Boroughs, M.N.; Roy, A.F. Evaluation of microbial culture techniques for the isolation of Pythium insidiosum from equine tissues. J. Vet. Diagn. Investig. 2002, 14, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Krajaejun, T.; Kunakorn, M.; Niemhom, S.; Chongtrakool, P.; Pracharktam, R. Development and evaluation of an in-house enzyme-linked immunosorbent assay for early diagnosis and monitoring of human pythiosis. Clin. Diagn. Lab. Immunol. 2002, 9, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Kaufman, L.; Mandy, W.; Glass, R. Serodiagnosis of human and animal pythiosis using an enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 1997, 4, 715–718. [Google Scholar] [CrossRef]
- Jindayok, T.; Piromsontikorn, S.; Srimuang, S.; Khupulsup, K.; Krajaejun, T. Hemagglutination test for rapid serodiagnosis of human pythiosis. Clin. Vaccine Immunol. 2009, 16, 1047–1051. [Google Scholar] [CrossRef]
- Mendoza, L.; Kaufman, L.; Standard, P.G. Immunodiffusion test for diagnosing and monitoring pythiosis in horses. J. Clin. Microbiol. 1986, 23, 813–816. [Google Scholar] [CrossRef]
- Pracharktam, R.; Changtrakool, P.; Sathapatayavongs, B.; Jayanetra, P.; Ajello, L. Immunodiffusion test for diagnosis and monitoring of human pythiosis insidiosi. J. Clin. Microbiol. 1991, 29, 2661–2662. [Google Scholar] [CrossRef] [PubMed]
- Keeratijarut, A.; Lohnoo, T.; Yingyong, W.; Sriwanichrak, K.; Krajaejun, T. A peptide ELISA to detect antibodies against Pythium insidiosum based on predicted antigenic determinants of exo-1,3-ß-glucanase. Southeast Asian J. Trop. Med. Public Health 2013, 44, 672–680. [Google Scholar] [PubMed]
- Supabandhu, J.; Vanittanakom, P.; Laohapensang, K.; Vanittanakom, N. Application of immunoblot assay for rapid diagnosis of human pythiosis. J. Med. Assoc. Thail. 2009, 92, 1063–1071. [Google Scholar]
- Vanittanakom, N.; Supabandhu, J.; Khamwan, C.; Praparattanapan, J.; Thirach, S.; Prasertwitayakij, N.; Louthrenoo, W.; Chiewchanvit, S.; Tananuvat, N. Identification of emerging human-pathogenic Pythium insidiosum by serological and molecular assay-based methods. J. Clin. Microbiol. 2004, 42, 3970–3974. [Google Scholar] [CrossRef] [PubMed]
- Keeratijarut, A.; Lohnoo, T.; Yingyong, W.; Nampoon, U.; Lerksuthirat, T.; Onpaew, P.; Chongtrakool, P.; Krajaejun, T. PCR amplification of a putative gene for exo-1,3-ß-glucanase to identify the pathogenic oomycete Pythium insidiosum. Asian Biomed. 2014, 8, 637–644. [Google Scholar] [CrossRef]
- Badenoch, P.R.; Coster, D.J.; Wetherall, B.L.; Brettig, H.T.; Rozenbilds, M.A.; Drenth, A.; Wagels, G. Pythium insidiosum keratitis confirmed by DNA sequence analysis. Br. J. Ophthalmol. 2001, 85, 502–503. [Google Scholar] [CrossRef] [PubMed]
- Botton, S.A.; Pereira, D.I.; Costa, M.M.; Azevedo, M.I.; Argenta, J.S.; Jesus, F.P.K.; Alves, S.H.; Santurio, J.M. Identification of Pythium insidiosum by nested PCR in cutaneous lesions of Brazilian horses and rabbits. Curr. Microbiol. 2011, 62, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Krajaejun, T.; Kunakorn, M.; Pracharktam, R.; Chongtrakool, P.; Sathapatayavongs, B.; Chaiprasert, A.; Vanittanakom, N.; Chindamporn, A.; Mootsikapun, P. Identification of a Novel 74-Kilodalton Immunodominant Antigen of Pythium insidiosum Recognized by Sera from Human Patients with Pythiosis. J. Clin. Microbiol. 2006, 44, 1674–1680. [Google Scholar] [CrossRef]
