Application of Real-Time PCR for the Detection and Quantification of Oomycetes in Ornamental Nursery Stock
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples and Oomycete Isolation and Identification
2.2. Extraction of Environmental DNA (eDNA)
2.3. Detection and Quantification of Pathogen Load in eDNA Extractions from Nursery Plants Using Real-Time PCR
2.4. Internal Positive Control (IPC) to Detect Inhibitors in eDNA
3. Results
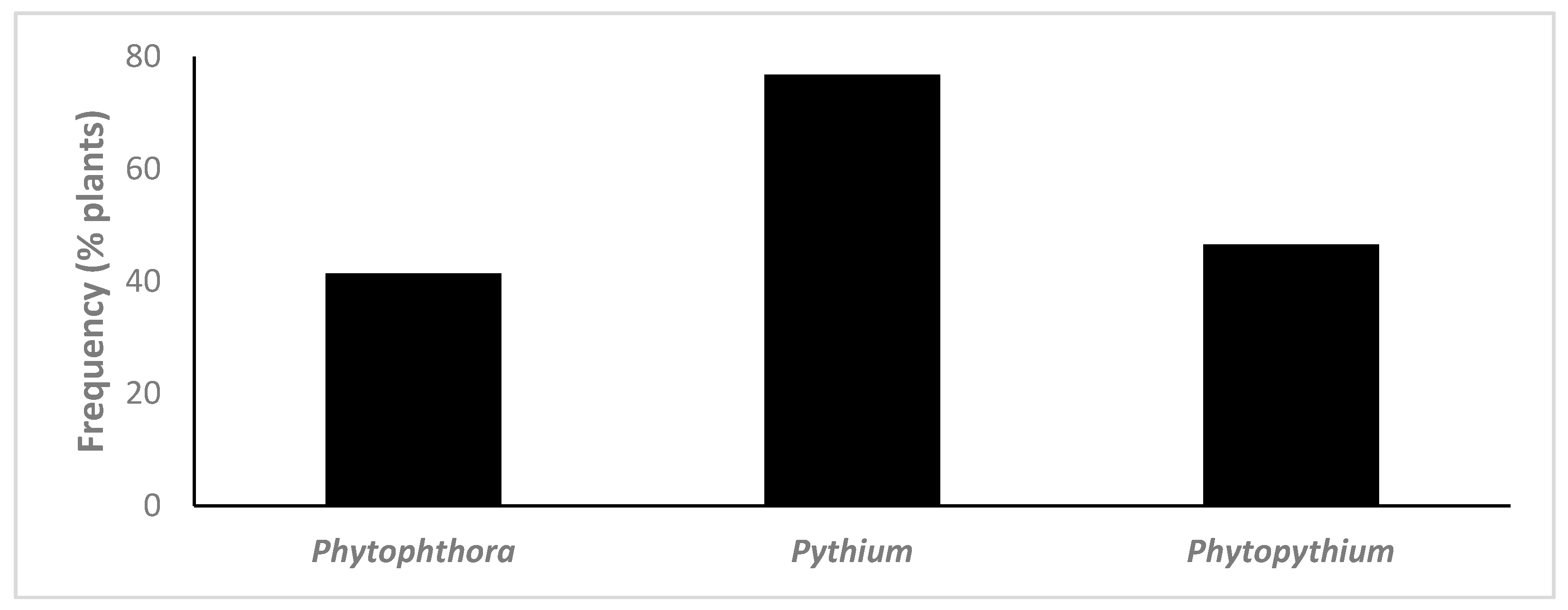
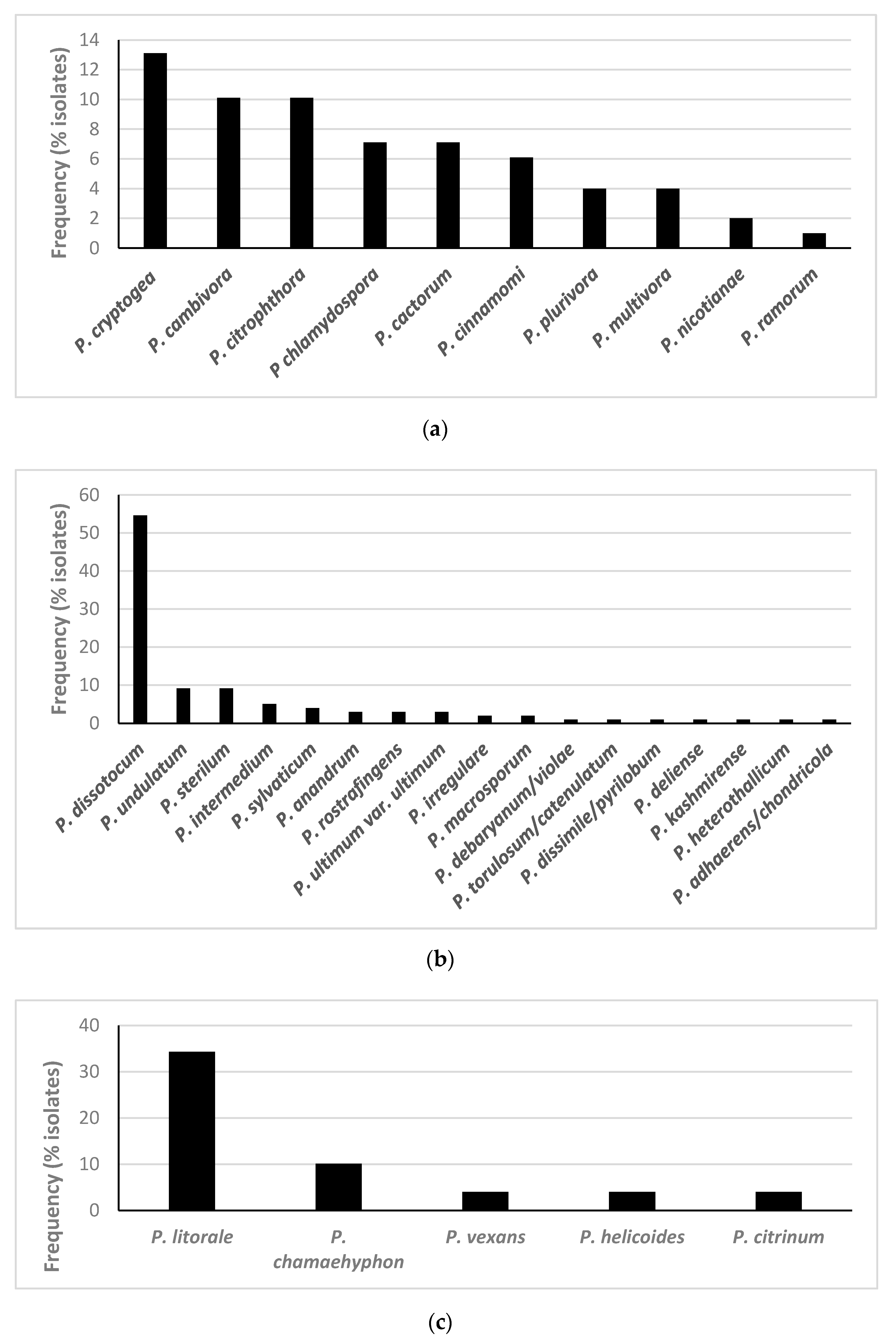
3.1. Isolation and Identification of Oomycetes
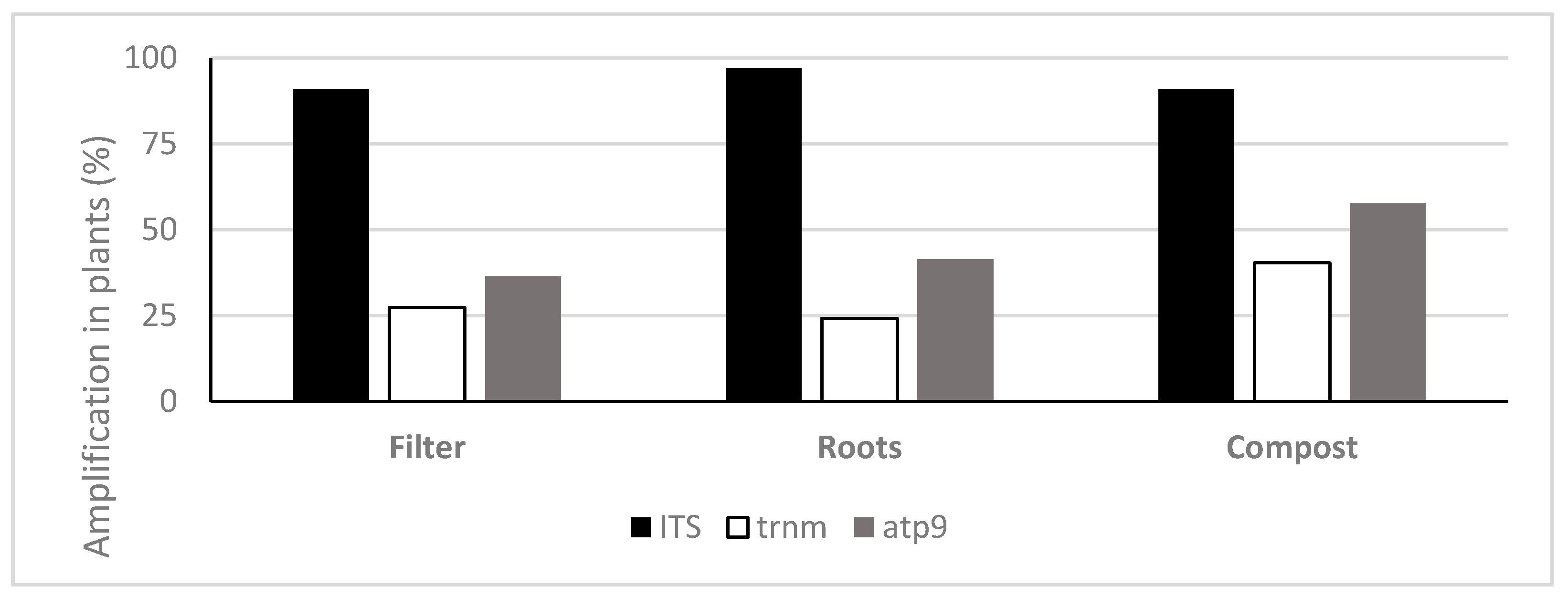
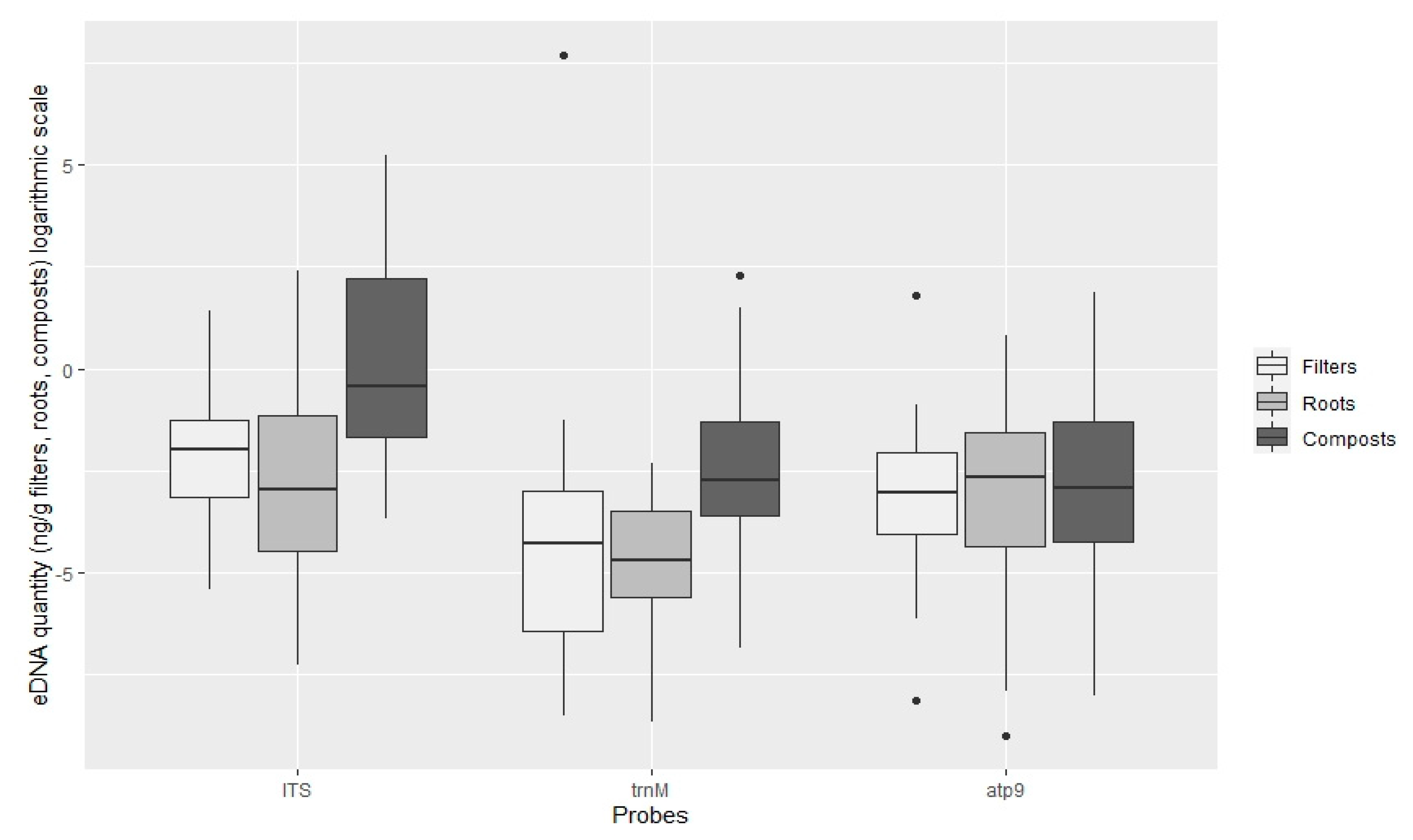
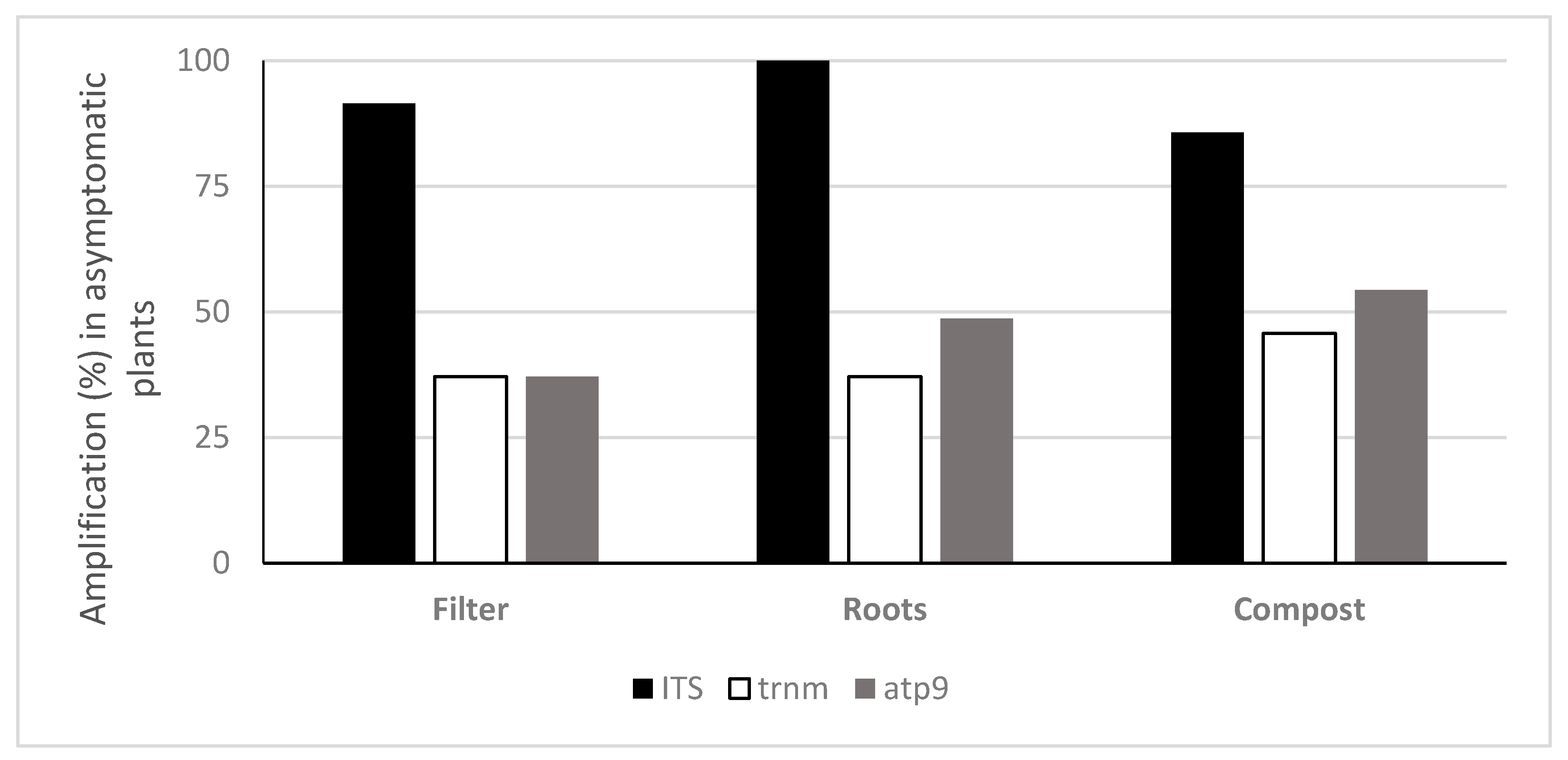
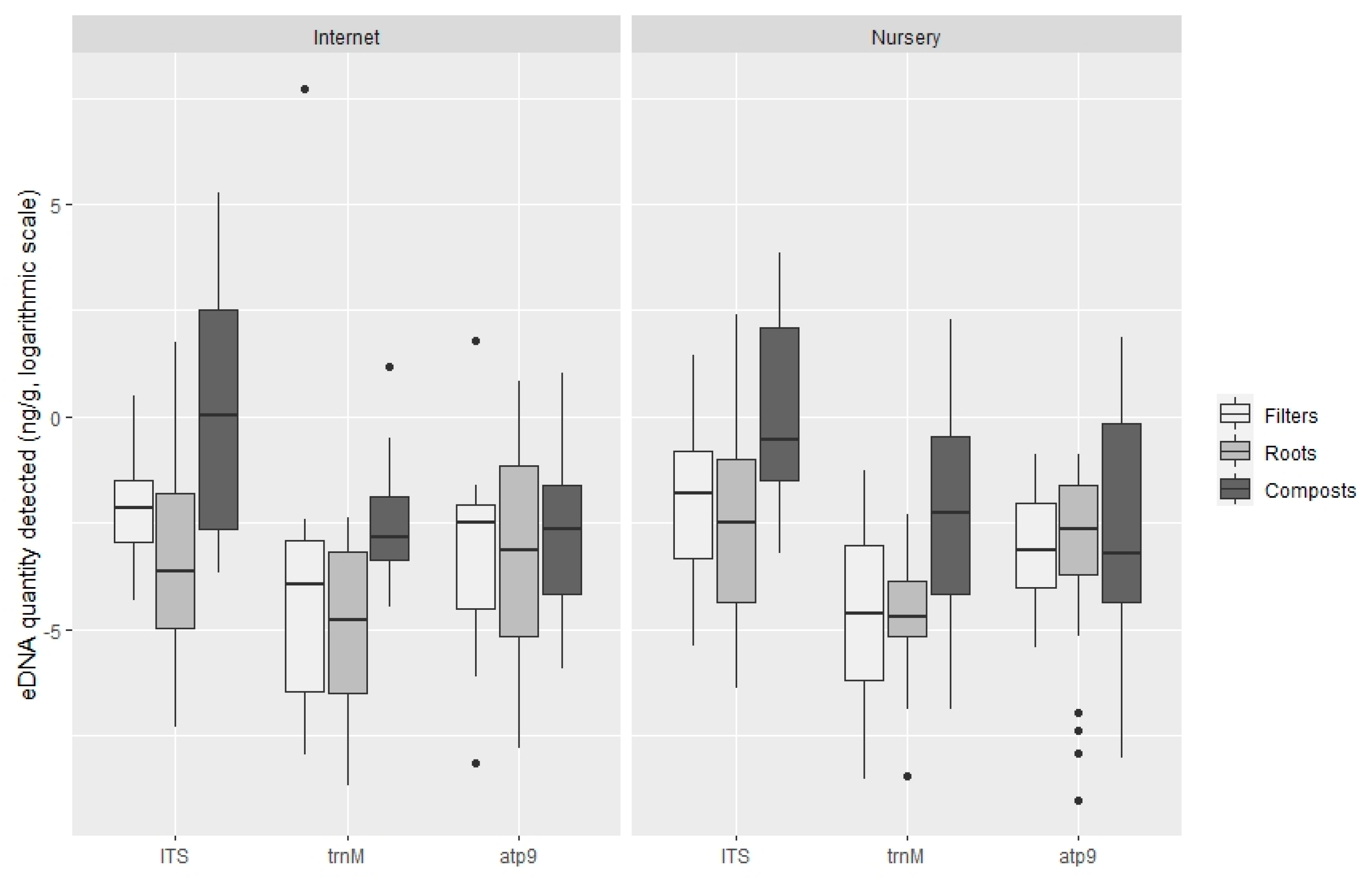
3.2. Quantification of Oomycota with TaqMan PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drew, J.; Anderson, N.; Andow, D. Conundrums of a complex vector for invasive species control: A detailed examination of the horticultural industry. Biol. Inv. 2010, 12, 2837–2851. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.M.; Jung, T. Phytophthora in woody ornamental nurseries. In Phytophthora. A Global Perspective; Lamour, K., Ed.; CABI Plant Protection Series; CAB International: Knoxville, TN, USA, 2013; pp. 166–177. [Google Scholar]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguín Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavarriaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- Dehnen-Schmutz, K.; Holdenrieder, O.; Jeger, M.J.; Pautasso, M. Structural change in the international horticultural industry: Some implications for plant health. Sci. Hortic. 2010, 125, 1–15. [Google Scholar] [CrossRef]
- Freund, C.L.; Weinhold, D. The effect of the Internet on international trade. J. Intern. Econ. 2004, 62, 171–189. [Google Scholar] [CrossRef]
- Giltrap, N.; Eyre, D.; Reed, P. Internet sales of plants for planting an increasing trend and threat? EPPO Bull. 2009, 39, 168–170. [Google Scholar] [CrossRef]
- FAO. Impact of Global Trade and Human Mobility on the Health of Agricultural Crops and Forests in Europe and Central Asia. 2019, ECA/41/19/5: 13. Available online: http://www.fao.org/3/nb096en/nb096en.pdf (accessed on 15 June 2017).
