Fungal–Metal Interactions: A Review of Toxicity and Homeostasis
Abstract
1. Introduction
1.1. Fungal–Metal Interactions
| Metal | Transport Type | Yeast Transporters | Reference | Filamentous Fungi Transporters | Reference |
|---|---|---|---|---|---|
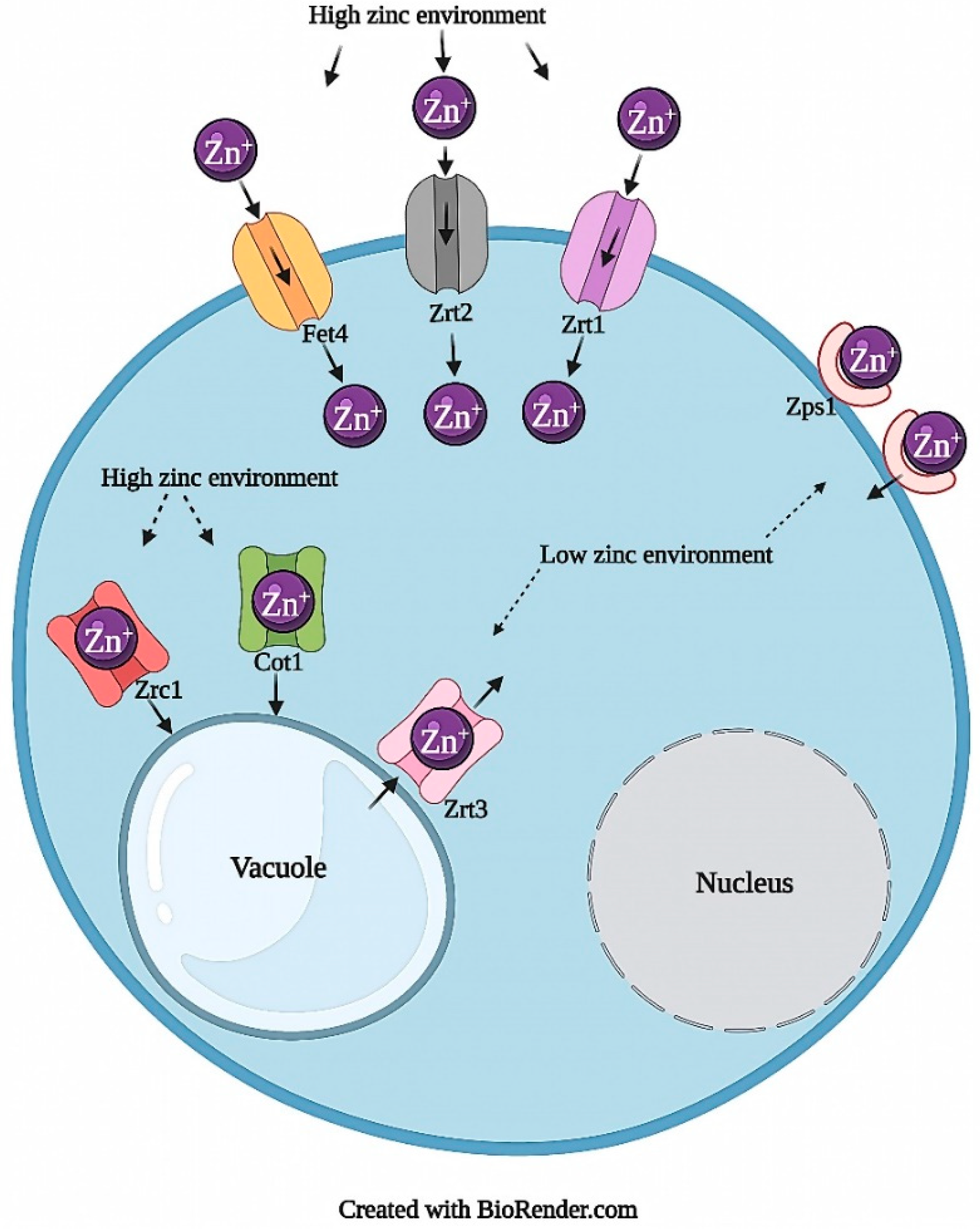
| Zinc | Import | Zrt1, Zrt2 | [15,22] | zrfA/B/C, UmZRT1/2, Zip1/2 | [23,24,25,26,27] |
| Vacuolar | Cot1, Zrc1 | [20,28] | - | - | |
| Vacuole to Cytosol | Zrt3 | [29] | - | - | |
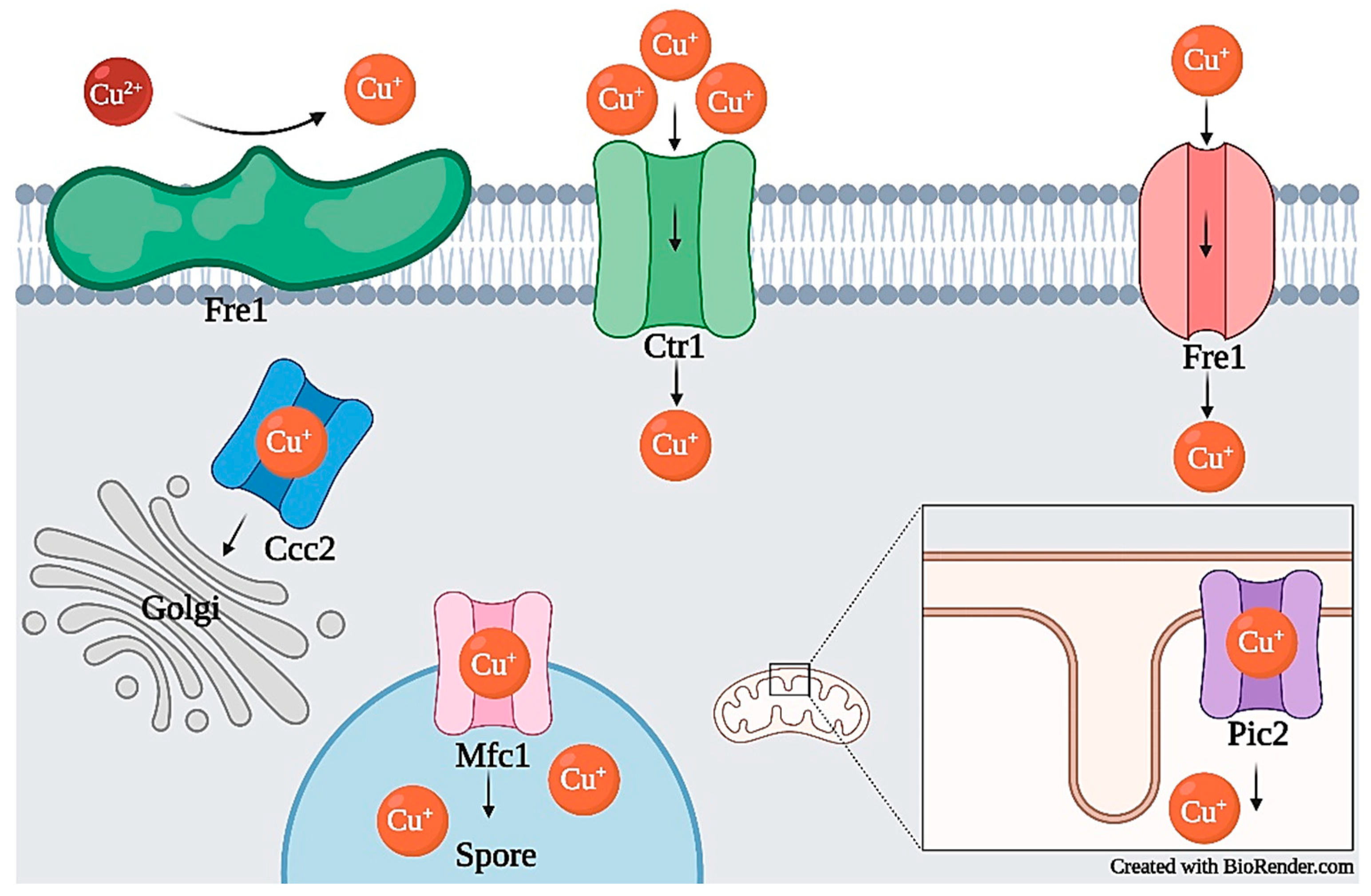
| Copper | Import | Ctr1, Ctr3, Fet4, Ctr4, Ctr5, Mfc1 | [30,31,32,33,34,35,36] | CtrA2, CtrC, Ctr1, PaCtr2 | [37,38,39] |
| Cytosol to Golgi | Atx1, Ccc2 | [34,40,41,42] | - | - | |
| Mitochondrial | Pic2, Cox17 | [21,43,44] | - | - | |
| Cytosol to Sod1 | Lys7, Pccs | [45,46] | - | - | |
| Mitochondrial Inner Membrane Space to Cytochrome c oxidase | Sco1, Sco2, Cox11 | [42,47,48] | - | - | |
| Export | - | - | CrpA | [49] | |
| Iron | Import | Fet4, Smf1, Fet3/Ftr1, Fip1, Str3, Shu1, Str1, Str2, Str3 | [50,51,52,53,54,55,56,57,58,59] | Fer2 | [60] |
| Within the Nucleus | Npb35, Nar1, Cfd1, Cia1 | [61,62] | - | - | |
| Vacuolar | Pcl1, Ccc1 | [63,64] | - | - | |
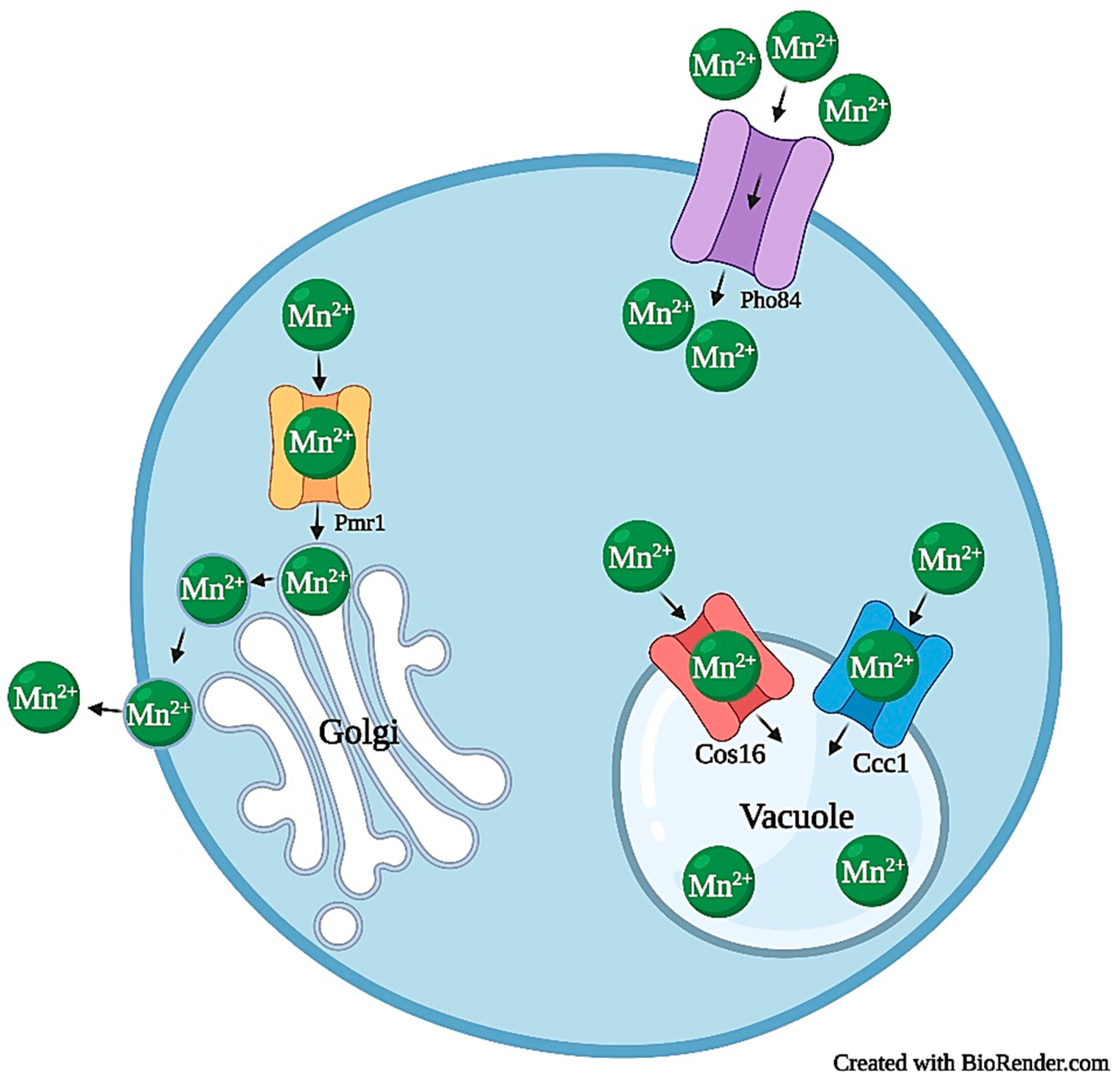
| Mangan-ese | Import | Smf1, Smf2, Pho85 | [52,65,66,67] | PcPho84, PcSmfs | [68] |
| Mitochondrial | Mtm1 | [69] | PcMtm1 | [68] | |
| Cytosol to Golgi Lumen | Pmr1, Gdt1 | [70,71,72] | - | - | |
| Cytosol to Endoplasmic Reticulum Lumen | Spf1 | [73] | - | - | |
| Vacuolar | Ccc1, Ypk9 | [64,74,75,76] | PcCCC1 | [68] | |
| Export | Pmr1, Hip1 | [77,78,79] | PcMnt | [68] | |
| Silver | Import | Ctr1 | [80,81] | - | - |
| Mitochondrial | Pic2 | [21] | - | - |
1.2. Metal Toxicity and Resistance
| Metal | Mechanism of Toxicity in Yeast | Reference | Mechanism of Toxicity in Filamentous Fungi | Reference |
|---|---|---|---|---|
| Zinc | Interference of synthesis of iron-sulfur clusters | [85,86] | increased chitin deposition within the cell wall, preventing hyphal extension | [87,88] |
| Interference in ergosterol biosynthesis | [83] | increased hyphal branching and apical swelling | [88] | |
| Cellular leakage, polarization, and increased membrane potential | [83] | interruption of conidia and conidiophore development (interference of reproduction) | [87] | |
| Reduced cell wall integrity | [83] | - | - | |
| Copper | Reduced ergosterol biosynthesis | [12,89] | Generation of reactive oxygen species | [90] |
| Reduced metallothionein activity | [84] | - | - | |
| Iron | Interference of vacuolar transport encoding gene CCC1 | [91,92] | Inability to acquire iron | [60,93] |
| Manganese | Down-regulation of HTB2, HTA1, HTA1, HTBI, HHF | [94,95] | potentially associated to reduced functioning of manganese peroxidase | [96,97,98] |
| Silver | Interference in ergosterol biosynthesis | [80,99,100] | - | - |
| Metal | Mechanism of Metal Resistance in Yeast | Reference | Mechanism of Metal Resistance in Filamentous Fungi | Reference |
|---|---|---|---|---|
| Zinc | Up-regulation of ZRC1 and COT1 | [83,105,106,107,108] | storage of excess zinc in vacuoles and cell walls of spores and hyphae | [109,110] |
| - | - | zinc efflux | [111] | |
| - | - | zinc metallothioneins | [112] | |
| Copper | Up-regulation of CUP1 and CRS5 | [113] | Up-regulation of crpA | [81,114,115,116] |
| Down-regulation of FRE1 and FRE7, and CTR1 | [113] | increased production of chelator copper oxalate | [117,118,119] | |
| Iron | Up-regulation of CCC1 | [64,120] | Unknown, but could associated with reduction of siderophore biosynthesis | [60,121] |
| Expression of plant ferritin genes | [122,123,124] | - | - | |
| Manganese | Up-regulation of MNR1 | [65,67,125,126] | Deletion of PcPHO84 | [68] |
| Down-regulation of PHO84, SMF1 | [67,125,126] | Expression of PcMNT | [68] | |
| Silver | Expression of CUP1-1, CUP1-2 | [81,115,116] | Expression of crpA | [90] |
| Down-regulation of PHO84 | [116] | - | - |
2. Fungal–Metal Interactions
2.1. Zinc
2.1.1. Zinc Transport and Homeostasis
2.1.2. Zinc Toxicity
2.1.3. Zinc Tolerance and Resistance
2.2. Copper
2.2.1. Copper Transport and Homeostasis
2.2.2. Copper Toxicity
2.2.3. Copper Tolerance and Resistance
2.3. Iron
2.3.1. Iron Transport and Homeostasis
2.3.2. Iron Toxicity
2.3.3. Iron Tolerance and Resistance
2.4. Manganese
2.4.1. Manganese Transport and Homeostasis
2.4.2. Manganese Toxicity
2.4.3. Manganese Tolerance and Resistance
2.5. Silver
2.5.1. Silver Transport and Homeostasis
2.5.2. Silver Toxicity
2.5.3. Silver Tolerance and Resistance
3. Omics and Metal Homeostasis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, A.; Mukherjee, P.; Senapati, S.; Mandal, D.; Khan, M.I.; Kumar, R.; Sastry, M. Extracellular Biosynthesis of Silver Nanoparticles Using the Fungus Fusarium oxysporum. Colloids Surf. B Biointerfaces 2003, 28, 313–318. [Google Scholar] [CrossRef]
- Birla, S.S.; Gaikwad, S.C.; Gade, A.K.; Rai, M.K. Rapid Synthesis of Silver Nanoparticles from Fusarium oxysporum by Optimizing Physicocultural Conditions. Sci. World J. 2013, 2013, 796018. [Google Scholar] [CrossRef] [PubMed]
- Naureen, B.; Miana, G.A.; Shahid, K.; Asghar, M.; Tanveer, S.; Sarwar, A. Iron (III) and Zinc (II) Monodentate Schiff Base Metal Complexes: Synthesis, Characterisation and Biological Activities. J. Mol. Struct. 2021, 1231, 129946. [Google Scholar] [CrossRef]
- Mani Chandrika, K.V.S.; Sharma, S. Promising Antifungal Agents: A Minireview. Bioorganic Med. Chem. 2020, 28, 115398. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.; Bhadauria, S.; Gaur, M.S. A Review: Biological Synthesis of Silver and Copper Nanoparticles. Nano Biomed. Eng. 2012, 4, 99–106. [Google Scholar] [CrossRef]
- Gudikandula, K.; Charya Maringanti, S. Synthesis of Silver Nanoparticles by Chemical and Biological Methods and Their Antimicrobial Properties. J. Exp. Nanosci. 2016, 11, 714–721. [Google Scholar] [CrossRef]
- Jamdagni, P.; Khatri, P.; Rana, J.S. Green Synthesis of Zinc Oxide Nanoparticles Using Flower Extract of Nyctanthes Arbor-tristis and Their Antifungal Activity. J. King Saud Univ. Sci. 2018, 30, 168–175. [Google Scholar] [CrossRef]
- Bundschuh, M.; Filser, J.; Lüderwald, S.; McKee, M.S.; Metreveli, G.; Schaumann, G.E.; Schulz, R.; Wagner, S. Nanoparticles in the Environment: Where Do We Come from, Where Do We Go to? Environ. Sci. Eur. 2018, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kittler, S.; Greulich, C.; Diendorf, J.; Koller, M.; Epple, M. Toxicity of Silver Nanoparticles Increases during Storage because of Slow Dissolution under Release of Silver Ions. Chem. Mater. 2010, 22, 4548–4554. [Google Scholar] [CrossRef]
- Mussin, J.E.; Roldán, M.V.; Rojas, F.; de los Ángeles Sosa, M.; Pellegri, N.; Giusiano, G. Antifungal Activity of Silver Nanoparticles in Combination with Ketoconazole against Malassezia Furfur. AMB Express 2019, 9, 131. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, Applications and Toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Cheong, Y.-K.; Arce, M.P.; Benito, A.; Chen, D.; Luengo Crisóstomo, N.; Kerai, L.V.; Rodríguez, G.; Valverde, J.L.; Vadalia, M.; Cerpa-Naranjo, A. Synergistic Antifungal Study of PEGylated Graphene Oxides and Copper Nanoparticles Against Candida Albicans. Nanomaterials 2020, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Mulenos, M.R.; Liu, J.; Lujan, H.; Guo, B.; Lichtfouse, E.; Sharma, V.K.; Sayes, C.M. Copper, Silver, and Titania Nanoparticles Do not Release Ions under Anoxic Conditions and Release Only Minute Ion Levels Under Oxic Conditions in Water: Evidence for the Low Toxicity of Nanoparticles. Environ. Chem. Lett. 2020, 18, 1319–1328. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, Y.; He, D.; Fang, X.; Xu, J.; Lee, Y.-W.; Keller, N.P.; Shi, J. Copper Tolerance Mediated by FgAceA and FgCrpA in Fusarium graminearum. Front. Microbiol. 2020, 11, 1392. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Eide, D. The Yeast ZRT1 Gene Encodes the Zinc Transporter Protein of a High-affinity uptake System Induced by Zinc Limitation. Proc. Natl. Acad. Sci. USA 1996, 93, 2454–2458. [Google Scholar] [CrossRef]
- Moore, R.E.; Kim, Y.; Philpott, C.C. The Mechanism of Ferrichrome Transport through Arn1p and Its Metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 5664–5669. [Google Scholar] [CrossRef]
- Voß, B.; Kirschhöfer, F.; Brenner-Weiß, G.; Fischer, R. Alternaria alternata Uses Two Siderophore Systems for Iron Acquisition. Sci. Rep. 2020, 10, 3587. [Google Scholar] [CrossRef] [PubMed]
- Butt, T.R.; Sternberg, E.; Herd, J.; Crooke, S.T. Cloning and Expression of a Yeast Copper Metallothionein Gene. Gene 1984, 27, 23–33. [Google Scholar] [CrossRef]
- Perinelli, M.; Tegoni, M.; Freisinger, E. Different Behavior of the Histidine Residue toward Cadmium and Zinc in a Cadmium-Specific Metallothionein from an Aquatic Fungus. Inorg. Chem. 2020, 59, 16988–16997. [Google Scholar] [CrossRef]
- Cho, M.; Hu, G.; Caza, M.; Horianopoulos, L.C.; Kronstad, J.W.; Jung, W.H. Vacuolar Zinc Transporter Zrc1 is Required for Detoxification of Excess Intracellular Zinc in the Human Fungal Pathogen Cryptococcus neoformans. J. Microbiol. 2018, 56, 65–71. [Google Scholar] [CrossRef]
- Vest, K.E.; Leary, S.C.; Winge, D.R.; Cobine, P.A. Copper Import into the Mitochondrial Matrix in Saccharomyces cerevisiae is Mediated by Pic2, a Mitochondrial Carrier Family Protein. J. Biol. Chem. 2013, 288, 23884–23892. [Google Scholar] [CrossRef]
