Establishment of Agrobacterium tumefaciens-Mediated Transformation of Cladonia macilenta, a Model Lichen-Forming Fungus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Sensitivity of the C. macilenta Mycobiont to Hygromycin B
2.3. Optimization of ATMT
2.4. Statistical Analysis
2.5. Genomic DNA Extraction
2.6. Real-Time PCR Conditions for T-DNA Copy Number Variation Analysis
2.7. Relative Quantification of T-DNA Copy Number
2.8. TAIL-PCR and Sequencing
2.9. Microscopy
2.10. Data Availability
3. Results
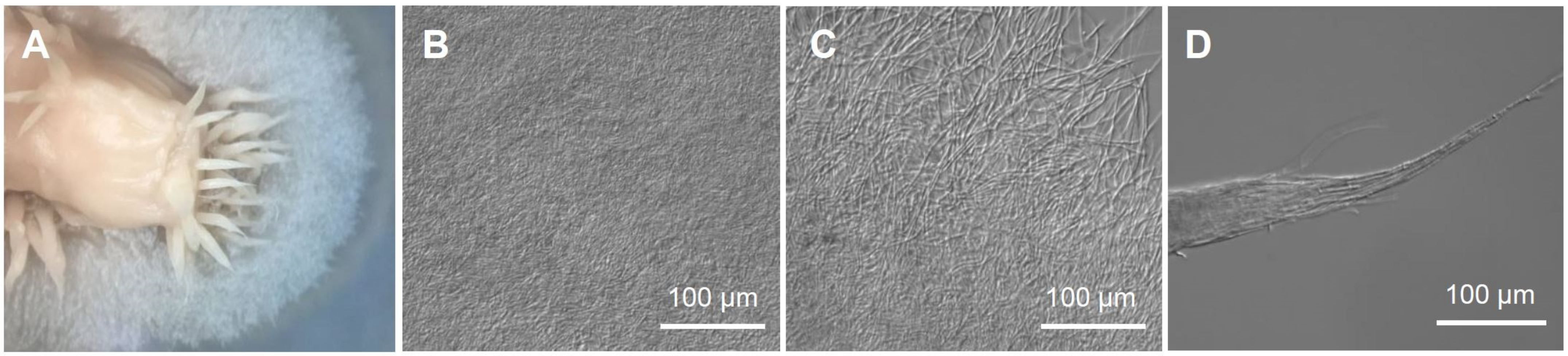
3.1. The Mycobiont of C. macilienta
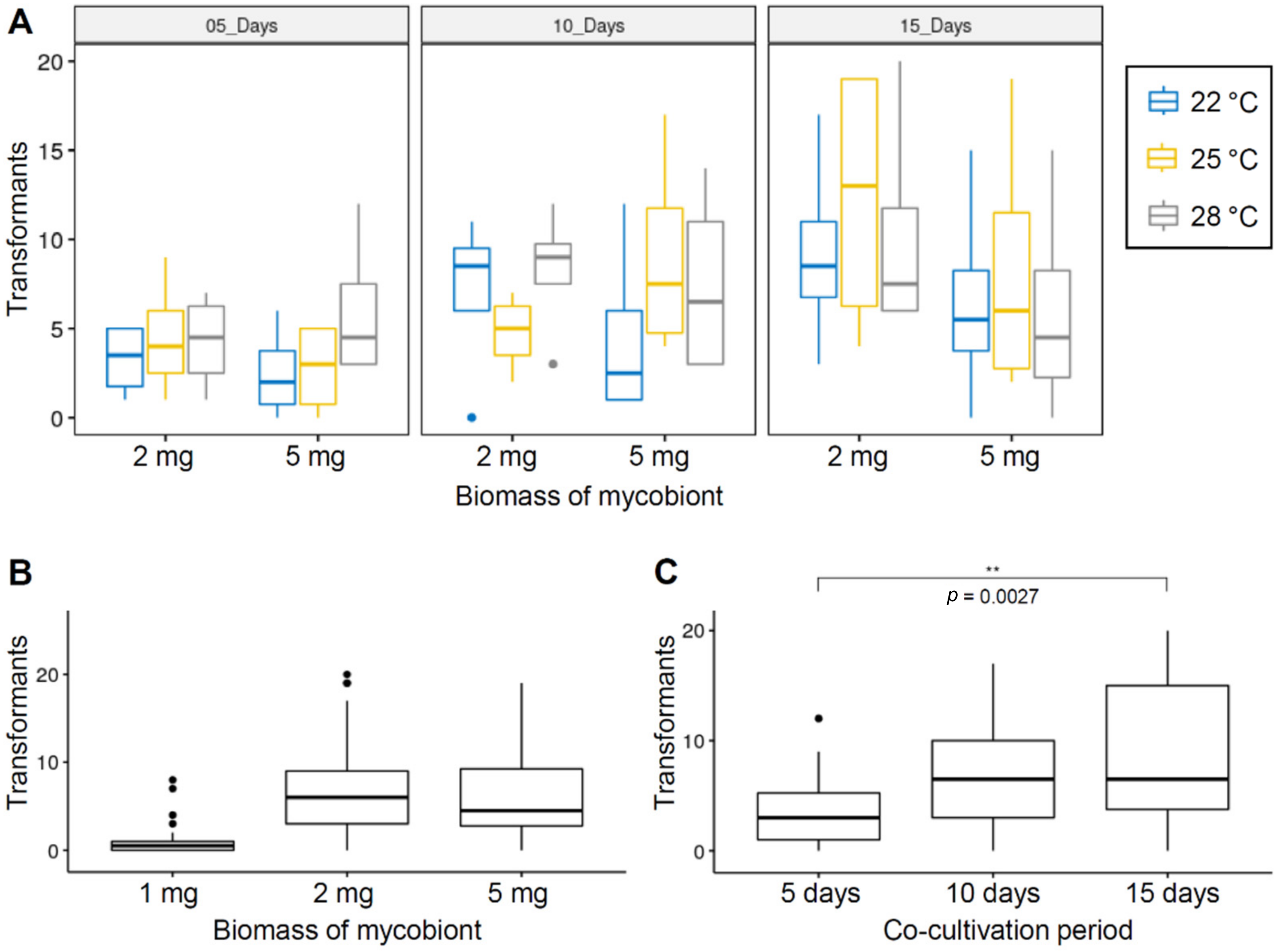
3.2. Optimization of ATMT Method for C. macilienta
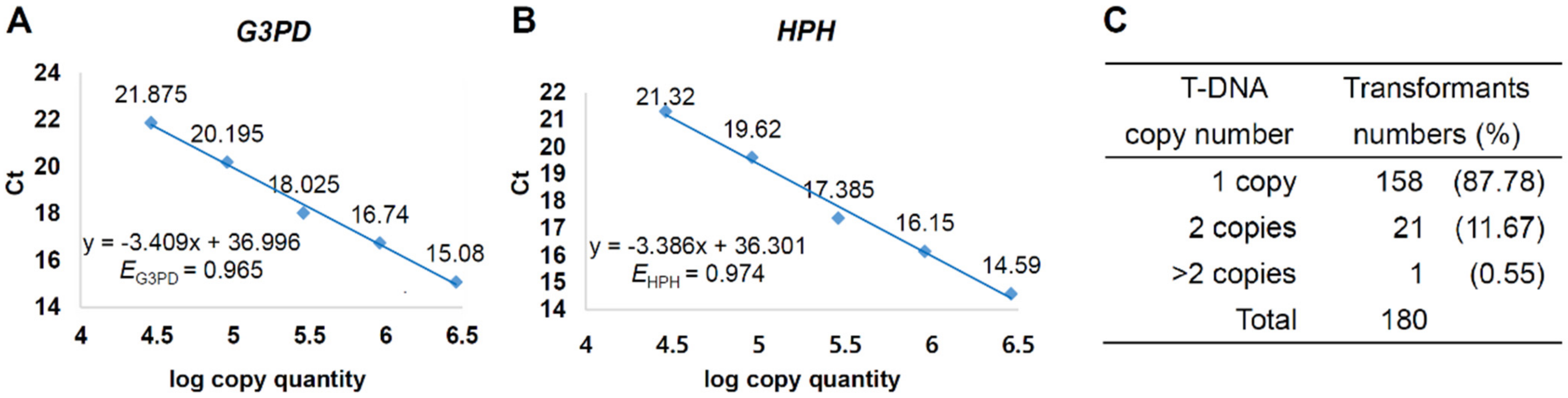
3.3. T-DNA Copy Number Variation of Transformants
3.4. Identification of T-DNA Insertion Sites
3.5. Phenotyping of C. macilenta Transformants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grube, M.; Wedin, M. Lichenized fungi and the evolution of symbiotic organization. In The Fungal Kingdom, 1st ed.; Heitman, J., Howlett, B.J., Crous, P.W., Stukenbrock, E.H., James, T.Y., Gow, N.A.R., Eds.; ASM Press: Washington, DC, USA, 2017; pp. 749–765. [Google Scholar]
- Honegger, R. The symbiotic phenotype of lichen-forming ascomycetes and their endo-and epibionts. In Fungal Associations; Hock, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 9, pp. 287–339. [Google Scholar]
- Meeßen, J.; Sánchez, F.J.; Brandt, A.; Balzer, E.-M.; de la Torre, R.; Sancho, L.G.; de Vera, J.-P.; Ott, S. Extremotolerance and resistance of lichens: Comparative studies on five species used in astrobiological research I. Morphological and anatomical characteristics. Orig. Life Evol. Biosph. 2013, 43, 283–303. [Google Scholar] [CrossRef]
- De la Torre Noetzel, R.; Ortega García, M.V.; Miller, A.Z.; Bassy, O.; Granja, C.; Cubero, B.; Jordão, L.; Martínez Frías, J.; Rabbow, E.; Backhaus, T.; et al. Lichen vitality after a space flight on board the expose-r2 facility outside the international space station: Results of the biology and mars experiment. Astrobiology 2020, 20, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Feuerer, T.; Hawksworth, D.L. Biodiversity of lichens, including a world-wide analysis of checklist data based on Takhtajan’s floristic regions. Biodivers. Conserv. 2007, 16, 85–98. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Kirk, P.M.; Sutton, B.C.; Pegler, D.N. Ainsworth & Bisby’s Dictionary of the Fungi, 8th ed.; CAB International: Wallingford, UK, 1995; p. 616. [Google Scholar]