- Krajaejun, T.; Keeratijarut, A.; Sriwanichrak, K.; Lowhnoo, T.; Rujirawat, T.; Petchthong, T.; Yingyong, W.; Kalambaheti, T.; Smittipat, N.; Juthayothin, T.; et al. The 74-Kilodalton Immunodominant Antigen of the Pathogenic Oomycete Pythium insidiosum Is a Putative Exo-1,3-ß-Glucanase. Clin. Vaccine Immunol. 2010, 17, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Lerksuthirat, T.; Lohnoo, T.; Inkomlue, R.; Rujirawat, T.; Yingyong, W.; Khositnithikul, R.; Phaonakrop, N.; Roytrakul, S.; Sullivan, T.D.; Krajaejun, T. The elicitin-like glycoprotein, ELI025, is secreted by the pathogenic oomycete Pythium insidiosum and evades host antibody responses. PLoS ONE 2015, 10, e0118547. [Google Scholar] [CrossRef]
- Dal Ben, V.; Oliveira, R.S.; Borchardt, J.L.; Valente, J.S.S.; Brasil, C.L.; Zambrano, C.G.; Leite, F.P.L.; Botton, S.A.; Pereira, D.I.B. Protein profile of Brazilian Pythium insidiosum isolates. Med. Mycol. 2018, 56, 485–492. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Rodrigues, A.M.; Kubitschek-Barreira, P.H.; Fernandes, G.F.; de Almeida, S.R.; Lopes-Bezerra, L.M.; de Camargo, Z.P. Two-dimensional gel electrophoresis data for proteomic profiling of Sporothrix yeast cells. Data Brief 2015, 2, 32–38. [Google Scholar] [CrossRef]
- Bradford, M.M. Quantificação de microgramas de proteína utilizando o princípio da proteína de ligação de corante. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chitasombat, M.N.; Jongkhajornpong, P.; Lekhanont, K.; Krajaejun, T. Recent update in diagnosis and treatment of human pythiosis. PeerJ 2020, 8, e8555. [Google Scholar] [CrossRef] [PubMed]
- Rujirawat, T.; Sridapan, T.; Lohnoo, T.; Yingyong, W.; Kumsang, Y.; Sae-Chew, P.; Tonpitak, W.; Krajaejun, T. Single nucleotide polymorphism-based multiplex PCR for identification and genotyping of the oomycete Pythium insidiosum from humans, animals and the environment. Infect. Genet. Evol. 2017, 54, 429–436. [Google Scholar] [CrossRef]
- Lohnoo, T.; Jongruja, N.; Rujirawat, T.; Yingyon, W.; Lerksuthirat, T.; Nampoon, U.; Kumsang, Y.; Onpaew, P.; Chongtrakool, P.; Keeratijarut, A.; et al. Efficiency comparison of three methods for extracting genomic DNA of the pathogenic oomycete Pythium insidiosum. J. Med. Assoc. Thail. 2014, 97, 342–348. [Google Scholar]
- Görg, A.; Obermaier, C.; Boguth, G.; Harder, A.; Scheibe, B.; Wildgruber, R.; Weiss, W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000, 21, 1037–1053. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Lima, D.B.; Leprevost, F.V.; Santos, M.D.M.; Fischer, J.S.G.; Aquino, P.F.; Moresco, J.J.; Yates, J.R., III; Barbosa, V.C. PatternLab for proteomics 4.0: A one-stop shop for analyzing shotgun proteomic data. Nat. Protoc. 2016, 11, 102–117. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, 158–169. [Google Scholar] [CrossRef]
- Oliveros, J.C. NoVENNY. An Interactive Tool for Comparing Lists with Venn Diagrams Title. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 19 November 2019).
- Venny 2.1. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 19 November 2019).
- STRING. Available online: https://string-db.org (accessed on 22 November 2019).
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef]
- SCRATCH Protein Predictor. Available online: http://scratch.proteomics.ics.uci.edu (accessed on 5 December 2019).