- Zhao, H.; Eide, D. The ZRT2 Gene Encodes the Low Affinity Zinc Transporter in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 23203–23210. [Google Scholar] [CrossRef]
- Amich, J.; Vicentefranqueira, R.; Mellado, E.; Ruiz-Carmuega, A.; Leal, F.; Calera, J.A. The ZrfC Alkaline Zinc Transporter is Required for Aspergillus fumigatus Virulence and Its Growth in the Presence of the Zn/Mn-chelating Protein Calprotectin. Cell. Microbiol. 2014, 16, 548–564. [Google Scholar] [CrossRef]
- Do, E.; Hu, G.; Caza, M.; Kronstad, J.W.; Jung, W.H. The ZIP Family Zinc Transporters Support the Virulence of Cryptococcus neoformans. Med. Mycol. 2016, 54, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, F.M.; Piffer, A.C.; Schneider, R.O.; Ribeiro, N.S.; Garcia, A.W.A.; Schrank, A.; Kmetzsch, L.; Vainstein, M.H.; Staats, C.C. Alterations of Zinc Homeostasis in Response to Cryptococcus neoformans in a Murine Macrophage Cell Line. Future Microbiol. 2017, 12, 491–504. [Google Scholar] [CrossRef]
- Martha-Paz, A.M.; Eide, D.; Mendoza-Cózatl, D.; Castro-Guerrero, N.A.; Aréchiga-Carvajal, E.T. Zinc Uptake in the Basidiomycota: Characterization of Zinc Transporters in Ustilago maydis. Mol. Membr. Biol. 2019, 35, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Vicentefranqueira, R.; Amich, J.; Laskaris, P.; Ibrahim-Granet, O.; Latgé, J.P.; Toledo, H.; Leal, F.; Calera, J.A. Targeting Zinc Homeostasis to combat Aspergillus fumigatus Infections. Front. Microbiol. 2015, 6, 160. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Milanick, M.A.; Eide, D.J. Induction of the ZRC1 Metal Tolerance Gene in Zinc-limited Yeast Confers Resistance to Zinc Shock. J. Biol. Chem. 2003, 278, 15065–15072. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.; Gaither, L.; Eide, D. Zinc Transporters That Regulate Vacuolar Zinc Storage in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2845–2855. [Google Scholar] [CrossRef]
- Gross, C.; Kelleher, M.; Iyer, V.R.; Brown, P.O.; Winge, D.R. Identification of the Copper Regulon in Saccharomyces cerevisiae by DNA Microarrays. J. Biol. Chem. 2000, 275, 32310–32316. [Google Scholar] [CrossRef]
- Hassett, R.; Dix, D.R.; Eide, D.J.; Kosman, D.J. The Fe(II) Permease Fet4p Functions as a Low Affinity Copper Transporter and Supports Normal Copper Trafficking in Saccharomyces cerevisiae. Biochem. J. 2000, 351 Pt 2, 477–484. [Google Scholar] [CrossRef]
- Pena, M.M.; Puig, S.; Thiele, D.J. Characterization of the Saccharomyces cerevisiae High Affinity Copper Transporter Ctr3. J. Biol. Chem. 2000, 275, 33244–33251. [Google Scholar] [CrossRef]
- Peña, M.M.O.; Koch, K.A.; Thiele, D.J. Dynamic Regulation of Copper Uptake and Detoxification Genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 2514–2523. [Google Scholar] [CrossRef]
- Beaudoin, J.; Ekici, S.; Daldal, F.; Ait-Mohand, S.; Guérin, B.; Labbé, S. Copper Transport and Regulation in Schizosaccharomyces pombe. Biochem. Soc. Trans. 2013, 41, 1679–1686. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beaudoin, J.; Ioannoni, R.; Mailloux, S.; Plante, S.; Labbé, S. Transcriptional Regulation of the Copper Transporter Mfc1 in Meiotic Cells. Eukaryot. Cell 2013, 12, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Thiele, D.J. Identification of a Novel High Affinity Copper Transport Complex in the Fission Yeast Schizosaccharomyces pombe. J. Biol. Chem. 2001, 276, 20529–20535. [Google Scholar] [CrossRef]
- Park, Y.-S.; Lian, H.; Chang, M.; Kang, C.-M.; Yun, C.-W. Identification of High-affinity Copper Transporters in Aspergillus fumigatus. Fungal Genet. Biol. 2014, 73, 29–38. [Google Scholar] [CrossRef]
- Peñas, M.M.; Azparren, G.; Domínguez, Á.; Sommer, H.; Ramírez, L.; Pisabarro, A.G. Identification and Functional Characterisation of Ctr1, a Pleurotus ostreatus Gene Coding for a Copper Transporter. Mol. Genet. Genom. 2005, 274, 402–409. [Google Scholar] [CrossRef]
- Raffa, N.; Osherov, N.; Keller, N. Copper Utilization, Regulation, and Acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 2019, 20, 1980. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-J.; Pufahl, R.A.; Dancis, A.; O’Halloran, T.V.; Culotta, V.C. A Role for the Saccharomyces cerevisiae ATX1 Gene in Copper Trafficking and Iron Transport. J. Biol. Chem. 1997, 272, 9215–9220. [Google Scholar] [CrossRef]
- Yuan, D.S.; Dancis, A.; Klausner, R.D. Restriction of Copper Export in Saccharomyces cerevisiae to a Late Golgi or Post-Golgi Compartment in the Secretory Pathway. J. Biol. Chem. 1997, 272, 25787–25793. [Google Scholar] [CrossRef]
- Peter, C.; Laliberté, J.; Beaudoin, J.; Labbé, S. Copper Distributed by Atx1 is Available to Copper Amine Oxidase 1 in Schizosaccharomyces pombe. Eukaryot. Cell 2008, 7, 1781–1794. [Google Scholar] [CrossRef]
- Beers, J.; Glerum, D.M.; Tzagoloff, A. Purification, Characterization, and Localization of Yeast Cox17p, a Mitochondrial Copper Shuttle. J. Biol. Chem. 1997, 272, 33191–33196. [Google Scholar] [CrossRef]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. Characterization of COX17, a Yeast Gene Involved in Copper Metabolism and Assembly of Cytochrome Oxidase. J. Biol. Chem. 1996, 271, 14504–14509. [Google Scholar] [CrossRef]
- Culotta, V.C.; Klomp, L.W.J.; Strain, J.; Casareno, R.L.B.; Krems, B.; Gitlin, J.D. The Copper Chaperone for Superoxide Dismutase. J. Biol. Chem. 1997, 272, 23469–23472. [Google Scholar] [CrossRef] [PubMed]
- Laliberté, J.; Whitson, L.J.; Beaudoin, J.; Holloway, S.P.; Hart, P.J.; Labbé, S. The Schizosaccharomyces pombe Pccs Protein Functions in Both Copper Trafficking and Metal Detoxification Pathways. J. Biol. Chem. 2004, 279, 28744–28755. [Google Scholar] [CrossRef] [PubMed]
- Carr, H.S.; George, G.N.; Winge, D.R. Yeast Cox11, a Protein Essential for Cytochrome cOxidase Assembly, Is a Cu(I)-binding Protein. J. Biol. Chem. 2002, 277, 31237–31242. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Logeman, B.L.; Thiele, D.J. Copper Acquisition and Utilization in Fungi. Annu. Rev. Microbiol. 2017, 71, 597–623. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Perevitsky, A.; Lim, F.Y.; Shadkchan, Y.; Knox, B.P.; Landero Figueora, J.A.; Choera, T.; Niu, M.; Steinberger, A.J.; Wüthrich, M.; et al. Aspergillus fumigatus Copper Export Machinery and Reactive Oxygen Intermediate Defense Counter Host Copper-Mediated Oxidative Antimicrobial Offense. Cell Rep. 2017, 19, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Askwith, C.; Kaplan, J. An Oxidase-permease-based Iron Transport System in Schizosaccharomyces pombe and Its Expression in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 401–405. [Google Scholar] [CrossRef]
- Chen, X.-Z.; Peng, J.-B.; Cohen, A.; Nelson, H.; Nelson, N.; Hediger, M.A. Yeast SMF1 Mediates H+-coupled Iron Uptake with Concomitant Uncoupled Cation Currents. J. Biol. Chem. 1999, 274, 35089–35094. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Nelson, H.; Nelson, N. The Family of SMF Metal Ion Transporters in Yeast Cells. J. Biol. Chem. 2000, 275, 33388–33394. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.M.; Askwith, C.C.; Eide, D.; Kaplan, J. The FET3 Gene Product Required for High Affinity Iron Transport in Yeast Is a Cell Surface Ferroxidase. J. Biol. Chem. 1995, 270, 1098–1101. [Google Scholar] [PubMed]
- Dix, D.; Bridgham, J.; Broderius, M.; Eide, D. Characterization of the FET4 Protein of Yeast Evidence for a Direct Role in the Transport of Iron. J. Biol. Chem. 1997, 272, 11770–11777. [Google Scholar] [CrossRef] [PubMed]
- Dix, D.R.; Bridgham, J.T.; Broderius, M.A.; Byersdorfer, C.A.; Eide, D.J. The FET4 Gene Encodes the Low Affinity Fe(II) Transport Protein of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 26092–26099. [Google Scholar] [CrossRef]
- Mourer, T.; Jacques, J.-F.; Brault, A.; Bisaillon, M.; Labbé, S. Shu1 Is a Cell-surface Protein Involved in Iron Acquisition from Heme in Schizosaccharomyces pombe. J. Biol. Chem. 2015, 290, 10176–10190. [Google Scholar] [CrossRef]
- Normant, V.; Mourer, T.; Labbé, S. The Major Facilitator Transporter Str3 is Required for Low-affinity Heme Acquisition in Schizosaccharomyces pombe. J. Biol. Chem. 2018, 293, 6349–6362. [Google Scholar] [CrossRef]
- Stearman, R.; Yuan, D.S.; Yamaguchi-Iwai, Y.; Klausner, R.D.; Dancis, A. A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast. Science 1996, 271, 1552–1557. [Google Scholar] [CrossRef]
- Pelletier, B.; Beaudoin, J.; Philpott, C.C.; Labbé, S. Fep1 Represses Expression of the Fission Yeast Schizosaccharomyces pombe Siderophore-iron Transport System. Nucleic Acids Res. 2003, 31, 4332–4344. [Google Scholar] [CrossRef][Green Version]
- Eichhorn, H.; Lessing, F.; Winterberg, B.; Schirawski, J.; Kämper, J.; Müller, P.; Kahmann, R. A Ferroxidation/permeation Iron Uptake System is Required for Virulence in Ustilago maydis. Plant Cell 2006, 18, 3332–3345. [Google Scholar] [CrossRef]
- Lindahl, P.A. A Comprehensive Mechanistic Model of Iron Metabolism in Saccharomyces cerevisiae. Met. Integr. Biometal Sci. 2019, 11, 1779–1799. [Google Scholar] [CrossRef]