- Hantula, J.; Müller, M.M.; Uusivuori, J. International plant trade associated risks: Laissez-faire or novel solutions. Envion. Sci. Policy 2014, 37, 158–160. [Google Scholar] [CrossRef]
- Ferguson, A.J.; Jeffers, S.N. Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Dis. 1999, 83, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Brasier, C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008, 57, 792–808. [Google Scholar] [CrossRef]
- EPPO. EPPO Study on the Risk of Imports of Plants for Planting; Technical Document No. 1061; EPPO: Paris, France, 2012. [Google Scholar]
- Migliorini, D.; Ghelardini, L.; Tondini, E.; Luchi, N.; Santini, A. The potential of symptomless potted plants for carrying invasive soil-borne plant pathogens. Divers. Distrib. 2015, 21, 1218–1229. [Google Scholar] [CrossRef]
- Prospero, S.; Vercauteren, A.; Heungens, K.; Belbahri, L.; Rigling, D. Phytophthora diversity and the population structure of Phytophthora ramorum in Swiss ornamental nurseries. Plant Pathol. 2013, 62, 1063–1071. [Google Scholar] [CrossRef]
- Blaya, J.; Lacasa, C.; Lacasa, A.; Martínez, V.; Santísima-Trinidad, A.B.; Pascual, J.A.; Ros, M. Characterization of Phytophthora nicotianae isolates in southeast Spain and their detection and quantification through a real-time TaqMan, P.C.R. J. Sci. Food Agric. 2015, 95, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Blaya, J.; Lloret, E.; Santísima-Trinidad, A.B.; Ros, M.; Pascual, J.A. Molecular methods (digital PCR and real-time PCR) for the quantification of low copy DNA of Phytophthora nicotianae in environmental samples. Pest Manag. Sci. 2016, 72, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Filion, M.; St-Arnaud, M.; Jabaji-Hare, S.H. Direct quantification of fungal DNA from soil substrate using real-time, P.C.R. J. Microbiol. Methods 2003, 53, 67–76. [Google Scholar] [CrossRef]
- Hayden, K.; Ivors, K.; Wilkinson, C.; Garbelotto, M. TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods. Phytopathology 2006, 96, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.J.D.; Tomlinson, J.A.; Griffin, R.L.; Boonham, N.; Inman, A.J.; Lane, C.R. Development of a one-step real-time polymerase chain reaction assay for diagnosis of Phytophthora ramorum. Phytopathology 2006, 96, 975–981. [Google Scholar] [CrossRef]