- Rujirawat, T.; Patumcharoenpol, P.; Lohnoo, T.; Yingyong, W.; Lerksuthirat, T.; Tangphatsornruang, S.; Suriyaphol, P.; Grenville-Briggs, L.J.; Garg, G.; Kittichotirat, W.; et al. Draft Genome Sequence of the Pathogenic Oomycete Pythium insidiosum Strain Pi-S, Isolated from a Patient with Pythiosis. Genome Announc. 2015, 3, e00574-15. [Google Scholar] [CrossRef]
- Sweredoski, M.J.; Baldi, P. COBEpro: A novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 2009, 22, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Pollastri, G.; Przybylski, D.; Rost, B.; Baldi, P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 2002, 47, 228–235. [Google Scholar] [CrossRef]
- Yao, B.; Zhang, L.; Liang, S.; Zhang, C. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE 2012, 7, e45152. [Google Scholar] [CrossRef] [PubMed]
- Chechi, J.L.; Franckin, T.; Barbosa, L.N.; Alves, F.C.B.; Leite, A.L.; Buzalaf, M.A.R.; dos Santos, L.D.; Bosco, S.M.G. Infering putative virulence factors for Pythium insidiosum by proteomic approach. Med. Mycol. 2019, 57, 92–100. [Google Scholar] [CrossRef]
- Wang, J.; Du, X.J.; Lu, X.N.; Wang, S. Immunoproteomic identification of immunogenic proteins in Cronobacter sakazakii strain BAA-894. Appl. Microbiol. Biotechnol. 2013, 97, 2077–2091. [Google Scholar] [CrossRef]
- González-Aravena, M.; Calfio, C.; Mercado, L.; Morales-Lange, B.; Bethke, J.; De Lorgeril, J.; Cárdenas, C.A. HSP70 from the Antarctic sea urchin Sterechinus neumayeri: Molecular characterization and expression in response to heat stress. Biol. Res. 2018, 51, 8. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Kregel, K.C. Heat shock proteins: Modifying factors in physiological stress responses and acquired thermotolerance. J. Appl. Physiol. 2002, 92, 2177–2186. [Google Scholar] [CrossRef]
- Tiwari, S.; Thakur, R.; Shankar, J. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnol. Res. Internat. 2015, 2015, 132635. [Google Scholar] [CrossRef] [PubMed]
- Ohba, M. A 70-kDa heat shock cognate protein suppresses the defects caused by a proteasome mutation in Saccharomyces cerevisiae. FEBS Lett. 1994, 351, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Bisio, L.C.; Silva, S.P.; Pereira, I.S.; Xavier, M.A.S.; Venancio, E.J.; Puccia, R.; Soares, C.M.A.; Felipe, M.S.S. A new Paracoccidioides brasiliensis 70-kDa heat shock protein reacts with sera from paracoccidioidomycosis patients. Med. Mycol. 2005, 43, 495–503. [Google Scholar] [CrossRef]
- Keeratijarut, A.; Lohnoo, T.; Rujirawat, T.; Yingyong, W.; Kalambaheti, T.; Miller, S.; Phuntumart, V.; Krajaejun, T. The Immunoreactive Exo-1,3-β-Glucanase from the Pathogenic Oomycete Pythium insidiosum Is Temperature Regulated and Exhibits Glycoside Hydrolase Activity. PLoS ONE 2015, 10, e0135239. [Google Scholar] [CrossRef]