- Lutzoni, F.; Pagel, M.; Reeb, V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature 2001, 411, 937–940. [Google Scholar] [CrossRef]
- Calcott, M.J.; Ackerley, D.F.; Knight, A.; Keyzers, R.A.; Owen, J.G. Secondary metabolism in the lichen symbiosis. Chem. Soc. Rev. 2018, 47, 1730–1760. [Google Scholar] [CrossRef]
- Yang, Y.; Bhosle, S.R.; Yu, Y.H.; Park, S.-Y.; Zhou, R.; Taş, İ.; Gamage, C.D.; Kim, K.K.; Pereira, I.; Hur, J.-S. Tumidulin, a lichen secondary metabolite, decreases the stemness potential of colorectal cancer cells. Molecules 2018, 23, 2968. [Google Scholar] [CrossRef]
- Harikrishnan, A.; Veena, V.; Lakshmi, B.; Shanmugavalli, R.; Theres, S.; Prashantha, C.; Shah, T.; Oshin, K.; Togam, R.; Nandi, S.; et al. Atranorin, an antimicrobial metabolite from lichen Parmotrema rampoddense exhibited in vitro anti-breast cancer activity through interaction with Akt activity. J. Biomol. Struct. Dyn. 2020, 1248–1258. [Google Scholar] [CrossRef]
- Goncu, B.; Sevgi, E.; Kizilarslan Hancer, C.; Gokay, G.; Ozten, N. Differential anti-proliferative and apoptotic effects of lichen species on human prostate carcinoma cells. PLoS ONE 2020, 15, e0238303. [Google Scholar]
- Gambichler, T.; Skrygan, M.; Tigges, C.; Kobus, S.; Gläser, R.; Kreuter, A. Significant upregulation of antimicrobial peptides and proteins in lichen sclerosus. Br. J. Dermatol. 2009, 161, 1136–1142. [Google Scholar] [CrossRef]
- Manojlovic, N.; Rankovic, B.; Kosanic, M.; Vasiljevic, P.; Stanojkovic, T. Chemical composition of three Parmelia lichens and antioxidant, antimicrobial and cytotoxic activities of some their major metabolites. Phytomedicine 2012, 19, 1166–1172. [Google Scholar] [CrossRef]
- Mitrović, T.; Stamenković, S.; Cvetković, V.; Tošić, S.; Stanković, M.; Radojević, I.; Stefanović, O.; Čomić, L.; Đačić, D.; Ćurčić, M. Antioxidant, antimicrobial and antiproliferative activities of five lichen species. Int. J. Mol. Sci. 2011, 12, 5428–5448. [Google Scholar] [CrossRef]