- Schena, L.; Hughes, K.J.D.; Cooke, D.E.L. Detection and quantification of Phytophthora ramorum, P. kernoviae, P. citricola and P. quercina in symptomatic leaves by multiplex real-time, P.C.R. Mol. Plant Pathol. 2006, 7, 365–379. [Google Scholar] [CrossRef]
- Hughes, K.J.D.; Tomlinson, J.A.; Giltrap, P.M.; Barton, V.; Hobden, E.; Boonham, N.; Lane, C.R. Development of a real-time PCR assay for detection of Phytophthora kernoviae and comparison of this method with a conventional culturing technique. Eur. J. Plant Pathol. 2011, 131, 695–703. [Google Scholar] [CrossRef]
- Bilodeau, G.J.; Martin, F.N.; Coffey, M.D.; Blomquist, C.L. Development of a multiplex assay for genus- and species-specific detection of Phytophthora based on differences in mitochondrial gene order. Phytopathology 2014, 104, 733–748. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Jeffers, S.N.; Martin, S.B. Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 1986, 70, 1038–1043. [Google Scholar] [CrossRef]
- Themann, K.; Werres, S.; Diener, H.A.; Lüttmann, R. Comparison of different methods to detect Phytophthora spp. in recycling water from nurseries. J. Plant Pathol. 2002, 84, 41–50. [Google Scholar]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Valverde-Corredor, A.; Jiménez-Díaz, R.M. Molecular variability within and among Verticillium dahliae vegetative compatibility groups determined by fluorescent amplified fragment length polymorphism and polymerase chain reaction markers. Phytopathology 2006, 96, 485–495. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal. Genet. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Català, S.; Pérez-Sierra, A.; Abad-Campos, P. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Català, S.; Berbegal, M.; Pérez-Sierra, A.; Abad-Campos, P. Metabarcoding and development of new real-time specific assays reveal Phytophthora species diversity in holm oak forests in eastern Spain. Plant Pathol. 2016, 66, 115–123. [Google Scholar] [CrossRef]
- Taberlet, P.; Prud’Homme, S.M.; Campione, E.; Roy, J.; Miquel, C.; Shehzad, W.; Gielly, L.; Rioux, D.; Choler, P.; Clément, J.; et al. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 2012, 21, 1816–1820. [Google Scholar] [CrossRef]