- Garfoot, A.L.; Dearing, K.L.; Vanschoiack, A.D.; Wysocki, V.H.; Rappleye, C.A. Eng1 and Exg8 Are the Major ß-Glucanases Secreted by the Fungal Pathogen Histoplasma capsulatum. J. Biol. Chem. 2017, 292, 4801–4810. [Google Scholar] [CrossRef]
- Holbrook, E.D.; Edwards, J.A.; Youseff, B.H.; Rappleye, C.A. Definition of the Extracellular Proteome of Pathogenic-Phase Histoplasma capsulatum. J. Proteome Res. 2011, 10, 1929–1943. [Google Scholar] [CrossRef]
- Moreira, A.L.E.; Oliveira, M.A.P.; Silva, L.O.S.; Inácio, M.M.; Bailão, A.M.; Parente-Rocha, J.A.; Cruz-Leite, V.R.M.; Paccez, J.D.; Soares, C.M.A.; Weber, S.S.; et al. Immunoproteomic Approach of Extracellular Antigens from Paracoccidioides Species Reveals Exclusive B-Cell Epitopes. Front. Microbiol. 2020, 10, 2968. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar] [CrossRef] [PubMed]
- Smits, G.J.; Van Den Ende, H.; Klis, F.M. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 2001, 147, 781–794. [Google Scholar] [CrossRef]
- Fontaine, T.; Hartland, R.P.; Beauvais, A.; Diaquin, M.; Latge, J.P. Purification, characterization of an endo-1,3-betaglucanase from Aspergillus fumigatus. Eur. J. Biochem. 1997, 243, 315–321. [Google Scholar] [CrossRef]
- Martin, K.; McDougall, B.M.; McIlroy, S.; Chen, J.; Seviour, R.J. Biochemistry and molecular biology of exo cellular fungal ß-(1,3)-and ß-(1,6)-glucanases. FEMS Microbiol. Rev. 2007, 31, 168–192. [Google Scholar] [CrossRef] [PubMed]
- Labeé, G.; Bezaire, J.; Groot, S.; How, C.; Rasmusson, T.; Yaeck, J.; Jervis, E.; Dmitrienko, G.I.; Guillemette, J.G. High level production of the Magnaporthe grisea fructose 1,6-biphosphate aldolase enzyme in Escherichia coli using a small volume bench-top fermentor. Protein Expr. Purif. 2007, 51, 110–119. [Google Scholar] [CrossRef]
- Cooper, S.J.; Leonard, G.A.; McSweeney, S.M.; Thompson, A.W.; Naismith, J.H.; Qamar, S.; Plater, A.; Berry, A.; Hunter, W.N. The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. Structure 1996, 4, 1303–1315. [Google Scholar] [CrossRef]
- Labbé, G.; Groot, S.; Rasmusson, T.; Milojevic, G.; Dmitrienko, G.I.; Guillemette, J.G. Evaluation of four microbial class II fructose 1,6-bisphosphate aldolase enzymes for use as biocatalysts. Protein Expr. Purif. 2011, 80, 224–233. [Google Scholar] [CrossRef]
- Han, X.; Zhu, X.; Zhu, S.; Wei, L.; Hong, Z.; Guo, L.; Chen, H.; Chi, B.; Liu, Y.; Feng, L.; et al. A Rational Design, Synthesis, Biological Evaluation and Structure−Activity Relationship Study of Novel Inhibitors against Cyanobacterial Fructose-1,6-bisphosphate Aldolase. J. Chem. Inf. Model. 2016, 56, 73–81. [Google Scholar] [CrossRef]