- Netz, D.; Pierik, A.; Stümpfig, M.; Muhlenhoff, U.; Lill, R. The Cfd1-Nbp35 Complex Acts as a Scaffold for Iron-sulfur Protein Assembly in the Yeast Cytosol. Nat. Chem. Biol. 2007, 3, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Labbé, S.; Khan, M.G.M.; Jacques, J.-F. Iron Uptake and Regulation in Schizosaccharomyces pombe. Curr. Opin. Microbiol. 2013, 16, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, O.S.; Ward, D.M.; Kaplan, J. CCC1 is a Transporter That Mediates Vacuolar Iron Storage in Yeast. J. Biol. Chem. 2001, 276, 29515–29519. [Google Scholar] [CrossRef]
- Supek, F.; Supekova, L.; Nelson, H.; Nelson, N. A Yeast Manganese Transporter Related to the Macrophage Protein Involved in Conferring Resistance to Mycobacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 5105–5110. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Supekova, L.; Nelson, H.; Nelson, N. Function of Metal-ion Homeostasis in the Cell Division Cycle, Mitochondrial Protein Processing, Sensitivity to Mycobacterial Infection and Brain Function. J. Exp. Biol. 1997, 200, 321–330. [Google Scholar] [PubMed]
- Jensen, L.T.; Ajua-Alemanji, M.; Culotta, V.C. The Saccharomyces cerevisiae High Affinity Phosphate Transporter Encoded by PHO84 also Functions in Manganese Homeostasis. J. Biol. Chem. 2003, 278, 42036–42040. [Google Scholar] [CrossRef]
- Diss, L.; Blaudez, D.; Gelhaye, E.; Chalot, M. Genome-wide Analysis of Fungal Manganese Transporters, with an Emphasis on Phanerochaete chrysosporium. Environ. Microbiol. Rep. 2011, 3, 367–382. [Google Scholar] [CrossRef]
- Luk, E.; Carroll, M.; Baker, M.; Culotta, V.C. Manganese Activation of Superoxide Dismutase 2 in Saccharomyces cerevisiae Requires MTM1, a Member of the Mitochondrial Carrier Family. Proc. Natl. Acad. Sci. USA 2003, 100, 10353–10357. [Google Scholar] [CrossRef] [PubMed]
- Dulary, E.; Yu, S.-Y.; Houdou, M.; de Bettignies, G.; Decool, V.; Potelle, S.; Duvet, S.; Krzewinski-Recchi, M.-A.; Garat, A.; Matthijs, G.; et al. Investigating the Function of Gdt1p in Yeast Golgi glycosylation. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 394–402. [Google Scholar] [CrossRef]
- Lapinskas, P.J.; Cunningham, K.W.; Liu, X.F.; Fink, G.R.; Culotta, V.C. Mutations in PMR1 Suppress Oxidative Damage in Yeast Cells Lacking Superoxide Dismutase. Mol. Cell. Biol. 1995, 15, 1382–1388. [Google Scholar] [CrossRef]
- Thines, L.; Deschamps, A.; Sengottaiyan, P.; Savel, O.; Stribny, J.; Morsomme, P. The Yeast Protein Gdt1p Transports Mn(2+) Ions and Thereby Regulates Manganese Homeostasis in the Golgi. J. Biol. Chem. 2018, 293, 8048–8055. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Megyeri, M.; Chen, O.C.W.; Condomitti, G.; Riezman, I.; Loizides-Mangold, U.; Abdul-Sada, A.; Rimon, N.; Riezman, H.; Platt, F.M.; et al. The Yeast P5 Type ATPase, Spf1, Regulates Manganese Transport into the Endoplasmic Reticulum. PLoS ONE 2014, 8, e85519. [Google Scholar] [CrossRef] [PubMed]
- Gitler, A.D.; Chesi, A.; Geddie, M.L.; Strathearn, K.E.; Hamamichi, S.; Hill, K.J.; Caldwell, K.A.; Caldwell, G.A.; Cooper, A.A.; Rochet, J.-C.; et al. Alpha-synuclein is Part of a Diverse and Highly Conserved Interaction Network That Includes PARK9 and Manganese Toxicity. Nat. Genet. 2009, 41, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Lapinskas, P.J.; Lin, S.-J.; Culotta, V.C. The Role of the Saccharomyces cerevisiae CCC1 Gene in the Homeostasis of Manganese Ions. Mol. Microbiol. 1996, 21, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Wolfe, D.M.; Stiller, B.; Pearce, D.A. Cd2+, Mn2+, Ni2+ and Se2+ Toxicity to Saccharomyces cerevisiae Lacking YPK9p the Orthologue of Human ATP13A2. Biochem. Biophys. Res. Commun. 2009, 383, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Culotta, V.C.; Yang, M.; Hall, M.D. Manganese Transport and Trafficking: Lessons Learned from Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1159–1165. [Google Scholar] [CrossRef]
- Farcasanu, I.; Mizunuma, M.; Hirata, D.; Miyakawa, T. Involvement of Histidine Permease (Hip1p) in Manganese Transport in Saccharomyces cerevisiae. Mol. Gen. Genetics MGG 1998, 259, 541–548. [Google Scholar] [CrossRef]
- Ton, V.-K.; Mandal, D.; Vahadji, C.; Rao, R. Functional Expression in Yeast of the Human Secretory Pathway Ca2+, Mn2+-ATPase Defective in Hailey-Hailey Disease. J. Biol. Chem. 2002, 277, 6422–6427. [Google Scholar] [CrossRef]
- Horstmann, C.; Campbell, C.; Kim, D.S.; Kim, K. Transcriptome Profile with 20 nm Silver Nanoparticles in Yeast. FEMS Yeast Res. 2019, 19. [Google Scholar] [CrossRef]
- Ruta, L.L.; Banu, M.A.; Neagoe, A.D.; Kissen, R.; Bones, A.M.; Farcasanu, I.C. Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins. Cells 2018, 7, 266. [Google Scholar] [CrossRef] [PubMed]
- B, A.W. Oligodynamic Phenomena of Living Cells. Nature 1893, 48, 331. [Google Scholar] [CrossRef]
- Galván Márquez, I.; Ghiyasvand, M.; Massarsky, A.; Babu, M.; Samanfar, B.; Omidi, K.; Moon, T.W.; Smith, M.L.; Golshani, A. Zinc Oxide and Silver Nanoparticles Toxicity in the Baker’s Yeast, Saccharomyces cerevisiae. PLoS ONE 2018, 13, e0193111. [Google Scholar] [CrossRef] [PubMed]
- Sinisi, V.; Pelagatti, P.; Carcelli, M.; Migliori, A.; Mantovani, L.; Righi, L.; Leonardi, G.; Pietarinen, S.; Hubsch, C.; Rogolino, D. A Green Approach to Copper-Containing Pesticides: Antimicrobial and Antifungal Activity of Brochantite Supported on Lignin for the Development of Biobased Plant Protection Products. ACS Sustain. Chem. Eng. 2019, 7, 3213–3221. [Google Scholar] [CrossRef]
- Pasquet, J.; Chevalier, Y.; Pelletier, J.; Couval, E.; Bouvier, D.; Bolzinger, M.A. The Contribution of Zinc Ions to the Antimicrobial Activity of Zinc Oxide. Colloids Surf. A Physicochem. Eng. Asp. 2014, 457, 263–274. [Google Scholar] [CrossRef]
- Xue, J.; Moyer, A.; Peng, B.; Wu, J.; Hannafon, B.N.; Ding, W.-Q. Chloroquine is a Zinc Ionophore. PLoS ONE 2014, 9, e109180. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, Y.; Mustapha, A.; Lin, M. Antifungal Activity of Zinc Oxide Nanoparticles against Botrytis cinerea and Penicillium expansum. Microbiol. Res. 2011, 166, 207–215. [Google Scholar] [CrossRef]
- Lanfranco, L.; Balsamo, R.; Martino, E.; Perotto, S.; Bonfante, P. Zinc Ions Alter Morphology and Chitin Deposition in an Ericoid Fungus. Eur. J. Histochem. 2002, 46, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.; Shah, Z.; Javed, A.; Ali, A.; Hussain, S.B.; Zafar, S.; Ali, H.; Muhammad, S.A. A Fungal Based Synthesis Method for Copper Nanoparticles with the Determination of Anticancer, Antidiabetic and Antibacterial Activities. J. Microbiol. Methods 2020, 174, 105966. [Google Scholar] [CrossRef] [PubMed]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. Copper Resistance in Aspergillus nidulans Relies on the PI-Type ATPase CrpA, Regulated by the Transcription Factor AceA. Front. Microbiol. 2017, 8, 912. [Google Scholar] [CrossRef]
- Li, L.; Ward, D.M. Iron Toxicity in Yeast: Transcriptional Regulation of the Vacuolar Iron Importer Ccc1. Curr. Genet. 2018, 64, 413–416. [Google Scholar] [CrossRef]
- Ward, P.P.; Conneely, O.M. Lactoferrin: Role in Iron Homeostasis and Host Defense against Microbial Infection. Biometals 2004, 17, 203–208. [Google Scholar] [CrossRef]
- Leal, S.M., Jr.; Roy, S.; Vareechon, C.; Carrion, S.D.; Clark, H.; Lopez-Berges, M.S.; diPietro, A.; Schrettl, M.; Beckmann, N.; Redl, B.; et al. Targeting Iron Acquisition Blocks Infection with the Fungal Pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. 2013, 9, e1003436. [Google Scholar] [CrossRef]
- Dollard, C.; Ricupero-Hovasse, S.L.; Natsoulis, G.; Boeke, J.D.; Winston, F. SPT10 and SPT21 are Required for Transcription of Particular Histone Genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994, 14, 5223–5228. [Google Scholar] [CrossRef][Green Version]
- Norris, D.; Osley, M.A. The Two Gene Pairs Encoding H2A and H2B Play Different Roles in the Saccharomyces cerevisiae Life Cycle. Mol. Cell. Biol. 1987, 7, 3473–3481. [Google Scholar] [CrossRef]
- Janusz, G.; Kucharzyk, K.H.; Pawlik, A.; Staszczak, M.; Paszczynski, A.J. Fungal Laccase, Manganese Peroxidase and Lignin Peroxidase: Gene Expression and Regulation. Enzym. Microb. Technol. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Manavalan, T.; Manavalan, A.; Heese, K. Characterization of Lignocellulolytic Enzymes from White-rot Fungi. Curr. Microbiol. 2015, 70, 485–498. [Google Scholar] [CrossRef]
- Xu, H.; Guo, M.-Y.; Gao, Y.-H.; Bai, X.-H.; Zhou, X.-W. Expression and Characteristics of Manganese Peroxidase from Ganoderma lucidum in Pichia pastoris and Its Application in the Degradation of Four Dyes and Phenol. BMC Biotechnol. 2017, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Ahmed, G. Silver Nanoparticles Damage Yeast Cell Wall. J. Biotechnol. 2012, 3, 36–39. [Google Scholar]
- Walker, C.; Ryu, S.; Trinh, C. Exceptional Solvent Tolerance in Yarrowia lipolytica Is Enhanced by Sterols. bioRxiv 2018, 324681. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Rice, L.B. Antifungal Agents: Mode of Action, Mechanisms of Resistance, and Correlation of These Mechanisms with Bacterial Resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef]
- Avery, S.V.; Howlett, N.G.; Radice, S. Copper Toxicity towards Saccharomyces cerevisiae: Dependence on Plasma Membrane Fatty Acid Composition. Appl. Environ. Microbiol. 1996, 62, 3960. [Google Scholar] [CrossRef] [PubMed]