- Zinger, L.; Chave, J.; Coissac, E.; Iribar, A.; Louisanna, E.; Manzi, S.; Schilling, V.; Schimann, H.; Sommeria-Klein, G.; Taberlet, P. Extracellular DNA extraction is a fast, cheap and reliable alternative for multi-taxa surveys based on soil, DNA. Soil Biol. Biochem. 2016, 96, 16–19. [Google Scholar] [CrossRef]
- Kox, L.F.F.; Van Brouwershaven, I.R.; Van De Vossenberg, B.T.L.H.; Van Den Beld, H.E.; Bonants, P.J.M.; De Gruyter, J. Diagnostic values and utility of immunological, morphological, and molecular methods for in planta detection of Phytophthora ramorum. Phytopathology 2007, 97, 1119–1129. [Google Scholar] [CrossRef]
- Waalwijk, C.; Van Der Heide, R.; De Vries, I.; Van Der Lee, T.; Schoen, C.; Costrel-de Corainville, G.; Häuser-Hahn, I.; Kastelein, P.; Köhl, J.; Lonnet, P.; et al. Quantitative detection of Fusarium species in wheat using TaqMan. Eur. J. Plant Pathol. 2004, 110, 481–494. [Google Scholar] [CrossRef]
- Hartman, L.J.; Coyne, S.R.; Norwood, D.A. Development of a novel internal positive control for TaqMan based assays. Mol. Cell. Probes 2005, 19, 51–59. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing; R Development Core Team: Vienna, Austria, 2008; ISBN 3-900051-07-0. Available online: www.R-project.org (accessed on 20 February 2017).
- Martin, F.N. Molecular identification of Phytophthora. In Phytophthora. A global perspective; Lamour, K., Ed.; CABI Plant Protection Series; CAB International: Knoxville, TN, USA, 2013; pp. 19–27. [Google Scholar]
- Maleszka, R.; Clark-Walker, G.D. Yeasts have a four-fold variation in ribosomal DNA copy number. Yeast 1993, 9, 53–58. [Google Scholar] [CrossRef]
- Bilodeau, G.J.; Koike, S.T.; Uribe, P.; Martin, F.N. Development of an assay for rapid detection and quantification of Verticillium dahliae in soil. Phytopathology 2012, 102, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Prigigallo, M.I.; Mosca, S.; Cacciola, S.O.; Cooke, D.E.L.; Schena, L. Molecular analysis of Phytophthora diversity in nursery-grown ornamental and fruit plants. Plant Pathol. 2015, 64, 1308–1319. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Abdelfattah, A.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Cooke, D.E.L.; Schena, L. Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 2016, 106, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Grünwald, N.J.; Goss, E.M.; Press, C.M. Phytophthora ramorum: A pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 2008, 9, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Vettraino, A.M.; Morel, O.; Perlerou, C.; Robin, C.; Diamandis, S.; Vannini, A. Occurrence and distribution of Phytophthora species in European chestnut stands, and their association with Ink Disease and crown decline. Eur. J. Plant Pathol. 2005, 111, 169–180. [Google Scholar] [CrossRef]
- Da Clara, M.I.E.; de Almeida Ribeiro, N.M.C. Decline of Mediterranean oak trees and its association with Phytophthora cinnamomi: A review. For. Pathol. 2013, 32, 411–432. [Google Scholar]
- Moricca, S.; Linaldeddu, B.T.; Ginetti, B.; Scanu, B.; Franceschini, A.; Ragazzi, A. Endemic and emerging pathogens threatening cork oak trees: Management options for conserving a unique forest ecosystem. Plant Dis. 2016, 100, 2184–2193. [Google Scholar] [CrossRef]
- Frisullo, S.; Lima, G.; Magnano di San Lio, G.; Camele, I.; Melissano, L.; Puglisi, I.; Pane, A.; Agosteo, G.E.; Prudente, L.; Cacciola, S.O. Phytophthora cinnamomi involved in the decline of holm oak (Quercus ilex) stands in southern Italy. For. Sci. 2018, 64, 290–298. [Google Scholar] [CrossRef]
- Weiland, J.E.; Beck, B.R.; Davis, A. Pathogenicity and virulence of Pythium species obtained from forest nursery soils on Douglas-fir seedlings. Plant Dis. 2013, 97, 744–748. [Google Scholar] [CrossRef]
- McGehee, C.; Raudales, R.E.; Elmer, W.H. First report of Pythium dissotocum causing Pythium root rot on hydroponically grown lettuce in Connecticut. Plant Dis. 2018, 102, 2043. [Google Scholar] [CrossRef]
- Del Castillo Múnera, J.; Hausbeck, M.K. Characterization of Pythium species associated with greenhouse floriculture crops in Michigan. Plant Dis. 2016, 100, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.; Bala, K. A new species of Pythium with inflated sporangia and coiled antheridia, isolated from India. FEMS Microbiol. Lett. 2008, 282, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Crocker, E.V.; Karp, M.A.; Nelson, E.B. Virulence of oomycete pathogens from Phragmites australis invaded and non-invaded soils to seedlings of wetland plant species. Ecol. Evol. 2015, 5, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Zitnick-Anderson, K.K.; Nelson, B.D., Jr. Identification and pathogenicity of Pythium on soybean in North Dakota. Plant Dis. 2015, 99, 31–38. [Google Scholar] [CrossRef]