- Rodaki, A.; Young, T.; Brown, A.J.P. Effects of Depleting the Essential Central Metabolic Enzyme Fructose-1,6-Bisphosphate Aldolase on the Growth and Viability of Candida albicans: Implications for Antifungal Drug Target Discovery. Eukaryot. Cell 2006, 5, 1371–1377. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Kubitschek-Barreira, P.H.; Pinheiro, B.G.; Teixeira-Ferreira, A.; Hahn, R.C.; Camargo, Z.P. Immunoproteomic analysis reveals novel candidate antigens for the diagnosis of paracoccidioidomycosis due to Paracoccidioides lutzii. J. Fungi 2020, 6, 357. [Google Scholar] [CrossRef] [PubMed]
- Tomazett, M.V.; Baeza, L.C.; Paccez, J.D.; Parente-Rocha, J.A.; Ribeiro-Dias, F.; de Almeida Soares, C.M. Identification and characterization of Paracoccidioides lutzii proteins interacting with macrophages. Microbes Infect. 2019, 21, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Dziadek, S.; Bundle, D.R.; Cutler, J.E. Synthetic glycopeptide vaccines combining ß-mannan and peptide epitopes induce protection against candidiasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13526–13531. [Google Scholar] [CrossRef]
- Xin, H.; Cutler, J.E. Vaccine and Monoclonal Antibody That Enhance Mouse Resistance to Candidiasis. Clin. Vaccine Immunol. 2011, 18, 1656–1667. [Google Scholar] [CrossRef]
- Stie, J.; Bruni, G.; Fox, D. Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion. PLoS ONE 2009, 4, e5780. [Google Scholar] [CrossRef] [PubMed]
- Chaves, E.G.; Weber, S.S.; Bao, S.N.; Pereira, L.A.; Bailao, A.M.; Borges, C.L.; Soares, C.M. Analysis of Paracoccidioides secreted proteins reveals fructose 1,6-bisphosphate aldolase as a plasminogen-binding protein. BMC Microbiol. 2015, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.B.; Yarden, O. Serine/Threonine Protein Kinases and Phosphatases in Filamentous Fungi. Fungal Genet. Biol. 1999, 26, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.N.; Masuda, C.A.; Ferreira-Pereira, A.; Carvajal, E.; Ghislain, M.; Montero-Lomelí, M. The serine/threonine protein phosphatase Sit4p activates multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 2010, 10, 674–686. [Google Scholar] [CrossRef]
- Leiter, É.; González, A.; Erdei, É.; Casado, C.; Kovács, L.; Ádám, C.; Oláh, J.; Miskei, M.; Molnar, M.; Farkas, I.; et al. Protein phosphatase Z modulates oxidative stress response in fungi. Fungal Genet. Biol. 2012, 49, 708–716. [Google Scholar] [CrossRef]
- Muszkieta, L.; Carrion, S.J.; Robinet, P.; Beau, R.; Elbim, C.; Pearlman, E.; Latgé, J.P. The protein phosphatase PhzA of A. fumigatus is involved in oxidative stress tolerance and fungal virulence. Fungal Genet. Biol. 2014, 66, 79–85. [Google Scholar] [CrossRef]
- Szabó, K.; Jakab, Á.; Póliska, S.; Petrényi, K.; Kovács, K.; Issa, L.H.B.; Emri, T.; Pócsi, I.; Dombrádi, V. Deletion of the fungus specific protein phosphatase Z1 exaggerates the oxidative stress response in Candida albicans. BMC Genom. 2019, 20, 873. [Google Scholar] [CrossRef]
- Dantas, A.S.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef]