- Noël, A.; Garnier, A.; Clément, M.; Rouaud, I.; Sauvager, A.; Bousarghin, L.; Vásquez-Ocmín, P.; Maciuk, A.; Tomasi, S. Lichen-associated bacteria transform antibacterial usnic acid to products of lower antibiotic activity. Phytochemistry 2020, 181, 112535. [Google Scholar] [CrossRef]
- Carpentier, C.; Queiroz, E.F.; Marcourt, L.; Wolfender, J.L.; Azelmat, J.; Grenier, D.; Boudreau, S.; Voyer, N. Dibenzofurans and pseudodepsidones from the lichen Stereocaulon paschale collected in Northern Quebec. J. Nat. Prod. 2017, 80, 210–214. [Google Scholar] [CrossRef]
- Tay, T.; Türk, A.Ö.; Yılmaz, M.; Türk, H.; Kıvanç, M. Evaluation of the antimicrobial activity of the acetone extract of the lichen Ramalina farinacea and its (+)-usnic acid, norstictic acid, and protocetraric acid constituents. Z. Naturforsch. C J. Biosci. 2004, 59, 384–388. [Google Scholar] [CrossRef]
- Bugni, T.S.; Andjelic, C.D.; Pole, A.R.; Rai, P.; Ireland, C.M.; Barrows, L.R. Biologically active components of a Papua New Guinea analgesic and anti-inflammatory lichen preparation. Fitoterapia 2009, 80, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Tanas, S.; Odabasoglu, F.; Halici, Z.; Cakir, A.; Aygun, H.; Aslan, A.; Suleyman, H. Evaluation of anti-inflammatory and antioxidant activities of Peltigera rufescens lichen species in acute and chronic inflammation models. J. Nat. Med. 2010, 64, 42. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.-K.; Kang, K.-Y.; Jang, H.-Y.; Hwang, Y.-H.; Hong, S.-G.; Kim, S.-J.; Cho, H.-W.; Chang, D.-J.; Hur, J.-S.; Yee, S.-T. Atraric acid exhibits anti-inflammatory effect in lipopolysaccharide-stimulated RAW264.7 cells and mouse models. Int. J. Mol. Sci. 2020, 21, 7070. [Google Scholar] [CrossRef]
- Gaya, E.; Fernández-Brime, S.; Vargas, R.; Lachlan, R.F.; Gueidan, C.; Ramírez-Mejía, M.; Lutzoni, F. The adaptive radiation of lichen-forming Teloschistaceae is associated with sunscreening pigments and a bark-to-rock substrate shift. Proc. Natl. Acad. Sci. USA 2015, 112, 11600–11605. [Google Scholar] [CrossRef] [PubMed]
- Gadea, A.; Charrier, M.; Fanuel, M.; Clerc, P.; Daugan, C.; Sauvager, A.; Rogniaux, H.; Boustie, J.; Le Lamer, A.-C.; Lohézic-Le Devehat, F. Overcoming deterrent metabolites by gaining essential nutrients: A lichen/snail case study. Phytochemistry 2019, 164, 86–93. [Google Scholar] [CrossRef]