- Babele, P.K.; Singh, A.K.; Srivastava, A. Bio-Inspired Silver Nanoparticles Impose Metabolic and Epigenetic Toxicity to Saccharomyces cerevisiae. Front. Pharmacol. 2019, 10, 1016. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, K.J.; Tobin, J.M.; Avery, S.V. Manganese Toxicity towards Saccharomyces cerevisiae: Dependence on Intracellular and Extracellular Magnesium Concentrations. Appl. Microbiol. Biotechnol. 1998, 49, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in Fungal Virulence. FEMS Microbiol. Rev. 2017, 42. [Google Scholar] [CrossRef] [PubMed]
- Simm, C.; Lahner, B.; Salt, D.; LeFurgey, A.; Ingram, P.; Yandell, B.; Eide, D.J. Saccharomyces cerevisiae Vacuole in Zinc Storage and Intracellular Zinc Distribution. Eukaryot. Cell 2007, 6, 1166. [Google Scholar] [CrossRef]
- Staats, C.; Kmetzsch, L.; Schrank, A.; Vainstein, M. Fungal Zinc Metabolism and Its Connections to Virulence. Front. Cell. Infect. Microbiol. 2013, 3, 65. [Google Scholar] [CrossRef]
- Wilson, D.; Citiulo, F.; Hube, B. Zinc Exploitation by Pathogenic Fungi. PLoS Pathog. 2012, 8, e1003034. [Google Scholar] [CrossRef]
- González Matute, R.; Serra, A.; Figlas, D.; Curvetto, N. Copper and Zinc Bioaccumulation and Bioavailability of Ganoderma lucidum. J. Med. Food 2011, 14, 1273–1279. [Google Scholar] [CrossRef]
- Ruytinx, J.; Nguyen, H.; Van Hees, M.; Op De Beeck, M.; Vangronsveld, J.; Carleer, R.; Colpaert, J.V.; Adriaensen, K. Zinc Export Results in Adaptive Zinc Tolerance in the Ectomycorrhizal Basidiomycete Suillus bovinus. Metallomics 2013, 5, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, T.; Khullar, S.; Agnihotri, R.; Reddy, M.S. Metal Induction of Two Metallothionein Genes in the Ectomycorrhizal Fungus Suillus himalayensis and Their Role in Metal Tolerance. Microbiology 2018, 164, 868–876. [Google Scholar] [CrossRef]
- Tucker, S.L.; Thornton, C.R.; Tasker, K.; Jacob, C.; Giles, G.; Egan, M.; Talbot, N.J. A Fungal Metallothionein is Required for Pathogenicity of Magnaporthe grisea. Plant Cell 2004, 16, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Yasokawa, D.; Murata, S.; Kitagawa, E.; Iwahashi, Y.; Nakagawa, R.; Hashido, T.; Iwahashi, H. Mechanisms of Copper Toxicity in Saccharomyces cerevisiae Determined by Microarray Analysis. Environ. Toxicol. 2008, 23, 599–606. [Google Scholar] [CrossRef]
- Hosiner, D.; Gerber, S.; Lichtenberg-Frate, H.; Glaser, W.; Schüller, C.; Klipp, E. Impact of Acute Metal Stress in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e83330. [Google Scholar] [CrossRef]
- Niazi, J.H.; Sang, B.-I.; Kim, Y.S.; Gu, M.B. Global Gene Response in Saccharomyces cerevisiae Exposed to Silver Nanoparticles. Appl. Biochem. Biotechnol. 2011, 164, 1278–1291. [Google Scholar] [CrossRef]
- Terzioğlu, E.; Alkım, C.; Arslan, M.; Balaban, B.G.; Holyavkin, C.; Kısakesen, H.İ.; Topaloğlu, A.; Yılmaz Şahin, Ü.; Gündüz Işık, S.; Akman, S.; et al. Genomic, Transcriptomic and Physiological Analyses of Silver-resistant Saccharomyces cerevisiae Obtained by Evolutionary Engineering. Yeast 2020, 37, 413–426. [Google Scholar] [CrossRef]
- Akgul, A.; Akgul, A. Mycoremediation of Copper: Exploring the Metal Tolerance of Brown Rot Fungi. Bioresources 2018, 13, 7155–7171. [Google Scholar]
- Ohno, K.M.; Clausen, C.A.; Green, F.; Diehl, S.V. Insights into the Mechanism of Copper-Tolerance in Fibroporia radiculosa: The Biosynthesis of Oxalate. Int. Biodeterior. Biodegrad. 2015, 105, 90–96. [Google Scholar] [CrossRef]
- Tang, J.D.; Parker, L.A.; Perkins, A.D.; Sonstegard, T.S.; Schroeder, S.G.; Nicholas, D.D.; Diehl, S.V. Gene Expression Analysis of Copper Tolerance and Wood Decay in the Brown Rot Fungus Fibroporia radiculosa. Appl. Environ. Microbiol. 2013, 79, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bagley, D.; Ward, D.M.; Kaplan, J. Yap5 Is an Iron-Responsive Transcriptional Activator That Regulates Vacuolar Iron Storage in Yeast. Mol. Cell. Biol. 2008, 28, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Mei, B.; Budde, A.D.; Leong, S.A. sid1, a Gene Initiating Siderophore Biosynthesis in Ustilago maydis: Molecular Characterization, Regulation by Iron, and Role in Phytopathogenicity. Proc. Natl. Acad. Sci. USA 1993, 90, 903–907. [Google Scholar] [CrossRef]
- de Llanos, R.; Martínez-Garay, C.A.; Fita-Torró, J.; Romero, A.M.; Martínez-Pastor, M.T.; Puig, S. Soybean Ferritin Expression in Saccharomyces cerevisiae Modulates Iron Accumulation and Resistance to Elevated Iron Concentrations. Appl. Environ. Microbiol. 2016, 82, 3052–3060. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Kim, H.-M.; Kim, J.-H.; Ryu, K.-S.; Park, S.-M.; Jahng, K.-Y.; Yang, M.-S.; Kim, D.-H. Expression of Heteropolymeric Ferritin Improves Iron Storage in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 1999–2005. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.-M.; Kwon, T.-H.; Kim, K.-S.; Chae, K.-S.; Kim, D.-H.; Kim, J.-H.; Yang, M.-S. Enhanced Iron Uptake of Saccharomyces cerevisiae by Heterologous Expression of a Tadpole Ferritin Gene. Appl. Environ. Microbiol. 2001, 67, 1280–1283. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, L.; Osaba, L.; Corchero, J.; Jimenez, A. A Single Nucleotide Change in the MNR1 (VCX1/HUM1) Gene Determines Resistance to Manganese in Saccharomyces cerevisiae. Yeast 1999, 15, 371–375. [Google Scholar] [CrossRef]
- Luk, E.E.-C.; Culotta, V.C. Manganese Superoxide Dismutase in Saccharomyces cerevisiae Acquires Its Metal Co-factor through a Pathway Involving the Nramp Metal Transporter, Smf2p. J. Biol. Chem. 2001, 276, 47556–47562. [Google Scholar] [CrossRef]
- Reza, M.H.; Shah, H.; Manjrekar, J.; Chattoo, B.B. Magnesium Uptake by CorA Transporters Is Essential for Growth, Development and Infection in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2016, 11, e0159244. [Google Scholar] [CrossRef][Green Version]
- Mendel, R.R. Molybdenum: Biological Activity and Metabolism. Dalton Trans. 2005, 21, 3404–3409. [Google Scholar] [CrossRef]
- Novotny, J.A.; Peterson, C.A. Molybdenum. Adv. Nutr. 2018, 9, 272–273. [Google Scholar] [CrossRef]
- Clarance, P.; Luvankar, B.; Sales, J.; Khusro, A.; Agastian, P.; Tack, J.C.; Al Khulaifi, M.M.; Al-Shwaiman, H.A.; Elgorban, A.M.; Syed, A.; et al. Green Synthesis and Characterization of Gold Nanoparticles Using Endophytic Fungi Fusarium solani and Its in vitro Anticancer and Biomedical Applications. Saudi J. Biol. Sci. 2020, 27, 706–712. [Google Scholar] [CrossRef]
- Sharma, K.; Giri, R.; Sharma, R. Lead, Cadmium and Nickel Removal Efficiency of White-rot Fungus Phlebia brevispora. Lett. Appl. Microbiol. 2020, 71, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.H.; Wuehler, S.E.; Peerson, J.M. The Importance of Zinc in Human Nutrition and Estimation of the Global Prevalence of Zinc Deficiency. Food Nutr. Bull. 2001, 22, 113–125. [Google Scholar] [CrossRef]
- Feldmann, H.; Branduardi, P. Yeast: Molecular and Cell Biology; Wiley-Blackwell: Weinheim, Germany, 2012; p. 464. [Google Scholar]
- Zhang, C.; Huang, H.; Deng, W.; Li, T. Genome-Wide Analysis of the Zn(II)₂Cys₆ Zinc Cluster-Encoding Gene Family in Tolypocladium guangdongense and Its Light-Induced Expression. Genes 2019, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Andrianopoulos, A. Evolution of a Fungal Regulatory Gene Family: The Zn(II)2Cys6 Binuclear Cluster DNA Binding Motif. Fungal Genet. Biol. 1997, 21, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Grotz, N.; Fox, T.; Connolly, E.; Park, W.; Guerinot, M.L.; Eide, D. Identification of a Family of Zinc Transporter Genes from Arabidopsis That Respond to Zinc Deficiency. Proc. Natl. Acad. Sci. USA 1998, 95, 7220–7224. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging Fungal Threats to Animal, Plant and Ecosystem Health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.-N.; Hollier, C. Crop Losses Due to Diseases and Their Implications for Global Food Production Losses and Food Security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide Emergence of Resistance to Antifungal Drugs Challenges Human Health and Food Security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- Fravel, D.; Olivain, C.; Alabouvette, C. Fusarium oxysporum and Its Biocontrol. New Phytol. 2003, 157, 493–502. [Google Scholar] [CrossRef]
- Sadasivam, N.; Sivaraj, R. Biogenic ZnO Nanoparticles Synthesized Using L. Aculeata Leaf Extract and Their Antifungal Activity against Plant Fungal Pathogens. Bull. Mater. Sci. 2016, 39, 1–5. [Google Scholar]
- Sharma, R. Pathogenecity of Aspergillus niger in Plants. Cibtech J. Microbiol. 2012, 1, 47–51. [Google Scholar]
- WIlliamson, B.; Tudzynski, B.; Tudzynski, P.; Van Kan, J.A.L. Botrytis cinerea: The Cause of Grey Mould Disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef]
- Yehia, R.; Ahmed, O. In vitro Study of the Antifungal Efficacy of Zinc Oxide Nanoparticles against Fusarium oxysporum and Penicillium expansum. Afr. J. Microbiol. Res. 2013, 7, 1917–1923. [Google Scholar]
- Bryden, W.L. Mycotoxin Contamination of the Feed Supply Chain: Implications for Animal Productivity and Feed Security. Anim. Feed Sci. Technol. 2012, 173, 134–158. [Google Scholar] [CrossRef]
- Flores-Flores, M.E.; Lizarraga, E.; López de Cerain, A.; González-Peñas, E. Presence of Mycotoxins in Animal Milk: A Review. Food Control 2015, 53, 163–176. [Google Scholar] [CrossRef]