- EPPO. Phytophthora ramorum. EPPO Bull. 2006, 36, 145–155. [Google Scholar] [CrossRef]
- Veal, C.D.; Freeman, P.J.; Jacobs, K.; Lancaster, O.; Jamain, S.; Leboyer, M.; Albanes, D.; Vaghela, R.R.; Gut, I.; Chanock, S.J.; et al. A mechanistic basis for amplification differences between samples and between genome regions. BMC Genom. 2012, 13, 455. [Google Scholar] [CrossRef]
- Lamarche, J.; Potvin, A.; Pelletier, G.; Stewart, D.; Feau, N.; Alayon, D.I.O.; Dale, A.L.; Coelho, A.; Uzunovic, A.; Bilodeau, G.J.; et al. Molecular detection of 10 most unwanted alien forest pathogens in Canada using real-time PCR. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]

| Locus | Probe/Primers | Fluorophore/Quencher of the Probe | Sequence (5′ to 3′) | Annealing Temperature (°C) | Ref |
|---|---|---|---|---|---|
| ITS | All-phy Probe | FAM/BHQ1 | TTGCTATCTAGTTAAAAGCA | 60 | [32] |
| FITS_15Ph | TGCGGAAAGGATCATTACCACACC | ||||
| RITS_279Ph | GCGAGCCTAGACATCCACTG | ||||
| trnM-trnP-trnM | TrnM_PhyG_probe2 | FAM/BHQ1 | ATRTTGTAGGTTCAARTCCTAYCATCAT | 62 | [21] |
| PhyG-F2 | CGTGGG AATCATAATCCT | ||||
| PhyG-Rb | CAGATTATGAGCCTGATAAG | ||||
| atp9-nad9 | ATP9_PhyG2_probeR | FAM/BHQ1 | AAAGCCATCATTAAACARAATAAAGC | 57 | [21] |
| PhyG_ATP9_2FTail | AATAAATCATAACCTTCTTTACAACAAGAATTAATG | ||||
| PhyG-R6_Tail | AATAAATCATAAATACATAATTCATTTTTATA | ||||
| PLRV (Internal Positive Control) | PLRV-P-HEX | HEX/BHQ1 | CGAAGACGCAGAAGAGGAGCCAAT | 60 | [33] |
| IntConF (PLRV-F) | AAGAGGCGAAGAAGGCAATCC | ||||
| IntConR (PLRV-R) | GCACTGATCCTCAGAAGAATCG |
| Nursery and Country of Purchase | Plant Species | Plant Codes | Oomycetes Species Isolated | ||
|---|---|---|---|---|---|
| Phytophthora spp. | Pythium spp. | Phytopythium spp. | |||
| Nursery 1—UK (I) | Viburnum x bodnantense ‘Dawn’ | P1 | P. ramorum (R, S), P. cryptogea (B, R) | P. dissotocum (B, R), P. lutarium/diclinum (B) | P. litorale (B, S) |
| Acer palmatum ‘Dissectum Filigree’ | P21, P22, P23, P24 | P. cambivora (B, S), P. plurivora (B) | P. dissotocum (B, R, S), P. lutarium (B), P. dissimile/pyrilobum (R), P. intermedium (S), P. perplexum (R) | NI | |