- Solhaug, K.A.; Gauslaa, Y. Secondary lichen compounds as protection against excess solar radiation and herbivores. In Progress in Botany 73, 1st ed.; Lüttge, U., Beyschlag, W., Büdel, B., Francis, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 283–304. [Google Scholar]
- Daboussi, M.; Djeballi, A.; Gerlinger, C.; Blaiseau, P.; Bouvier, I.; Cassan, M.; Lebrun, M.; Parisot, D.; Brygoo, Y. Transformation of seven species of filamentous fungi using the nitrate reductase gene of Aspergillus nidulans. Curr. Genet. 1989, 15, 453–456. [Google Scholar] [CrossRef]
- Leung, H.; Lehtinen, U.; Karjalainen, R.; Skinner, D.; Tooley, P.; Leong, S.; Ellingboe, A. Transformation of the rice blast fungus Magnaporthe grisea to hygromycin B resistance. Curr. Genet. 1990, 17, 409–411. [Google Scholar] [CrossRef]
- Gay, G.; Debaud, J.-C.; Casselton, L.A. Genetic transformation of the symbiotic basidiomycete fungus Hebeloma cylindrosporum. Curr. Genet. 1992, 22, 41–45. [Google Scholar]
- Kinoshita, Y. Protoplast isolation from lichen mycobionts. In Protocols in Lichenology, 1st ed.; Kranner, I.C., Beckett, R.P., Varma, A.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 61–64. [Google Scholar]
- Kinoshita, Y.; Hayase, S.; Higuchi, M.; Yamamoto, Y.; Ahmadjian, V.; Yoshimura, I.; Yamada, Y. Improvement of protoplast isolation from lichen mycobionts. Agric. Biol. Chem. 1991, 55, 1891–1892. [Google Scholar]
- Bundock, P.; den Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef]
- De Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Liu, N.; Chen, G.-Q.; Ning, G.-A.; Shi, H.-B.; Zhang, C.-L.; Lu, J.-P.; Mao, L.-J.; Feng, X.-X.; Liu, X.-H.; Su, Z.-Z.; et al. Agrobacterium tumefaciens-mediated transformation: An efficient tool for insertional mutagenesis and targeted gene disruption in Harpophora oryzae. Microbiol. Res. 2016, 182, 40–48. [Google Scholar] [CrossRef]
- Sugui, J.A.; Chang, Y.C.; Kwon-Chung, K.J. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: An efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 2005, 71, 1798–1802. [Google Scholar] [CrossRef]
- Chen, X.; Stone, M.; Schlagnhaufer, C.; Romaine, C.P. A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl. Environ. Microbiol. 2000, 66, 4510–4513. [Google Scholar] [CrossRef]