- Appell, M.; Jackson, M.A.; Wang, L.C.; Ho, C.-H.; Mueller, A. Determination of Fusaric Acid in Maize Using Molecularly Imprinted SPE Clean-up. J. Sep. Sci. 2014, 37, 281–286. [Google Scholar] [CrossRef]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine Hydroxylase and Regulation of Dopamine Synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Rahman, M.K.; Rahman, F.; Rahman, T.; Kato, T. Dopamine-β-Hydroxylase (DBH), Its Cofactors and Other Biochemical Parameters in the Serum of Neurological Patients in Bangladesh. Int. J. Biomed. Sci. IJBS 2009, 5, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Cuero, R.; Ouellet, T. Metal Ions Modulate Gene Expression and Accumulation of the Mycotoxins Aflatoxin and Zearalenone. J. Appl. Microbiol. 2005, 98, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Savi, G.D.; Bortoluzzi, A.J.; Scussel, V.M. Antifungal Properties of Zinc-compounds against Toxigenic Fungi and Mycotoxin. Int. J. Food Sci. Technol. 2013, 48, 1834–1840. [Google Scholar] [CrossRef]
- Eide, D.J. Zinc Transporters and the Cellular Trafficking of Zinc. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 711–722. [Google Scholar] [CrossRef]
- Gaither, L.A.; Eide, D.J. Eukaryotic Zinc Transporters and Their Regulation. Biometals 2001, 14, 251–270. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Eide, D.J. Combinatorial Control of Yeast FET4 Gene Expression by Iron, Zinc, and Oxygen. J. Biol. Chem. 2002, 277, 33749–33757. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.J. Multiple Regulatory Mechanisms Maintain Zinc Homeostasis in Saccharomyces cerevisiae. J. Nutr. 2003, 133, 1532S–1535S. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Butler, E.; Rodgers, J.; Spizzo, T.; Duesterhoeft, S.; Eide, D. Regulation of Zinc Homeostasis in Yeast by Binding of the ZAP1 Transcriptional Activator to Zinc-responsive Promoter Elements. J. Biol. Chem. 1998, 273, 28713–28720. [Google Scholar] [CrossRef]
- Wang, Y.; Weisenhorn, E.; MacDiarmid, C.W.; Andreini, C.; Bucci, M.; Taggart, J.; Banci, L.; Russell, J.; Coon, J.J.; Eide, D.J. The Cellular Economy of the Saccharomyces cerevisiae Zinc Proteome. Met. Integr. Biometal Sci. 2018, 10, 1755–1776. [Google Scholar] [CrossRef]
- Frey, A.G.; Eide, D.J. Zinc-responsive Coactivator Recruitment by the Yeast Zap1 Transcription Factor. Microbiologyopen 2012, 1, 105–114. [Google Scholar] [CrossRef]
- Frey, A.G.; Bird, A.J.; Evans-Galea, M.V.; Blankman, E.; Winge, D.R.; Eide, D.J. Zinc-Regulated DNA Binding of the Yeast Zap1 Zinc-Responsive Activator. PLoS ONE 2011, 6, e22535. [Google Scholar] [CrossRef]
- Guerinot, M.L. The ZIP Family of Metal Transporters. Biochim. Biophys. Acta Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef]
- Vicentefranqueira, R.; Moreno, M.Á.; Leal, F.; Calera, J.A. The zrfA and zrfB Genes of Aspergillus fumigatus Encode the Zinc Transporter Proteins of a Zinc Uptake System Induced in an Acid, Zinc-Depleted Environment. Eukaryot. Cell 2005, 4, 837–848. [Google Scholar] [CrossRef]
- Moreno, M.Á.; Ibrahim-Granet, O.; Vicentefranqueira, R.; Amich, J.; Ave, P.; Leal, F.; Latgé, J.-P.; Calera, J.A. The Regulation of Zinc Homeostasis by the ZafA Transcriptional Activator is Essential for Aspergillus fumigatus Virulence. Mol. Microbiol. 2007, 64, 1182–1197. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S. ZafA-Mediated Regulation of Zinc Homeostasis is Required for Virulence in the Plant Pathogen Fusarium oxysporum. Mol. Plant Pathol. 2020, 21, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Reeder, N.L.; Xu, J.; Youngquist, R.S.; Schwartz, J.R.; Rust, R.C.; Saunders, C.W. The Antifungal Mechanism of Action of Zinc Pyrithione. Br. J. Dermatol. 2011, 165, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Cho, Y.-J.; Lee, Y.W.; Jung, W.H. Understanding the Mechanism of Action of the Anti-Dandruff Agent Zinc Pyrithione against Malassezia restricta. Sci. Rep. 2018, 8, 12086. [Google Scholar] [CrossRef]
- Stehling, O.; Lill, R. The Role of Mitochondria in Cellular Iron-sulfur Protein Biogenesis: Mechanisms, Connected Processes, and Diseases. Cold Spring Harbor Perspect. Biol. 2013, 5, a011312. [Google Scholar] [CrossRef]
- Buchman, C.; Skroch, P.; Welch, J.; Fogel, S.; Karin, M. The CUP2 Gene Product, Regulator of Yeast Metallothionein Expression, is a Copper-activated DNA-binding Protein. Mol. Cell. Biol. 1989, 9, 4091–4095. [Google Scholar] [CrossRef]
- Dancis, A.; Haile, D.; Yuan, D.S.; Klausner, R.D. The Saccharomyces cerevisiae Copper Transport Protein (Ctr1p). Biochemical Characterization, Regulation by Copper, and Physiologic Role in Copper Uptake. J. Biol. Chem. 1994, 269, 25660–25667. [Google Scholar] [CrossRef]
- Crawford, A.C.; Lehtovirta-Morley, L.E.; Alamir, O.; Niemiec, M.J.; Alawfi, B.; Alsarraf, M.; Skrahina, V.; Costa, A.C.B.P.; Anderson, A.; Yellagunda, S.; et al. Biphasic Zinc Compartmentalisation in a Human Fungal Pathogen. PLoS Pathog. 2018, 14, e1007013. [Google Scholar] [CrossRef]
- Khouja, H.R.; Abbà, S.; Lacercat-Didier, L.; Daghino, S.; Doillon, D.; Richaud, P.; Martino, E.; Vallino, M.; Perotto, S.; Chalot, M.; et al. OmZnT1 and OmFET, Two Metal Transporters from the Metal-tolerant Strain Zn of the Ericoid Mycorrhizal Fungus Oidiodendron maius, Confer Zinc Tolerance in Yeast. Fungal Genet. Biol. 2013, 52, 53–64. [Google Scholar] [CrossRef] [PubMed]
- González-Guerrero, M.; Melville, L.H.; Ferrol, N.; Lott, J.N.A.; Azcón-Aguilar, C.; Peterson, R.L. Ultrastructural Localization of Heavy Metals in the Extraradical Mycelium and Spores of the Arbuscular Mycorrhizal Fungus Glomus intraradices. Can. J. Microbiol. 2008, 54, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, T.; Sácký, J.; Šimek, P.; Šantrůček, J.; Kotrba, P. Metallothionein-like Peptides Involved in Sequestration of Zn in the Zn-accumulating Ectomycorrhizal Fungus Russula atropurpurea. Metallomics 2014, 6, 1693–1701. [Google Scholar] [CrossRef]
- Sácký, J.; Leonhardt, T.; Borovička, J.; Gryndler, M.; Briksí, A.; Kotrba, P. Intracellular Sequestration of Zinc, Cadmium and Silver in Hebeloma mesophaeum and Characterization of Its Metallothionein Genes. Fungal Genet. Biol. 2014, 67, 3–14. [Google Scholar] [CrossRef]
- Ouda, S. Antifungal Activity of Silver and Copper Nanoparticles on Two Plant Pathogens, Alternaria alternata and Botrytis cinerea. Res. J. Microbiol. 2014, 9, 34–42. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, D.; Zhao, X.; Wei, D.; Wang, Y.; Zhu, X. Effects of CTR4 Deletion on Virulence and Stress Response in Cryptococcus neoformans. Antonie van Leeuwenhoek 2016, 109, 1081–1090. [Google Scholar] [CrossRef]
- Hassett, R.; Kosman, D.J. Evidence for Cu(II) Reduction as a Component of Copper Uptake by Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 128–134. [Google Scholar] [CrossRef]
- Zhu, Z.; Labbé, S.; Peña, M.M.O.; Thiele, D.J. Copper Differentially Regulates the Activity and Degradation of Yeast Mac1 Transcription Factor. J. Biol. Chem. 1998, 273, 1277–1280. [Google Scholar] [CrossRef]
- de Silva, D.; Davis-Kaplan, S.; Fergestad, J.; Kaplan, J. Purification and Characterization of Fet3 Protein, a Yeast Homologue of Ceruloplasmin. J. Biol. Chem. 1997, 272, 14208–14213. [Google Scholar] [CrossRef]
- Askwith, C.; Eide, D.; Van Ho, A.; Bernard, P.S.; Li, L.; Davis-Kaplan, S.; Sipe, D.M.; Kaplan, J. The FET3 Gene of S. Cerevisiae Encodes a Multicopper Oxidase Required for Ferrous Iron Uptake. Cell 1994, 76, 403–410. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic Response to Iron Deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Iwai, Y.; Dancis, A.; Klausner, R.D. AFT1: A Mediator of Iron Regulated Transcriptional Control in Saccharomyces cerevisiae. EMBO J. 1995, 14, 1231–1239. [Google Scholar] [CrossRef]
- Horng, Y.-C.; Cobine, P.A.; Maxfield, A.B.; Carr, H.S.; Winge, D.R. Specific Copper Transfer from the Cox17 Metallochaperone to Both Sco1 and Cox11 in the Assembly of Yeast Cytochrome c Oxidase. J. Biol. Chem. 2004, 279, 35334–35340. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Lledías, F.; Hansberg, W.; Covarrubias, A.A. Cu,Zn-superoxide Dismutase of Saccharomyces cerevisiae is Required for Resistance to Hyperosmosis. FEBS Lett. 2003, 539, 68–72. [Google Scholar] [CrossRef]
- Zyrina, A.N.; Smirnova, E.A.; Markova, O.V.; Severin, F.F.; Knorre, D.A. Mitochondrial Superoxide Dismutase and Yap1p Act as a Signaling Module Contributing to Ethanol Tolerance of the Yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83, e02759–e16. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide Anion Radical (O·2), Superoxide Dismutases, and Related Matters. J. Biol. Chem. 1997, 272, 18515–18517. [Google Scholar] [CrossRef]
- Imlay, J.A.; Fridovich, I. Suppression of Oxidative Envelope Damage by Pseudoreversion of a Superoxide Dismutase-deficient Mutant of Escherichia coli. J. Bacteriol. 1992, 174, 953. [Google Scholar] [CrossRef][Green Version]
- Lushchak, V.; Semchyshyn, H.; Mandryk, S.; Lushchak, O. Possible Role of Superoxide Dismutases in the Yeast Saccharomyces cerevisiae under Respiratory Conditions. Arch. Biochem. Biophys. 2005, 441, 35–40. [Google Scholar] [CrossRef]
- Seah, T.C.M.; Kaplan, J.G. Purification and Properties of the Catalase of Bakers’ Yeast. J. Biol. Chem. 1973, 248, 2889–2893. [Google Scholar] [CrossRef]
- Culotta, V.C.; Howard, W.R.; Liu, X.F. CRS5 Encodes a Metallothionein-like Protein in Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 25295–25302. [Google Scholar] [CrossRef]