- Brito, W.A.; Rezende, T.C.V.; Parente, A.F.; Ricart, C.A.O.; Sousa, M.V.; Báo, S.N.; Soares, C.M.A. Identification, characterization and regulation studies of the aconitase of Paracoccidioides brasiliensis. Fungal Biol. 2011, 115, 697–707. [Google Scholar] [CrossRef]
- Narahari, J.; Ma, R.; Wang, M.; Walden, W.E. The aconitase function of iron regulatory protein 1. Genetic studies in yeast implicate its role in iron-mediated redox regulation. J. Biol. Chem. 2000, 275, 16227–16234. [Google Scholar] [CrossRef]
- Villafranca, J.J.; Mildvan, A.S. The mechanism of aconitase action. Detection and properties of enzyme-metal-substrate and enzyme-metal-inhibitor bridge complexes with manganese (II) and iron (II). J. Biol. Chem. 1972, 247, 3454–3463. [Google Scholar] [CrossRef]
- Lian, T.; Simmer, M.I.; D’Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.J.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2005, 55, 1452–1472. [Google Scholar] [CrossRef]
- Almirón, M.; Martínez, M.; Sanjuan, N.; Ugalde, R.A. Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect. Immun. 2001, 69, 6225–6230. [Google Scholar] [CrossRef][Green Version]
- Ibrahim, A.S. Host cell invasion in mucormycosis: Role of iron. Curr. Opin. Microbiol. 2011, 14, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Schaible, U.E.; Kaufmann, S.H.E. Iron and microbial infection. Nat. Rev. Microbiol. 2004, 2, 946–953. [Google Scholar] [CrossRef]
- Krajaejun, T.; Khositnithikul, R.; Lerksuthirat, T.; Lowhnoo, T.; Rujirawat, T.; Petchthong, T.; Yingyong, W.; Suriyaphol, P.; Smittipat, N.; Juthayothin, T.; et al. Expressed sequence tags reveal genetic diversity and putative virulence factors of the pathogenic oomycete Pythium insidiosum. Fungal Biol. 2011, 115, 683–696. [Google Scholar] [CrossRef]
- Santurio, J.M.; Zanette, R.A.; Bitencourt, P.E.R.; Alves, S.H.; Fighera, R.A.; Flores, M.M.; Wolkmer, P.; Hecktheuer, P.A.; Thomas, L.R.; Pereira, P.L.; et al. Insights into the pathophysiology of iron metabolism in Pythium insidiosum infections. Vet. Microbiol. 2013, 162, 826–830. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Robatzek, S. 14-3-3 Proteins in Plant-Pathogen Interactions. Mol. Plant-Microbe Interact. 2015, 28, 511–518. [Google Scholar] [CrossRef]
- Gardino, A.K.; Yaffe, M.B. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell. Dev. Biol. 2011, 22, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Obsilova, V.; Kopecka, M.; Kosek, D.; Kacirova, M.; Kylarova, S.; Rezabkova, L.; Obsil, T. Mechanisms of the 14-3-3 protein function: Regulation of protein function through conformational modulation. Physiol. Res. 2014, 63, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Kafle, A.; Puchadapirom, P.; Plumworasawat, S.; Dontumprai, R.; Chan-On, W.; Buates, S.; Laha, T.; Sripa, B.; Suttiprapa, S. Identification and characterization of protein 14-3-3 in carcinogenic liver fluke Opisthorchis viverrini. Parasitol. Int. 2017, 66, 426–431. [Google Scholar] [CrossRef]