- Park, S.Y.; Jeong, M.H.; Wang, H.Y.; Kim, J.A.; Yu, N.H.; Kim, S.; Cheong, Y.H.; Kang, S.; Lee, Y.H.; Hur, J.S. Agrobacterium tumefaciens-mediated transformation of the lichen fungus, Umbilicaria muehlenbergii. PLoS ONE 2013, 8, e83896. [Google Scholar] [CrossRef]
- Tan, W.H.; Li, Y.P.; Xu, Y. Factors affect the formation and regeneration of protoplasts of microorganism. Xiandai Shipin Keji 2006, 22, 263–268. [Google Scholar]
- Eriksson, O.E. Outline of ascomycota. Myconet 2006, 12, 1–82. [Google Scholar]
- Trembley, M.L.; Ringli, C.; Honegger, R. Morphological and molecular analysis of early stages in the resynthesis of the lichen Baeomyces rufus. Mycol. Res. 2002, 106, 768–776. [Google Scholar] [CrossRef]
- Joneson, S.; Armaleo, D.; Lutzoni, F. Fungal and algal gene expression in early developmental stages of lichen-symbiosis. Mycologia 2011, 103, 291–306. [Google Scholar] [CrossRef]
- Armaleo, D.; Müller, O.; Lutzoni, F.; Andrésson, Ó.S.; Blanc, G.; Bode, H.B.; Collart, F.R.; Dal Grande, F.; Dietrich, F.; Grigoriev, I.V. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterochloris glomerata. BMC Genom. 2019, 20, 605. [Google Scholar] [CrossRef] [PubMed]
- Culberson, C.F.; Ahmadjian, V. Artificial reestablishment of lichens. II. Secondary products of resynthesized Cladonia cristatella and Lecanora chrysoleuca. Mycologia 1980, 72, 90–109. [Google Scholar] [CrossRef]
- Luo, H.; Li, C.; Kim, J.C.; Liu, Y.; Jung, J.S.; Koh, Y.J.; Hur, J.-S. Biruloquinone, an acetylcholinesterase inhibitor produced by lichen-forming fungus Cladonia macilenta. J. Microbiol. Biotechnol. 2013, 23, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Stenroos, S.; Hyvönen, J.; Myllys, L.; Thell, A.; Ahti, T. Phylogeny of the genus Cladonia s. lat. (Cladoniaceae, Ascomycetes) inferred from molecular, morphological, and chemical data. Cladistics 2002, 18, 237–278. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Plasmid Vectors. In Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; Volume 3, p. 626. [Google Scholar]
- Rho, H.-S.; Kang, S.; Lee, Y.-H. Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cells 2001, 12, 407–411. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, C.; Kim, J.; Shin, S.G.; Hwang, S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 2006, 123, 273–280. [Google Scholar] [CrossRef]