- George, G.N.; Byrd, J.; Winge, D.R. X-ray Absorption Studies of Yeast Copper Metallothionein. J. Biol. Chem. 1988, 263, 8199–8203. [Google Scholar] [CrossRef]
- Jensen, L.T.; Howard, W.R.; Strain, J.J.; Winge, D.R.; Culotta, V.C. Enhanced Effectiveness of Copper Ion Buffering by CUP1 Metallothionein Compared with CRS5 Metallothionein in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 18514–18519. [Google Scholar] [CrossRef]
- Nagakubo, T.; Kumano, T.; Ohta, T.; Hashimoto, Y.; Kobayashi, M. Copper Amine Oxidases Catalyze the Oxidative Deamination and Hydrolysis of Cyclic Imines. Nat. Commun. 2019, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. New Insights into Copper Homeostasis in Filamentous Fungi. Int. Microbiol. 2020, 23, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kusuya, Y.; Hagiwara, D.; Sakai, K.; Yaguchi, T.; Gonoi, T.; Takahashi, H. Transcription Factor Afmac1 Controls Copper Import Machinery in Aspergillus fumigatus. Curr. Genet. 2017, 63, 777–789. [Google Scholar] [CrossRef]
- Park, Y.-S.; Kang, S.; Seo, H.; Yun, C.-W. A copper Transcription Factor, AfMac1, Regulates Both Iron and Copper Homeostasis in the Opportunistic Fungal Pathogen Aspergillus fumigatus. Biochem. J. 2018, 475, 2831–2845. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-S.; Kim, T.-H.; Yun, C.-W. Functional Characterization of the Copper Transcription Factor AfMac1 from Aspergillus fumigatus. Biochem. J. 2017, 474, 2365–2378. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.D.; Perkins, A.D.; Sonstegard, T.S.; Schroeder, S.G.; Burgess, S.C.; Diehl, S.V. Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa. Appl. Environ. Microbiol. 2012, 78, 2272. [Google Scholar] [CrossRef]
- Bayat, N.; Rajapakse, K.; Marinsek-Logar, R.; Drobne, D.; Cristobal, S. The Effects of Engineered Nanoparticles on the Cellular Structure and Growth of Saccharomyces cerevisiae. Nanotoxicology 2014, 8, 363–373. [Google Scholar] [CrossRef]
- Kasemets, K.; Käosaar, S.; Vija, H.; Fascio, U.; Mantecca, P. Toxicity of Differently Sized and Charged Silver Nanoparticles to Yeast Saccharomyces cerevisiae BY4741: A Nano-biointeraction Perspective. Nanotoxicology 2019, 13, 1041–1059. [Google Scholar] [CrossRef]
- Jo, W.J.; Loguinov, A.; Chang, M.; Wintz, H.; Nislow, C.; Arkin, A.P.; Giaever, G.; Vulpe, C.D. Identification of Genes Involved in the Toxic Response of Saccharomyces cerevisiae against Iron and Copper Overload by Parallel Analysis of Deletion Mutants. Toxicol. Sci. 2007, 101, 140–151. [Google Scholar] [CrossRef]
- Kasemets, K.; Ivask, A.; Dubourguier, H.-C.; Kahru, A. Toxicity of Nanoparticles of ZnO, CuO and TiO2 to Yeast Saccharomyces cerevisiae. Toxicol. In Vitro 2009, 23, 1116–1122. [Google Scholar] [CrossRef]
- Giannousi, K.; Sarafidis, G.; Mourdikoudis, S.; Pantazaki, A.; Dendrinou-Samara, C. Selective Synthesis of Cu2O and Cu/Cu2O NPs: Antifungal Activity to Yeast Saccharomyces cerevisiae and DNA Interaction. Inorg. Chem. 2014, 53, 9657–9666. [Google Scholar] [CrossRef]
- Hitchcock, C.A.; Pye, G.W.; Troke, P.F.; Johnson, E.M.; Warnock, D.W. Fluconazole Resistance in Candida glabrata. Antimicrob. Agents Chemother. 1993, 37, 1962–1965. [Google Scholar] [CrossRef]
- Orozco, A.S.; Higginbotham, L.M.; Hitchcock, C.A.; Parkinson, T.; Falconer, D.; Ibrahim, A.S.; Ghannoum, M.A.; Filler, S.G. Mechanism of Fluconazole Resistance in Candida krusei. Antimicrob. Agents Chemother. 1998, 42, 2645–2649. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Nagy, E.; Dobiasova, S.; Rinaldi, M.; Barton, R.; Veselov, A. Global Antifungal Surveillance, G. Candida krusei, a Multidrug-resistant Opportunistic Fungal Pathogen: Geographic and Temporal Trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J. Clin. Microbiol. 2008, 46, 515–521. [Google Scholar] [CrossRef]
- Gomes da Silva Dantas, F.; Araújo de Almeida-Apolonio, A.; Pires de Araújo, R.; Regiane Vizolli Favarin, L.; Fukuda de Castilho, P.; de Oliveira Galvão, F.; Inez Estivalet Svidzinski, T.; Antônio Casagrande, G.; Mari Pires de Oliveira, K. A Promising Copper(II) Complex as Antifungal and Antibiofilm Drug against Yeast Infection. Molecules 2018, 23, 1856. [Google Scholar] [CrossRef]
- Coyle, B.; Kavanagh, K.; McCann, M.; Devereux, M.; Geraghty, M. Mode of Anti-fungal Activity of 1,10-phenanthroline and Its Cu(II), Mn(II) and Ag(I) Complexes. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2003, 16, 321–329. [Google Scholar] [CrossRef]
- Borgatta, J.; Ma, C.; Hudson-Smith, N.; Elmer, W.; Plaza Pérez, C.D.; De La Torre-Roche, R.; Zuverza-Mena, N.; Haynes, C.L.; White, J.C.; Hamers, R.J. Copper Based Nanomaterials Suppress Root Fungal Disease in Watermelon (Citrullus lanatus): Role of Particle Morphology, Composition and Dissolution Behavior. ACS Sustain. Chem. Eng. 2018, 6, 14847–14856. [Google Scholar] [CrossRef]
- Cai, Z.; Du, W.; Zhang, Z.; Guan, L.; Zeng, Q.; Chai, Y.; Dai, C.; Lu, L. The Aspergillus fumigatus Transcription Factor AceA is Involved not only in Cu but also in Zn Detoxification through Regulating Transporters CrpA and ZrcA. Cell. Microbiol. 2018, 20, e12864. [Google Scholar] [CrossRef]
- Ragasa, L.R.P.; Joson, S.E.A.; Bagay, W.L.R.; Perez, T.R.; Velarde, M.C. Transcriptome Analysis Reveals Involvement of Oxidative Stress Response in a Copper-tolerant Fusarium oxysporum Strain. Fungal Biol. 2021. [Google Scholar] [CrossRef]
- Bolm, C. A New Iron Age. Nat. Chem. 2009, 1, 420. [Google Scholar] [CrossRef]
- Weber, K.A.; Achenbach, L.A.; Coates, J.D. Microorganisms Pumping Iron: Anaerobic Microbial Iron Oxidation and Reduction. Nat. Rev. Microbiol. 2006, 4, 752–764. [Google Scholar] [CrossRef]
- Hissen, A.H.T.; Wan, A.N.C.; Warwas, M.L.; Pinto, L.J.; Moore, M.M. The Aspergillus fumigatus Siderophore Biosynthetic Gene sidA, Encoding l-Ornithine N5-Oxygenase, Is Required for Virulence. Infect. Immun. 2005, 73, 5493–5503. [Google Scholar] [CrossRef] [PubMed]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore Biosynthesis but Not Reductive Iron Assimilation Is Essential for Aspergillus fumigatus Virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S.; Capilla, J.; Turrà, D.; Schafferer, L.; Matthijs, S.; Jöchl, C.; Cornelis, P.; Guarro, J.; Haas, H.; Di Pietro, A. HapX-mediated Iron Homeostasis is Essential for Rhizosphere Competence and Virulence of the Soilborne Pathogen Fusarium oxysporum. Plant Cell 2012, 24, 3805–3822. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, M.; Ling, N.; Shen, Q.; Guo, S. Effects of Iron and Boron Combinations on the Suppression of Fusarium Wilt in Banana. Sci. Rep. 2016, 6, 38944. [Google Scholar] [CrossRef]
- Hashem, A.R. Influence of Iron on the Growth of the Tomato Wilt Pathogen, Fusarium oxysporum, Isolated in Saudi Arabia. J. Plant Dis. Prot. 1995, 102, 326–330. [Google Scholar]
- Philpott, C.C. Iron Uptake in Fungi: A System for Every Source. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 636–645. [Google Scholar] [CrossRef]
- Yun, C.-W.; Ferea, T.; Rashford, J.; Ardon, O.; Brown, P.O.; Botstein, D.; Kaplan, J.; Philpott, C.C. Desferrioxamine-mediated Iron Uptake in Saccharomyces cerevisiae: Evidence for Two Pathways of Iron Uptake. J. Biol. Chem. 2000, 275, 10709–10715. [Google Scholar] [CrossRef]
- Yun, C.-W.; Tiedeman, J.S.; Moore, R.E.; Philpott, C.C. Siderophore-Iron Uptake in Saccharomyces cerevisiae: Identification of Ferrichrome and Fusarinine Transporters J. Biol. Chem. 2000, 275, 16354–16359. [Google Scholar] [CrossRef]
- Georgatsou, E.; Alexandraki, D. Two distinctly Regulated Genes are Required for Ferric Reduction, the First Step of Iron Uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994, 14, 3065–3073. [Google Scholar] [CrossRef][Green Version]
- Martínez-Pastor, M.T.; Perea-García, A.; Puig, S. Mechanisms of Iron Sensing and Regulation in the Yeast Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 75. [Google Scholar] [CrossRef]
- Jensen, L.T.; Culotta, V.C. Regulation of Saccharomyces cerevisiae FET4 by Oxygen and Iron. J. Mol. Biol. 2002, 318, 251–260. [Google Scholar] [CrossRef]
- Lesuisse, E.; Raguzzi, F.; Crichton, R. Iron Uptake by the Yeast Saccharomyces cerevisiae: Involvement of a Reduction Step. J. Gen. Microbiol. 1987, 133, 3229–3236. [Google Scholar] [CrossRef]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The Yeast Fre1p/Fre2p Cupric Reductases Facilitate Copper Uptake and Are Regulated by the Copper-modulated Mac1p Activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef]
- Caetano, S.M.; Menezes, R.; Amaral, C.; Rodrigues-Pousada, C.; Pimentel, C. Repression of the Low Affinity Iron Transporter Gene FET4: A Novel Mechanism against Cadmium Toxicity Orchestrated by Yap1 via Rox1. J. Biol. Chem. 2015, 290, 18584–18595. [Google Scholar] [CrossRef]
- Liu, X.F.; Supek, F.; Nelson, N.; Culotta, V.C. Negative Control of Heavy Metal Uptake by the Saccharomyces cerevisiae BSD2 Gene. J. Biol. Chem. 1997, 272, 11763–11769. [Google Scholar] [CrossRef]
- Kim, Y.; Lampert, S.M.; Philpott, C.C. A Receptor Domain Controls the Intracellular Sorting of the Ferrichrome Transporter, ARN1. EMBO J. 2005, 24, 952–962. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, Y.; Yun, C.-W.; Philpott, C.C. Ferrichrome Induces Endosome to Plasma Membrane Cycling of the Ferrichrome Transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 2002, 21, 3632–3642. [Google Scholar] [CrossRef]
