Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Cultures
2.2. Separation of Extracellular Vesicles (EVs)
2.3. Heat-Treatment of Fgr Cultures
2.4. Preparation of Secretomes and Whole-Cell Lysates (WCL)
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Transmission-Electron Microscopy (TEM)
2.7. Mass Spectrometry (MS)
2.8. Computational Prediction of Effector Proteins in EV Samples
2.9. Gene Ontology (GO) Analysis
2.10. Maize Leaf Sheath Infection Assay
2.11. RNA Extraction from Infected Corn Tissue and Fgr Mycelium
2.12. Transcriptome Analysis
3. Results
3.1. Culture Optimization to Improve the Yield of EVs from Fgr
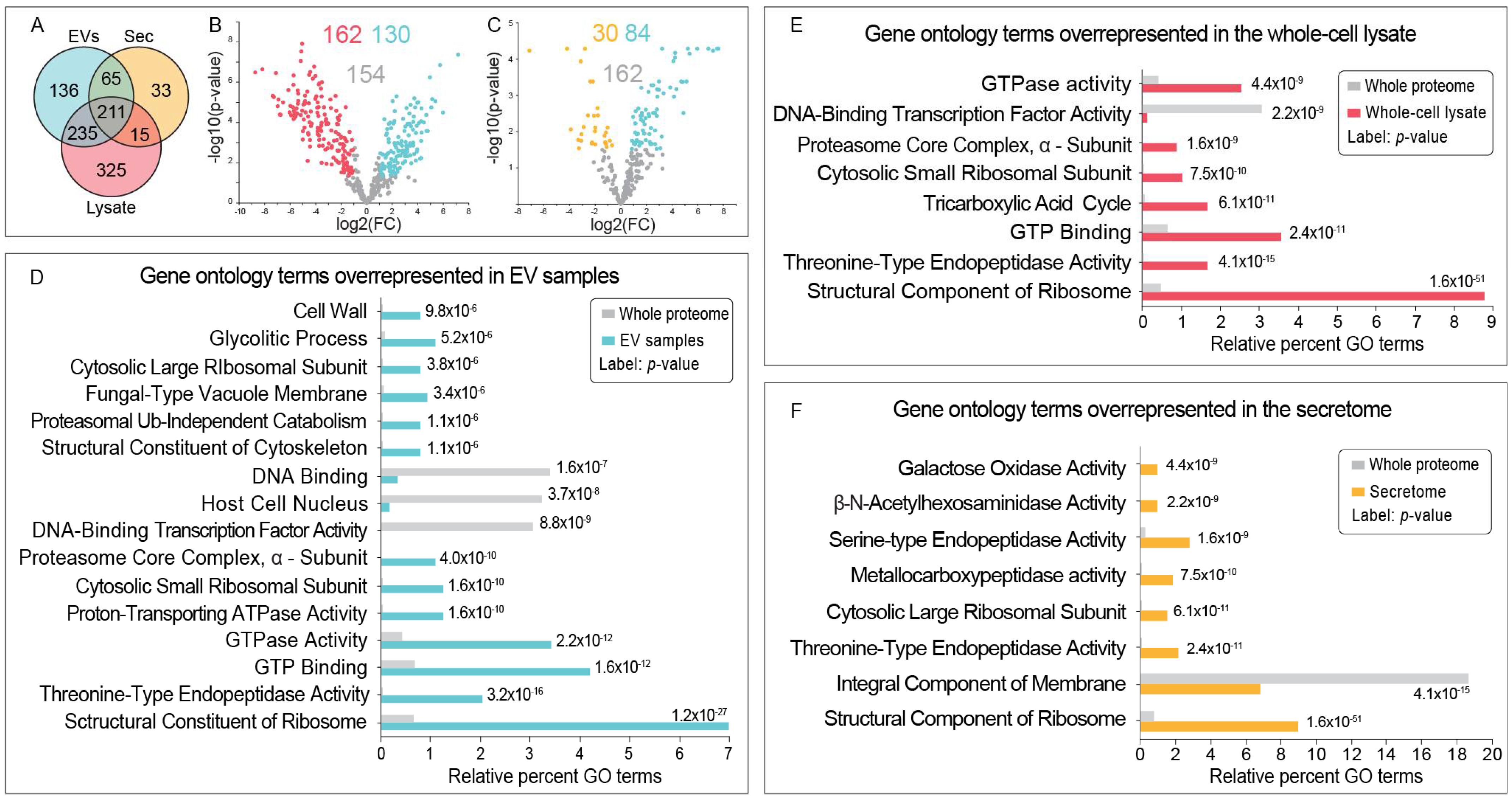
3.2. Fgr EV Samples Contain Putative Fungal EV Protein Markers and Proteins with Potential Roles in Toxin Synthesis, Cell Wall Modifications, and Virulence
3.3. The Secretome from Fgr Contains Proteins with Potential Roles in Carbohydrate Metabolism, Oxidoreduction and Pathogenesis
3.4. EV Samples from Fgr Contain Candidate Protein Effectors
3.5. Candidate Protein Effectors Detected in EV Samples Are Expressed In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oghenekaro, A.; Oviedo-Ludena, M.; Serajazari, M.; Wang, X.; Henriquez, M.; Wenner, N.; Kuldau, G.; Navabi, A.; Kutcher, H.; Fernando, W. Population Genetic Structure and Chemotype Diversity of Fusarium graminearum Populations from Wheat in Canada and North Eastern United States. Toxins 2021, 13, 180. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [Green Version]
- Munkvold, G.P.; Proctor, R.H.; Moretti, A. Mycotoxin Production in Fusarium According to Contemporary Species Concepts. Annu. Rev. Phytopathol. 2021, 59, 373–402. [Google Scholar] [CrossRef]
- de Castilla, P.E.M.; Tong, L.; Huang, C.; Sofias, A.M.; Pastorin, G.; Chen, X.; Storm, G.; Schiffelers, R.M.; Wang, J.-W. Extracellular vesicles as a drug delivery system: A systematic review of preclinical studies. Adv. Drug Deliv. Rev. 2021, 175, 113801. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Honorato, L.; Guimarães, A.J.; Rodrigues, M.L.; Reis, F.C.G.; Vale, A.M.; Ray, A.; Nosanchuk, J.D.; Nimrichter, L. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell. Microbiol. 2020, 22, e13238. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.; Wong, S.S.W.; Gazi, A.D.; Moyrand, F.; Chaze, T.; Commere, P.; Novault, S.; Matondo, M.; Péhau-Arnaudet, G.; Reis, F.C.G.; et al. Cryptococcus extracellular vesicles properties and their use as vaccine platforms. J. Extracell. Vesicles 2021, 10, e12129. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’Donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 2018, 9, 1556. [Google Scholar] [CrossRef] [Green Version]