| Viburnum plicatum ‘Lanarth’ | P25, P26, P27, P28, P29 | NI | P. dissotocum (B, R), P. intermedium (S), P. deliense (R), P. sterilum (B), P. kashmirense (B) | P. helicoides (S) | |
| Viburnum burkwoodii ‘Park Farm hybrid’ | P30, P31, P32, P33, P34, P45, P46, P47, P48, P49 | P. cryptogea (B, R, S), P. chlamydospora (B, R, S), P. cactorum (S) | P. dissotocum (B, R), P. anandrum (S), P. dissotocum/coloratum (B) | P. litorale (B, S) | |
| Viburnum tinus ‘Eve Price’ | P35, P36, P37, P38, P39 | P. citrophthora (B), P. cactorum (B, S), P. cryptogea (B) | P. dissotocum (B) | P. vexans (S), P. helicoides (R), P. chamaehyphon (S) | |
| Ceanothus thyrsiflorus ‘Southmead’ | P99 | P. cinnamomi (B), P. cambivora (B) | P. intermedium (R), P. dissotocum/diclinum (B) | NI | |
| Nursery 2—UK | Acer palmatum ‘Orange Dream’ | P2, P3, P4 | P. plurivora (B, S) | P. irregulare (B), P. debaryanum/violae (B) | P. chamaehyphon (S), P. citrinum (S) |
| Acer palmatum ‘Atropurpureum’ | P5, P6, P7 | NI | P. torulosum/catenulatum (B), P. undulatum (B), P. intermedium (B) | NI | |
| Hebe x franciscana ‘Variegata’ | *P8, *P9, *P10, *P11, *P12, *P13 | NI | P. dissotocum (B, R, S), P. diclinum/lutarium (B) | P. chamaehyphon (S), P. litorale (S), P. vexans (S) | |
| Nursery 3—UK | Rhododendron ‘Germania’ | *P14, *P15 | P. cinnamomi (B, R, S) | P. undulatum (B, S) | NI |
| Rhododendron ‘Marcel Menard’ | *P16, *P17 | NI | P. undulatum (B, S), P. macrosporum (B, S) | P. helicoides (B) | |
| Rhododendron ‘Percy Wiseman’ | *P18, *P19 | NI | P. undulatum (B, S) | NI | |
| Nursery 4—UK (I) | Camellia alba ‘Plena’ | *P40, *P41, *P42, *P43, *P44 | NI | P. heterothallicum (R), P. irregulare (B), P. intermedium (R) | NI |
| Nursery 5—NL (I) | Ceanothus thyrsiflorus ‘Repens’ | *P50, *P51, *P52, *P53, *P54 | P. nicotianae (B), P. cactorum (B, R, S), P. citrophthora (B, R, S), P. cinnamomi (B, S) | P. sylvaticum (R, S), P. sylvaticum/terrestris (S), P. dissotocum (B), P. lutarium/diclinum (B), P. rostratifingens (B), P. adhaerens/ chondricola (B) | NI |
| Euonymus fortunei ‘Emerald Gaiety’ | P55, P56, P57, P58, P59 | NI | P. dissotocum (B), P. rostratifingens like (R) | P. vexans (B) | |
| Nursery 6—NL | Ilex meserveae ‘Blue Maid’ | P60, P61, P62, P63 | P. plurivora (B), P. cinnamomi (B, R, S), P. cambivora (B), P. cryptogea (B, S) | P. dissotocum (B) | P. litorale (B, S), P. citrinum (B) |