- Andreotti, P.F.; Monteiro da Silva, J.L.; Bailão, A.M.; Soares, C.M.; Benard, G.; Soares, C.P.; Mendes-Giannini, M.J. Isolation and partial characterization of a 30 kDa adhesin from Paracoccidioides brasiliensis. Microbes Infect. 2005, 7, 875–881. [Google Scholar] [CrossRef]
- Marcos, C.M.; Silva, J.F.; Oliveira, H.C.; Assato, P.A.; Singulani, J.L.; Lopez, A.M.; Tamayo, D.P.; Hernandez-Ruiz, O.; McEwen, J.G.; Mendes-Giannini, M.J.; et al. Decreased expression of 14-3-3 in Paracoccidioides brasiliensis confirms its involvement in fungal pathogenesis. Virulence 2016, 7, 72–84. [Google Scholar] [CrossRef]
- Marcos, C.M.; Oliveira, H.C.; Assato, P.A.; Andrade, C.R.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Paracoccidioides brasiliensis 14-3-3 protein is important for virulence in a murine model. Med. Mycol. 2018, 57, 900–904. [Google Scholar] [CrossRef]
- Scorzoni, L.; de Paula e Silva, A.C.A.; Oliveira, H.C.; Santos, C.T.; Singulani, J.L.; Assato, P.A.; Marcos, C.M.; Oliveira, L.T.; Fregonezi, N.F.; Rossi, D.C.P.; et al. In Vitro and In Vivo Effect of Peptides Derived from 14-3-3 Paracoccidioides spp. Protein. J. Fungi 2021, 7, 52. [Google Scholar] [CrossRef]
- Champer, J.; Ito, J.I.; Clemons, K.V.; Stevens, D.A.; Kalkum, M. Proteomic Analysis of Pathogenic Fungi Reveals Highly Expressed Conserved Cell Wall Proteins. J. Fungi 2016, 2, 6. [Google Scholar] [CrossRef] [PubMed]
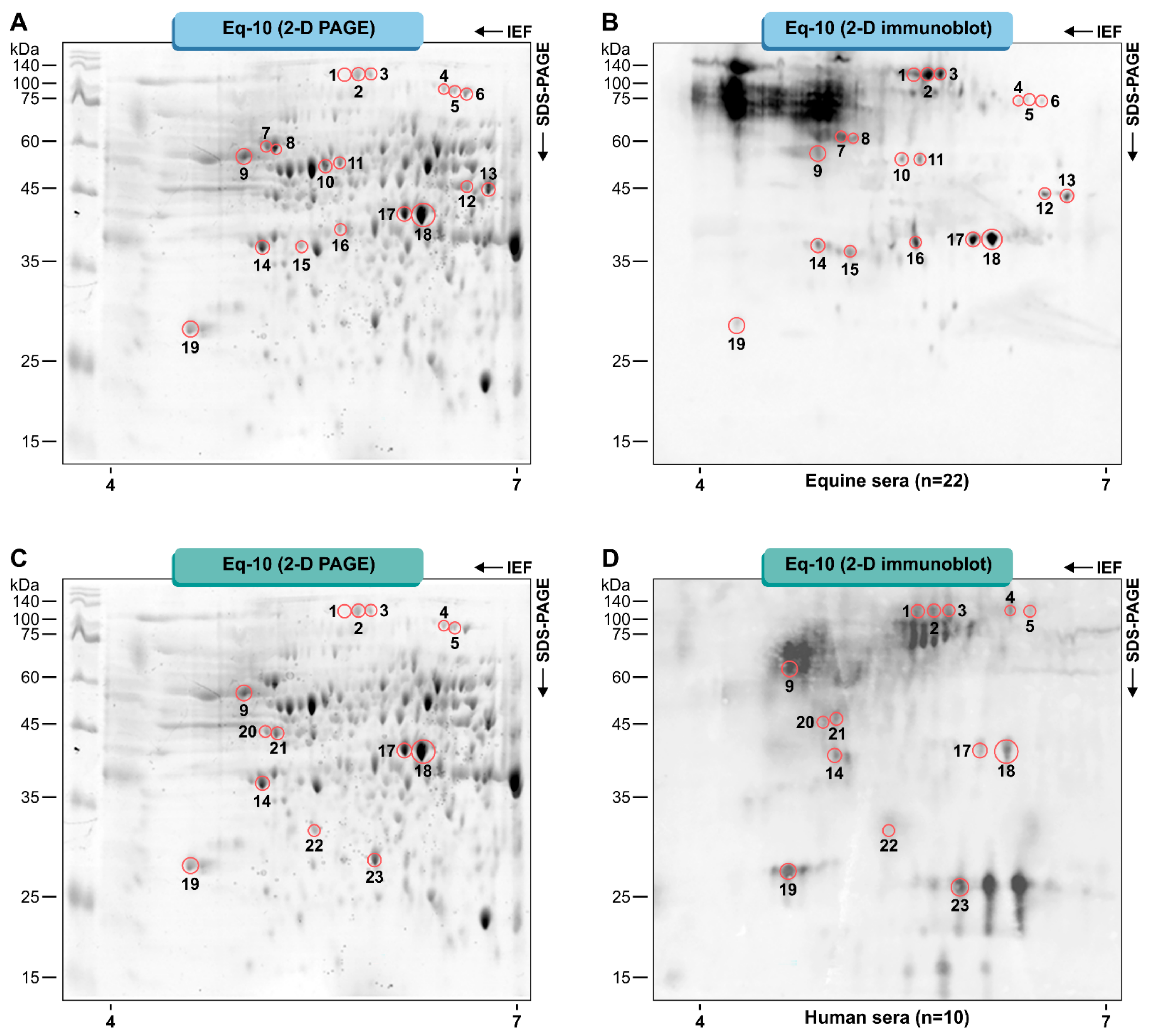

| Spot No | Protein Name | Accession No (Uniprot) | Gene | Pred MW (KDa) | pI | Protein Score | Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | Heat shock cognate 70 kDa protein | D0NHI7 | PITG_11913 | 86 | 5.54 | 66.239 | 16 |
| 2 | Heat shock cognate 70 kDa protein | D0NHI7 | PITG_11913 | 86 | 5.65 | 54.232 | 14 |
| Glucan 1,3-beta-glucosidase | D0NEL9 | PITG_10218 | 69 | 5.65 | 40.664 | 7 | |
| 3 | Heat shock cognate 70 kDa protein | D0NHI7 | PITG_11913 | 86 | 5.74 | 41.397 | 14 |
| Glucan 1,3-beta-glucosidase | D0NEL9 | PITG_10218 | 69 | 5.74 | 17.895 | 5 | |
| 4 | Aconitate hydratase | D0NY26 | PITG_18048 | 88 | 6.26 | 122.175 | 23 |
| 5 | Aconitate hydratase | D0NY26 | PITG_18048 | 88 | 6.35 | 191.649 | 27 |
| 6 | Aconitate hydratase | D0NY26 | PITG_18048 | 88 | 6.47 | 164.533 | 26 |
| 7 | Chaperonin CPN60-1 | D0NHM8 | PITG_11966 | 63 | 5.07 | 293.798 | 34 |
| 8 | Chaperonin CPN60-1 | D0NHM8 | PITG_11966 | 63 | 5.12 | 359.499 | 35 |
| 9 | Heat shock 70 kDa protein | D0NSJ5 | PITG_15786 | 68 | 4.90 | 12.61 | 8 |
| 10 | 6-phosphogluconate dehydrogenase | D0NE49 | PITG_10032 | 53 | 5.46 | 82.972 | 28 |
| 11 | Vacuolar proton pump subunit B | D0N6F5 | PITG_06118 | 57 | 5.55 | 248.97 | 50 |