- Heymann, P.; Ernst, J.F.; Winkelmann, G. Identification of a Fungal Triacetylfusarinine C Siderophore Transport Gene (TAF1) in Saccharomyces cerevisiae as a Member of the Major Facilitator Superfamily. Biometals 1999, 12, 301–306. [Google Scholar] [CrossRef]
- Lesuisse, E.; Blaiseau, P.-L.; Dancis, A.; Camadro, J.-M. Siderophore Uptake and Use by the Yeast Saccharomyces cerevisiae. Microbiology 2001, 147, 289–298. [Google Scholar] [CrossRef]
- Froissard, M.; Belgareh-Touzé, N.; Dias, M.; Buisson, N.; Camadro, J.M.; Haguenauer-Tsapis, R.; Lesuisse, E. Trafficking of Siderophore Transporters in Saccharomyces cerevisiae and Intracellular Fate of Ferrioxamine B Conjugates. Traffic 2007, 8, 1601–1616. [Google Scholar] [CrossRef]
- Kosman, D.J. Molecular Mechanisms of Iron Uptake in Fungi. Mol. Microbiol. 2003, 47, 1185–1197. [Google Scholar] [CrossRef]
- Kohlhaw, G.B. Leucine Biosynthesis in Fungi: Entering Metabolism through the Back Door. Microbiol. Mol. Biol. Rev. 2003, 67, 1–15. [Google Scholar] [CrossRef]
- Pokharel, S.; Campbell, J.L. Cross Talk between the Nuclease and Helicase Activities of Dna2: Role of an Essential Iron-sulfur Cluster Domain. Nucleic Acids Res. 2012, 40, 7821–7830. [Google Scholar] [CrossRef]
- Labbé, S.; Pelletier, B.; Mercier, A. Iron Homeostasis in the Fission Yeast Schizosaccharomyces pombe. Biometals 2007, 20, 523–537. [Google Scholar] [CrossRef]
- Schrettl, M.; Winkelmann, G.; Haas, H. Ferrichrome in Schizosaccharomyces pombe–an Iron Transport and Iron Storage Compound. Biometals 2004, 17, 647–654. [Google Scholar] [CrossRef]
- Roman, D.G.; Dancis, A.; Anderson, G.J.; Klausner, R.D. The Fission Yeast Ferric Reductase Gene frp1+ is Required for Ferric Iron Uptake and Encodes a Protein That is Homologous to the gp91-phox Subunit of the Human NADPH Phagocyte Oxidoreductase. Mol. Cell. Biol. 1993, 13, 4342–4350. [Google Scholar] [CrossRef]
- Pouliot, B.; Jbel, M.; Mercier, A.; Labbé, S. abc3 + Encodes an Iron-Regulated Vacuolar ABC-Type Transporter in Schizosaccharomyces pombe. Eukaryot. Cell 2010, 9, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Lowry, C.V.; Zitomer, R.S. ROX1 Encodes a Heme-induced Repression Factor Regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 1988, 8, 4651–4658. [Google Scholar] [CrossRef]
- Hickman, M.J.; Winston, F. Heme Levels Switch the Function of Hap1 of Saccharomyces cerevisiae between Transcriptional Activator and Transcriptional Repressor. Mol. Cell. Biol. 2007, 27, 7414–7424. [Google Scholar] [CrossRef]
- Santos, R.; Buisson, N.; Knight, S.; Dancis, A.; Camadro, J.-M.; Lesuisse, E. Haemin Uptake and Use as an Iron Source by Candida albicans: Role of CaHMX1-encoded Haem Oxygenase. Microbiology 2003, 149, 579–588. [Google Scholar] [CrossRef]
- Tsiftsoglou, A.S.; Tsamadou, A.I.; Papadopoulou, L.C. Heme as Key Regulator of Major Mammalian Cellular Functions: Molecular, Cellular, and Pharmacological Aspects. Pharmacol. Ther. 2006, 111, 327–345. [Google Scholar] [CrossRef]
- Mercier, A.; Pelletier, B.; Labbé, S. A Transcription Factor Cascade Involving Fep1 and the CCAAT-Binding Factor Php4 Regulates Gene Expression in Response to Iron Deficiency in the Fission Yeast Schizosaccharomyces pombe. Eukaryot. Cell 2006, 5, 1866–1881. [Google Scholar] [CrossRef]
- Weissman, Z.; Shemer, R.; Conibear, E.; Kornitzer, D. An Endocytic Mechanism for Haemoglobin-iron Acquisition in Candida albicans. Mol. Microbiol. 2008, 69, 201–217. [Google Scholar] [CrossRef]
- Ardon, O.; Nudelman, R.; Caris, C.; Libman, J.; Shanzer, A.; Chen, Y.; Hadar, Y. Iron Uptake in Ustilago maydis: Tracking the Iron Path. J. Bacteriol. 1998, 180, 2021–2026. [Google Scholar] [CrossRef]
- Wang, J.; Budde, A.D.; Leong, S.A. Analysis of Ferrichrome Biosynthesis in the Phytopathogenic Fungus Ustilago maydis: Cloning of an Ornithine-N5-oxygenase Gene. J. Bacteriol. 1989, 171, 2811–2818. [Google Scholar] [CrossRef]
- Lin, H.; Li, L.; Jia, X.; Ward, D.M.; Kaplan, J. Genetic and Biochemical Analysis of High Iron Toxicity in Yeast: Iron Toxicity is Due to the Accumulation of Cytosolic Iron and Occurs under Both Aerobic and Anaerobic Conditions. J. Biol. Chem. 2011, 286, 3851–3862. [Google Scholar] [CrossRef]
- Berthelet, S.; Usher, J.; Shulist, K.; Hamza, A.; Maltez, N.; Johnston, A.; Fong, Y.; Harris, L.J.; Baetz, K. Functional Genomics Analysis of the Saccharomyces cerevisiae Iron Responsive Transcription Factor Aft1 Reveals Iron-independent Functions. Genetics 2010, 185, 1111–1128. [Google Scholar] [CrossRef]
- van Bakel, H.; Strengman, E.; Wijmenga, C.; Holstege, F.C.P. Gene Expression Profiling and Phenotype Analyses of S. Cerevisiae in Response to Changing Copper Reveals Six Genes with New Roles in Copper and Iron Metabolism. Physiol. Genom. 2005, 22, 356–367. [Google Scholar] [CrossRef]
- Adamczyk, Z.; Oćwieja, M.; Mrowiec, H.; Walas, S.; Lupa, D. Oxidative Dissolution of Silver Nanoparticles: A New Theoretical Approach. J. Colloid Interface Sci. 2016, 469, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Elsharkawy, M.M.; Shimizu, M.; Hyakumachi, M. Control of Root Rot and Wilt Diseases of Roselle under Field Conditions. Mycobiology 2014, 42, 376–384. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Adhilakshmi, M.; Karthikeyan, M.; Devadason, A. Effect of Combination of Bio-agents and Mineral Nutrients for the Management of Alfalfa Wilt Pathogen Fusarium oxysporum f. sp. medicaginis. Arch. Phytopathol. Plant Prot. 2008, 47, 514–525. [Google Scholar] [CrossRef]
- Clark, H.L.; Jhingran, A.; Sun, Y.; Vareechon, C.; de Jesus Carrion, S.; Skaar, E.P.; Chazin, W.J.; Calera, J.A.; Hohl, T.M.; Pearlman, E. Zinc and Manganese Chelation by Neutrophil S100A8/A9 (Calprotectin) Limits Extracellular Aspergillus fumigatus Hyphal Growth and Corneal Infection. J. Immunol. 2016, 196, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Isikhuemhen, O.S.; Mikiashvilli, N.A. Lignocellulolytic Enzyme Activity, Substrate Utilization, and Mushroom Yield by Pleurotus ostreatus Cultivated on Substrate Containing Anaerobic Digester Solids. J. Ind. Microbiol. Biotechnol. 2009, 36, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Isikhuemhen, O.S.; Nerud, F. Preliminary Studies on the Ligninolytic Enzymes Produced by the Tropical Fungus Pleurotus tuber-regium (Fr.) Sing. Antonie van Leeuwenhoek 1999, 75, 257–260. [Google Scholar] [CrossRef]
- Portnoy, M.E.; Liu, X.F.; Culotta, V.C. Saccharomyces cerevisiae Expresses Three Functionally Distinct Homologues of the Nramp Family of Metal Transporters. Mol. Cell. Biol. 2000, 20, 7893–7902. [Google Scholar] [CrossRef]
- Luk, E.; Yang, M.; Jensen, L.T.; Bourbonnais, Y.; Culotta, V.C. Manganese Activation of Superoxide Dismutase 2 in the Mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 22715–22720. [Google Scholar] [CrossRef]
- Van Loon, A.; Pesold-Hurt, B.; Schatz, G. A Yeast Mutant Lacking Mitochondrial Manganese-superoxide Dismutase is Hypersensitive to Oxygen. Proc. Natl. Acad. Sci. USA 1986, 83, 3820–3824. [Google Scholar] [CrossRef] [PubMed]
- Reddi, A.R.; Jensen, L.T.; Naranuntarat, A.; Rosenfeld, L.; Leung, E.; Shah, R.; Culotta, V.C. The Overlapping Roles of Manganese and Cu/Zn SOD in Oxidative Stress Protection. Free Radic. Biol. Med. 2009, 46, 154–162. [Google Scholar] [CrossRef]
- Liu, X.F.; Culotta, V.C. Post-translation Control of Nramp Metal Transport in Yeast: Role of Metal Ions and the BSD2 Gene. J. Biol. Chem. 1999, 274, 4863–4868. [Google Scholar] [CrossRef]
- Stimpson, H.E.; Lewis, M.J.; Pelham, H.R. Transferrin Receptor-like Proteins Control the Degradation of a Yeast Metal Transporter. EMBO J. 2006, 25, 662–672. [Google Scholar] [CrossRef]
- Jensen, L.T.; Carroll, M.C.; Hall, M.D.; Harvey, C.J.; Beese, S.E.; Culotta, V.C. Down-regulation of a Manganese Transporter in the Face of Metal Toxicity. Mol. Biol. Cell 2009, 20, 2810–2819. [Google Scholar] [CrossRef]
- Yang, B.; Kumar, S. Nedd4 and Nedd4-2: Closely Related Ubiquitin-protein Ligases with Distinct Physiological Functions. Cell Death Differ. 2010, 17, 68–77. [Google Scholar] [CrossRef]
- Bun-Ya, M.; Nishimura, M.; Harashima, S.; Oshima, Y. The PHO84 Gene of Saccharomyces cerevisiae Encodes an Inorganic Phosphate Transporter. Mol. Cell. Biol. 1991, 11, 3229–3238. [Google Scholar] [CrossRef]
- Wykoff, D.D.; Rizvi, A.H.; Raser, J.M.; Margolin, B.; O’Shea, E.K. Positive Feedback Regulates Switching of Phosphate Transporters in S. cerevisiae. Mol. Cell 2007, 27, 1005–1013. [Google Scholar] [CrossRef]
- Reddi, A.R.; Jensen, L.T.; Culotta, V.C. Manganese Homeostasis in Saccharomyces cerevisiae. Chem. Rev. 2009, 109, 4722–4732. [Google Scholar] [CrossRef]
- Rayner, J.C.; Munro, S. Identification of the MNN2 and MNN5Mannosyltransferases Required for Forming and Extending the Mannose Branches of the Outer Chain Mannans of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26836–26843. [Google Scholar] [CrossRef]
- Romero, P.A.; Herscovics, A. Glycoprotein Biosynthesis in Saccharomyces cerevisiae. Characterization of Alpha-1,6-mannosyltransferase Which Initiates outer Chain Formation. J. Biol. Chem. 1989, 264, 1946–1950. [Google Scholar] [CrossRef]
- Wiggins, C.A.; Munro, S. Activity of the Yeast MNN1 Alpha-1,3-mannosyltransferase Requires a Motif Conserved in Many Other Families of Glycosyltransferases. Proc. Natl. Acad. Sci. USA 1998, 95, 7945–7950. [Google Scholar] [CrossRef]