| Ilex aquifolium ‘Argentea’ | *P64, P65, P66,* P67 | P. chlamydospora (R) | P. anandrum (R, S), P. dissotocum (B) | P. litorale (B, R) | |
| Ilex x altaclerensis ‘Golden King’ | *P68, P69, P70, P71 | P. cambivora (B) | NI | P. litorale/sterilum (B, R, S) | |
| Pinus mugo | *P72, *P73, *P74, *P75, *P76, *P77 | P. cambivora (S) | P. dissotocum (R), P. litorale/sterilum (R) | P. litorale (B) | |
| Nursery 7—NL | Camellia japonica | P78, P79, P80, P81 | NI | P. dissotocum (B), P. lutarium/diclinum (B), P. anandrum (S) | P. litorale (B, S) |
| Buxus sempervirens | P82, P83, P84, P85, P86 | P. multivora (B) | P. sylvaticum (B), P. ultimum var. ultimum (S), P. dissotocum (B), P. dissotocum/lutarium (B), P. rostratifingens (R), P. rostratifingens/camurandrum (R) | P. litorale (B) | |
| Rhododendron obtusum ‘Anouk’ | P87, P88, P89, P90 | NI | P. undulatum/ultimum (S) | P. litorale (B, S) | |
| Nursery 8—NL | Euonymus fortunei ‘Arlequin’ | *P91, *P92, *P93, *P94, P95, P96, P97, P98 | P. multivora (B), P. citrophthora/colocasiae (B), P. citrophthora (B), P. plurivora (B) | P. dissotocum/lutarium (B, R), P. undulatum/ultimum (B) | P. litorale (B), P. citrinum/helicoides (B), P. citrinum/chamaehyphon (B), P. vexans (S) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puertolas, A.; Bonants, P.J.M.; Boa, E.; Woodward, S. Application of Real-Time PCR for the Detection and Quantification of Oomycetes in Ornamental Nursery Stock. J. Fungi 2021, 7, 87. https://doi.org/10.3390/jof7020087
Puertolas A, Bonants PJM, Boa E, Woodward S. Application of Real-Time PCR for the Detection and Quantification of Oomycetes in Ornamental Nursery Stock. Journal of Fungi. 2021; 7(2):87. https://doi.org/10.3390/jof7020087
Chicago/Turabian StylePuertolas, Alexandra, Peter J. M. Bonants, Eric Boa, and Steve Woodward. 2021. "Application of Real-Time PCR for the Detection and Quantification of Oomycetes in Ornamental Nursery Stock" Journal of Fungi 7, no. 2: 87. https://doi.org/10.3390/jof7020087
APA StylePuertolas, A., Bonants, P. J. M., Boa, E., & Woodward, S. (2021). Application of Real-Time PCR for the Detection and Quantification of Oomycetes in Ornamental Nursery Stock. Journal of Fungi, 7(2), 87. https://doi.org/10.3390/jof7020087