- Choi, J.; Park, J.; Jeon, J.; Chi, M.H.; Goh, J.; Yoo, S.Y.; Park, J.; Jung, K.; Kim, H.; Park, S.Y. Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae. Mol. Microbiol. 2007, 66, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y. Discharge and germination of lichen ascospores in the laboratory. Lichenology 2002, 1, 11–22. [Google Scholar]
- Park, S.-Y.; Choi, J.; Kim, J.A.; Jeong, M.-H.; Kim, S.; Lee, Y.-H.; Hur, J.-S. Draft genome sequence of Cladonia macilenta KoLRI003786, a lichen-forming fungus producing biruloquinone. Genome Announc. 2013, 1, e00695-13. [Google Scholar] [CrossRef] [PubMed]
- Abad, S.; Kitz, K.; Hörmann, A.; Schreiner, U.; Hartner, F.S.; Glieder, A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol. J. 2010, 5, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Mullins, E.D.; Chen, X.; Romaine, P.; Raina, R.; Geiser, D.M.; Kang, S. Agrobacterium-mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology 2001, 91, 173–180. [Google Scholar] [CrossRef]
- Islam, M.N.; Nizam, S.; Verma, P.K. A highly efficient Agrobacterium mediated transformation system for chickpea wilt pathogen Fusarium oxysporum f. sp ciceri using DsRed-Express to follow root colonisation. Microbiol. Res. 2012, 167, 332–338. [Google Scholar] [CrossRef]
- Neville, M.J.; Johnstone, E.C.; Walton, R.T. Identification and characterization of ANKK1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum. Mutat. 2004, 23, 540–545. [Google Scholar] [CrossRef]
- Tang, K.S.; Fersht, A.R.; Itzhaki, L.S. Sequential unfolding of ankyrin repeats in tumor suppressor p16. Structure 2003, 11, 67–73. [Google Scholar] [CrossRef][Green Version]
- Miller, M.K.; Bang, M.-L.; Witt, C.C.; Labeit, D.; Trombitas, C.; Watanabe, K.; Granzier, H.; McElhinny, A.S.; Gregorio, C.C.; Labeit, S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 2003, 333, 951–964. [Google Scholar] [CrossRef]


| Parameter | d.f. | F Statistics | p-Value | GES 1 |
|---|---|---|---|---|
| Biomass input (i) | 1 | 0.980 | 0.327 | 0.018 |
| Co-cultivation period (ii) | 2 | 5.522 | 0.007 * | 0.170 |
| Incubation temperature (iii) | 2 | 0.671 | 0.515 | 0.024 |
| (i):(ii) | 2 | 1.069 | 0.351 | 0.038 |
| (i):(iii) | 2 | 0.131 | 0.878 | 0.005 |