| 12 | Isocitrate dehydrogenase [NADP] | D0N755 | PITG_07056 | 48 | 6.47 | 188.754 | 43 |
| 13 | Isocitrate dehydrogenase [NADP] | D0N755 | PITG_07056 | 48 | 6.60 | 183.697 | 44 |
| 14 | Serine/threonine-protein phosphatase | D0MXY7 | PITG_03574 | 35 | 5.06 | 10.346 | 23 |
| 15 | 40S ribosomal protein SA | D0MUE8 | PITG_01922 | 31 | 5.22 | 72.538 | 26 |
| 16 | Transaldolase | D0N3B2 | PITG_05636 | 37 | 5.74 | 24.121 | 16 |
| 17 | Fructose-bisphosphate aldolase | D0MX78 | PITG_02785 | 43 | 6.00 | 25.961 | 4 |
| 18 | Fructose-bisphosphate aldolase | D0MX78 | PITG_02785 | 43 | 6.14 | 246.075 | 26 |
| 19 | 14-3-3 protein epsilon | D0NYS2 | PITG_19017 | 28 | 4.58 | 425.094 | 82 |
| 20 | Arginase | D0N266 | PITG_04851 | 38 | 5.00 | 59.674 | 13 |
| 21 | Arginase | D0N266 | PITG_04851 | 38 | 5.10 | 115.704 | 17 |
| 22 | Glycerol-3-phosphate dehydrogenase [NAD(+)] | D0N0G8 | PITG_04065 | 38 | 5.34 | 46.334 | 20 |
| 23 | Triosephosphate isomerase | D0MZB6 | PITG_03078 | 25 | 5.81 | 52.947 | 14 |
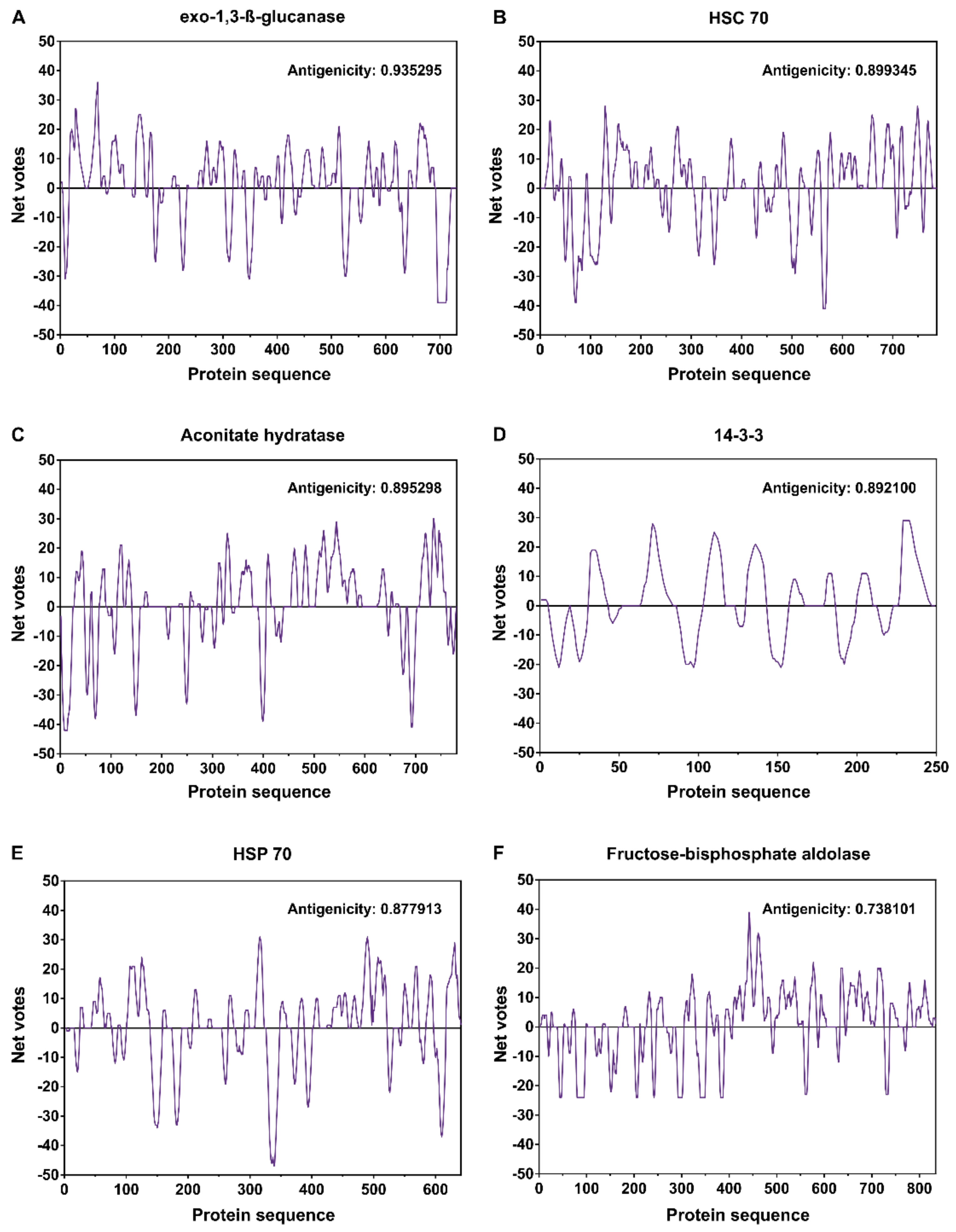
| Spot No | Protein | SCRATCH Score | Biological Process |
|---|---|---|---|
| 2 and 3 | Exo-1,3-ß-glucanase | 0.935295 | carbohydrate metabolic process |
| 1, 2, and 3 | HSC 70 | 0.899345 | stress response |
| 4 and 5 | Aconitate hydratase | 0.895298 | tricarboxylic acid cycle |
| 19 | 14-3-3 | 0.892100 | signaling protein ligands |
| 9 | HSP 70 | 0.877913 | protein folding and stress response |
| 17 and 18 | Fructose-bisphosphate aldolase | 0.738101 | glycolytic process |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chechi, J.L.; Rotchanapreeda, T.; da Paz, G.S.; Prado, A.C.; Oliveira, A.L.; Vieira, J.C.S.; Buzalaf, M.A.R.; Rodrigues, A.M.; Santos, L.D.d.; Krajaejun, T.; et al. Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis. J. Fungi 2021, 7, 423. https://doi.org/10.3390/jof7060423
Chechi JL, Rotchanapreeda T, da Paz GS, Prado AC, Oliveira AL, Vieira JCS, Buzalaf MAR, Rodrigues AM, Santos LDd, Krajaejun T, et al. Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis. Journal of Fungi. 2021; 7(6):423. https://doi.org/10.3390/jof7060423
Chicago/Turabian StyleChechi, Jéssica Luana, Tiwa Rotchanapreeda, Giselle Souza da Paz, Ana Carolina Prado, Alana Lucena Oliveira, José Cavalcante Souza Vieira, Marília Afonso Rabelo Buzalaf, Anderson Messias Rodrigues, Lucilene Delazari dos Santos, Theerapong Krajaejun, and et al. 2021. "Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis" Journal of Fungi 7, no. 6: 423. https://doi.org/10.3390/jof7060423
APA StyleChechi, J. L., Rotchanapreeda, T., da Paz, G. S., Prado, A. C., Oliveira, A. L., Vieira, J. C. S., Buzalaf, M. A. R., Rodrigues, A. M., Santos, L. D. d., Krajaejun, T., & Bosco, S. d. M. G. (2021). Prospecting Biomarkers for Diagnostic and Therapeutic Approaches in Pythiosis. Journal of Fungi, 7(6), 423. https://doi.org/10.3390/jof7060423