- Striebeck, A.; Robinson, D.A.; Schüttelkopf, A.W.; van Aalten, D.M.F. Yeast Mnn9 is Both a Priming Glycosyltransferase and an Allosteric Activator of Mannan Biosynthesis. Open Biol. 2013, 3, 130022. [Google Scholar] [CrossRef]
- Solá, R.J.; Griebenow, K. Effects of Glycosylation on the Stability of Protein Pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef]
- Xiang, M.; Mohamalawari, D.; Rao, R. A Novel Isoform of the Secretory Pathway Ca2+,Mn(2+)-ATPase, hSPCA2, Has Unusual Properties and is Expressed in the Brain. J. Biol. Chem. 2005, 280, 11608–11614. [Google Scholar] [CrossRef]
- Tanaka, J.; Fink, G.R. The Histidine Permease Gene (HIP1) of Saccharomyces cerevisiae. Gene 1985, 38, 205–214. [Google Scholar] [CrossRef]
- Bakshi, D.K.; Saha, S.; Sindhu, I.; Sharma, P. Use of Phanerochaete Chrysosporium Biomass for the Removal of Textile Dyes from a Synthetic Effluent. World J. Microbiol. Biotechnol. 2006, 22, 835–839. [Google Scholar] [CrossRef]
- Bending, G.D.; Friloux, M.; Walker, A. Degradation of Contrasting Pesticides by White Rot Fungi and Its Relationship with Ligninolytic Potential. FEMS Microbiol. Lett. 2002, 212, 59–63. [Google Scholar] [CrossRef]
- Kirk, T.K.; Connors, W.; Zeikus, J.G. Requirement for a Growth Substrate during Lignin Decomposition by Two Wood-rotting Fungi. Appl. Environ. Microbiol. 1976, 32, 192–194. [Google Scholar] [CrossRef]
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and Characterization of Two Extracelluar H2O2-dependent Oxidases from Ligninolytic Cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250. [Google Scholar] [CrossRef]
- Bonnarme, P.; Jeffries, T.W. Mn(II) Regulation of Lignin Peroxidases and Manganese-dependent Peroxidases from Lignin-degrading White Rot Fungi. Appl. Environ. Microbiol. 1990, 56, 210–217. [Google Scholar] [CrossRef]
- Isikhuemhen, O.S.; Mikiashvili, N.A.; Kelkar, V. Application of Solid Waste from Anaerobic Digestion of Poultry Litter in Agrocybe aegerita Cultivation: Mushroom Production, Lignocellulolytic Enzymes Activity and Substrate Utilization. Biodegradation 2009, 20, 351–361. [Google Scholar] [CrossRef]
- Isikhuemhen, O.S.; Mikiashvili, N.A.; Adenipekun, C.O.; Ohimain, E.I.; Shahbazi, G. The Tropical White Rot Fungus, Lentinus squarrosulus Mont.: Lignocellulolytic Enzymes Activities and Sugar Release from Cornstalks under Solid State Fermentation. World J. Microbiol. Biotechnol. 2012, 28, 1961–1966. [Google Scholar] [CrossRef]
- Isikhuemhen, O.S.; Mikiashvili, N.A.; Senwo, Z.N.; Ohimain, E.I. Biodegradation and Sugar Release from Canola Plant Biomass by Selected White Rot Fungi. Adv. Biol. Chem. 2014, 4, 395. [Google Scholar] [CrossRef]
- Zebulun, H.O.; Isikhuemhen, O.S.; Inyang, H. Decontamination of Anthracene-polluted Soil through White Rot Fungus-induced Biodegradation. Environmentalist 2011, 31, 11–19. [Google Scholar] [CrossRef]
- Mori, T.; Nagai, Y.; Kawagishi, H.; Hirai, H. Functional Characterization of the Manganese Transporter smf2 Homologue Gene, PsMnt, of Phanerochaete sordida YK-624 via Homologous Overexpression. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhou, B. Copper and Manganese Induce Yeast Apoptosis via Different Pathways. Mol. Biol. Cell 2007, 18, 4741–4749. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jensen, L.T.; Gardner, A.J.; Culotta, V.C. Manganese Toxicity and Saccharomyces cerevisiae Mam3p, a Member of the ACDP (Ancient Conserved Domain Protein) Family. Biochem. J. 2005, 386, 479–487. [Google Scholar] [CrossRef]
- Hereford, L.; Fahrner, K.; Woolford, J., Jr.; Rosbash, M.; Kaback, D.B. Isolation of Yeast Histone Genes H2A and H2B. Cell 1979, 18, 1261–1271. [Google Scholar] [CrossRef]
- Cunningham, K.W.; Fink, G.R. Calcineurin Inhibits VCX1-dependent H+/Ca2+ Exchange and Induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 2226–2237. [Google Scholar] [CrossRef]
- Pozos, T.C.; Sekler, I.; Cyert, M.S. The Product of HUM1, a Novel Yeast Gene, is Required for Vacuolar Ca2+/H+ Exchange and is Related to Mammalian Na+/Ca2+ Exchangers. Mol. Cell. Biol. 1996, 16, 3730–3741. [Google Scholar] [CrossRef]
- Bianchi, M.; Carbone, M.; Lucchini, G.; Magni, G. Mutants Resistant to Manganese in Saccharomyces cerevisiae. Curr. Genet. 1981, 4, 215–220. [Google Scholar] [CrossRef]
- Peters, R.J.B.; Bouwmeester, H.; Gottardo, S.; Amenta, V.; Arena, M.; Brandhoff, P.; Marvin, H.J.P.; Mech, A.; Moniz, F.B.; Pesudo, L.Q.; et al. Nanomaterials for Products and Application in Agriculture, Feed and Food. Trends Food Sci. Technol. 2016, 54, 155–164. [Google Scholar] [CrossRef]
- Wesley, A. History of the Medical Use of Silver. Surg. Infect. 2009, 10, 289–292. [Google Scholar]
- Jo, Y.-K.; Kim, B.H.; Jung, G. Antifungal Activity of Silver Ions and Nanoparticles on Phytopathogenic Fungi. Plant Dis. 2009, 93, 1037–1043. [Google Scholar] [CrossRef]
- Kaur, P.; Thakur, R.; Duhan, J.S.; Chaudhury, A. Management of Wilt Disease of Chickpea in vivo by Silver Nanoparticles Biosynthesized by Rhizospheric Microflora of Chickpea (Cicer arietinum). J. Chem. Technol. Biotechnol. 2018, 93, 3233–3243. [Google Scholar] [CrossRef]
- Farooq, M.; Ilyas, N.; Khan, I.; Saboor, A.; Khan, K.; Khan, M.N.; Qayum, A.; Ilyas, N.; Bakhtia, M. Antifungal Activity of Plant Extracts and Silver Nano Particles Against Citrus Brown Spot Pathogen (Alternaria citri). Int. J. Environ. Agric. Res. 2018, 4, 118–125. [Google Scholar]
- Gholami-Ahangaran, M.; Zia-Jahromi, N. Nanosilver Effects on Growth Parameters in Experimental Aflatoxicosis in Broiler Chickens. Toxicol. Ind. Health 2013, 29, 121–125. [Google Scholar] [CrossRef]
- Carbone, M.; Donia, D.T.; Sabbatella, G.; Antiochia, R. Silver Nanoparticles in Polymeric Matrices for Fresh Food Packaging. J. King Saud Univ. Sci. 2016, 28, 273–279. [Google Scholar] [CrossRef]
- Vest, K.E.; Wang, J.; Gammon, M.G.; Maynard, M.K.; White, O.L.; Cobine, J.A.; Mahone, W.K.; Cobine, P.A. Overlap of Copper and Iron Uptake Systems in Mitochondria in Saccharomyces cerevisiae. Open Biol. 2016, 6, 150223. [Google Scholar] [CrossRef]
- Vagabov, V.; Yu Ivanov, A.; Kulakovskaya, T.; Kulakovskaya, E.; Petrov, V.; Kulaev, I. Efflux of Potassium Ions from Cells and Spheroplasts of Saccharomyces cerevisiae Yeast Treated with Silver and Copper Ions. Biochemistry 2008, 73, 1224–1227. [Google Scholar] [CrossRef]
- Cyert, M.S.; Philpott, C.C. Regulation of Cation Balance in Saccharomyces cerevisiae. Genetics 2013, 193, 677–713. [Google Scholar] [CrossRef]
- Barros, D.; Pradhan, A.; Pascoal, C.; Cássio, F. Transcriptomics Reveals the Action Mechanisms and Cellular Targets of Citrate-coated Silver Nanoparticles in a Ubiquitous Aquatic Fungus. Environ. Pollut. 2021, 268, 115913. [Google Scholar] [CrossRef]
- Ball, B.; Langille, M.; Geddes-McAlister, J. Fun(gi)omics: Advanced and Diverse Technologies to Explore Emerging Fungal Pathogens and Define Mechanisms of Antifungal Resistance. Mbio 2020, 11, e01020–e20. [Google Scholar] [CrossRef]
- Chitarrini, G.; Riccadonna, S.; Zulini, L.; Vecchione, A.; Stefanini, M.; Larger, S.; Pindo, M.; Cestaro, A.; Franceschi, P.; Magris, G.; et al. Two-omics Data Revealed Commonalities and Differences between Rpv12- and Rpv3-mediated Resistance in Grapevine. Sci. Rep. 2020, 10, 12193. [Google Scholar] [CrossRef] [PubMed]
- Bazzicalupo, A.L.; Ruytinx, J.; Ke, Y.-H.; Coninx, L.; Colpaert, J.V.; Nguyen, N.H.; Vilgalys, R.; Branco, S. Fungal Heavy Metal Adaptation through Single Nucleotide Polymorphisms and Copy-number Variation. Mol. Ecol. 2020, 29, 4157–4169. [Google Scholar] [CrossRef] [PubMed]
- Libkind, D.; Peris, D.; Cubillos, F.A.; Steenwyk, J.L.; Opulente, D.A.; Langdon, Q.K.; Rokas, A.; Hittinger, C.T. Into the Wild: New Yeast Genomes from Natural Environments and New Tools for Their Analysis. FEMS Yeast Res. 2020, 20. [Google Scholar] [CrossRef]
- Samaras, A.; Ntasiou, P.; Myresiotis, C.; Karaoglanidis, G. Multidrug Resistance of Penicillium expansum to Fungicides: Whole Transcriptome Analysis of MDR Strains Reveals Overexpression of Efflux Transporter Genes. Int. J. Food Microbiol. 2020, 335, 108896. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-H.; Huang, Z.-X.; Luo, X.-H.; Chen, H.; Weng, B.-Q.; Wang, Y.-X.; Chen, L.-S. Comparative Transcriptome Analysis Reveals Candidate Genes Related to Cadmium Accumulation and Tolerance in Two Almond Mushroom (Agaricus brasiliensis) Strains with Contrasting Cadmium Tolerance. PLoS ONE 2020, 15, e0239617. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, J.R.; Isikhuemhen, O.S.; Anike, F.N. Fungal–Metal Interactions: A Review of Toxicity and Homeostasis. J. Fungi 2021, 7, 225. https://doi.org/10.3390/jof7030225
Robinson JR, Isikhuemhen OS, Anike FN. Fungal–Metal Interactions: A Review of Toxicity and Homeostasis. Journal of Fungi. 2021; 7(3):225. https://doi.org/10.3390/jof7030225
Chicago/Turabian StyleRobinson, Janelle R., Omoanghe S. Isikhuemhen, and Felicia N. Anike. 2021. "Fungal–Metal Interactions: A Review of Toxicity and Homeostasis" Journal of Fungi 7, no. 3: 225. https://doi.org/10.3390/jof7030225
APA StyleRobinson, J. R., Isikhuemhen, O. S., & Anike, F. N. (2021). Fungal–Metal Interactions: A Review of Toxicity and Homeostasis. Journal of Fungi, 7(3), 225. https://doi.org/10.3390/jof7030225