| (ii):(iii) | 4 | 0.297 | 0.879 | 0.022 |
| (i):(ii):(iii) | 4 | 0.551 | 0.699 | 0.039 |
| Strain | Flanking Sequence of T-DNA 1 | Length 2 (Identity) | Insertion Site 3 | Gene ID | Pfam Domain (E-Value) |
|---|---|---|---|---|---|
| CmT-2 | ATGATCATAGAAAGGATGCC | 134 (100) | 17:318521 | Cma_08524 | no domain found |
| CmT-5 | CAGGACGTCGATTGTAGCAC | 149 (99) | 5:324280 | intergenic | - |
| CmT-7 | CTGGAGGAGAATCAGGAGGT | 237 (100) | 2:2003077 | intergenic | - |
| CmT-9 | CGGCCGGGGAAAACCGTTCG | 133 (100) | 15:985193 | intergenic | - |
| CmT-41 | TCCGCTTTTTGGCAGGCTGC | 142 (99) | 21:390480 | intergenic | - |
| CmT-45 | AAGGAAACCTTACGCTAGTG | 130 (100) | 0:1782331 | intergenic | - |
| CmT-79 | TGGAGGGATGATCTCTATGG | 105 (100) | 16:437828 | intergenic | - |
| CmT-81 | ATTTTTTGTCCTGTCTGAGG | 61 (100) | 2:1031916 | Cma_01716 | PF00364 (7.9 × 10−16) |
| CmT-82 | TAGGAAATCAATGGGTTAAG | 527 (100) | 4:843652 | Cma_02877 | PF01812 (1.5 × 10−31) |
| CmT-83 | GTTCACCCCTCTTCGCAATA | 642 (100) | 20:650595 | Cma_09655 | PF12796 (3.8 × 10−23) |
| CmT-85 | TAGCGGATGCTTTAGGCGAA | 88 (100) | 14:378660 | intergenic | - |
| CmT-86 | CCTAGAGTAAGGTAGGTATG | 126 (100) | 20:271306 | intergenic | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Kim, W.; Paguirigan, J.A.; Jeong, M.-H.; Hur, J.-S. Establishment of Agrobacterium tumefaciens-Mediated Transformation of Cladonia macilenta, a Model Lichen-Forming Fungus. J. Fungi 2021, 7, 252. https://doi.org/10.3390/jof7040252
Liu R, Kim W, Paguirigan JA, Jeong M-H, Hur J-S. Establishment of Agrobacterium tumefaciens-Mediated Transformation of Cladonia macilenta, a Model Lichen-Forming Fungus. Journal of Fungi. 2021; 7(4):252. https://doi.org/10.3390/jof7040252
Chicago/Turabian StyleLiu, Rundong, Wonyong Kim, Jaycee Augusto Paguirigan, Min-Hye Jeong, and Jae-Seoun Hur. 2021. "Establishment of Agrobacterium tumefaciens-Mediated Transformation of Cladonia macilenta, a Model Lichen-Forming Fungus" Journal of Fungi 7, no. 4: 252. https://doi.org/10.3390/jof7040252
APA StyleLiu, R., Kim, W., Paguirigan, J. A., Jeong, M.-H., & Hur, J.-S. (2021). Establishment of Agrobacterium tumefaciens-Mediated Transformation of Cladonia macilenta, a Model Lichen-Forming Fungus. Journal of Fungi, 7(4), 252. https://doi.org/10.3390/jof7040252