- Brauer, V.S.; Pessoni, A.M.; Bitencourt, T.A.; de Paula, R.G.; Rocha, L.D.O.; Goldman, G.H.; Almeida, F. Extracellular Vesicles from Aspergillus flavus Induce M1 Polarization In Vitro. mSphere 2020, 5, 00190-20. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.; Pathan, M.; Zhao, K.; Ang, C.-S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridout, C.; Skamnioti, P.; Porritt, O.; Sacristan, S.; Jones, J.; Brown, J.K. Multiple Avirulence Paralogues in Cereal Powdery Mildew Fungi May Contribute to Parasite Fitness and Defeat of Plant Resistance. Plant Cell 2006, 18, 2402–2414. [Google Scholar] [CrossRef]
- Micali, C.O.; Neumann, U.; Grunewald, D.; Panstruga, R.; O’Connell, R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 2010, 13, 210–226. [Google Scholar] [CrossRef] [PubMed]
- Reindl, M.; Hänsch, S.; Weidtkamp-Peters, S.; Schipper, K. A Potential Lock-Type Mechanism for Unconventional Secretion in Fungi. Int. J. Mol. Sci. 2019, 20, 460. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ceron, D.; Dawson, C.S.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Size-exclusion chromatography allows the isolation of EVs from the filamentous fungal plant pathogen Fusarium oxysporum f. sp. vasinfectum (Fov). Proteomics 2021, 21, 2000240. [Google Scholar] [CrossRef] [PubMed]
- Dawson, C.S.; Garcia-Ceron, D.; Rajapaksha, H.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins. J. Extracell. Vesicles 2020, 9, 1750810. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Dodds, P.N.; Singh, K.B.; Taylor, J.M. ApoplastP: Prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2018, 217, 1764–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, K.; Grote, A.; Scheer, M.; Münch, R.; Jahn, D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004, 32, W375–W379. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Kiemer, L.; Fausbøll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef] [Green Version]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Götz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, Z.; Luo, M.; Peng, H.; Lin, H.; Qin, C.; Yuan, G.; Shen, Y.; Ding, H.; Zhao, M.; et al. Genome expression profile analysis of the maize sheath in response to inoculation to R. solani. Mol. Biol. Rep. 2014, 41, 2471–2483. [Google Scholar] [CrossRef] [Green Version]
- Altier, J.; Dahlbacka, I.; Herrmann, R.; Hunter-Cervera, J.; McCutchen, B.; Presnail, J.; Rice, J.; Schepers, E.; Simmons, C.; Torok, T.; et al. Antifungal Polypeptides. European Patent EP1763536B1, 8 September 2010. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Edwards, M.C. Genome-Wide Analysis of Small Secreted Cysteine-Rich Proteins Identifies Candidate Effector Proteins Potentially Involved in Fusarium graminearum−Wheat Interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.A.; Evans, J.; Mead, A.; Hammond-Kosack, K.E. A spatial temporal analysis of the Fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Mol. Plant Pathol. 2017, 18, 1295–1312. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.S.; Mitchell, T.K.; Dean, R.A. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 2007, 273, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Yuan, J.; Qiu, D. The Verticillium dahliae SnodProt1-Like Protein VdCP1 Contributes to Virulence and Triggers the Plant Immune System. Front. Plant Sci. 2017, 8, 1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, K.; Irieda, H.; Sugimoto, F.; Yoshioka, H.; Okuno, T.; Takano, Y. Cell Death of Nicotiana benthamiana Is Induced by Secreted Protein NIS1 of Colletotrichum orbiculare and Is Suppressed by a Homologue of CgDN3. Mol. Plant-Microbe Interact. 2012, 25, 625–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blümke, A.; Falter, C.; Herrfurth, C.; Sode, B.; Bode, R.; Schäfer, W.; Feussner, I.; Voigt, C. Secreted Fungal Effector Lipase Releases Free Fatty Acids to Inhibit Innate Immunity-Related Callose Formation during Wheat Head Infection. Plant Physiol. 2014, 165, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Piffer, A.C.; Kuczera, D.; Rodrigues, M.L.; Nimrichter, L. The paradoxical and still obscure properties of fungal extracellular vesicles. Mol. Immunol. 2021, 135, 137–146. [Google Scholar] [CrossRef]
- Garcia-Ceron, D.; Bleackley, M.R.; Anderson, M.A. Fungal Extracellular Vesicles in Pathophysiology. In New Frontiers: Extracellular Vesicles; Subcellular Biochemistry, 2021/03/30 ed.; Mathivanan, S., Fonseka, P., Nedeva, C., Atukorala, I., Eds.; Springer: Cham, Switzerland, 2021; Volume 97, pp. 151–177. [Google Scholar]
- Cleare, L.G.; Zamith, D.; Heyman, H.M.; Couvillion, S.P.; Nimrichter, L.; Rodrigues, M.L.; Nakayasu, E.S.; Nosanchuk, J.D. Media matters! Alterations in the loading and release of Histoplasma capsulatum extracellular vesicles in response to different nutritional milieus. Cell. Microbiol. 2020, 22, e13217. [Google Scholar] [CrossRef]
- Takegawa, K.; Satoh, K.; Ramli, N.; Jikibara, T.; Iwahara, S. Production and characterization of extracellular uronic acid-containing glycoproteins from Fusarium oxysporum. J. Ferment. Bioeng. 1997, 83, 197–200. [Google Scholar] [CrossRef]
- Giese, H.; Sondergaard, T.E.; Sørensen, J.L. The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 2013, 117, 814–821. [Google Scholar] [CrossRef]
- Koch, A.; Schlemmer, T.; Lischka, R. Elucidating the role of extracellular vesicles in the Barley-Fusarium interaction. Trillium Exctracell. Vesicles 2020, 2, 28–35. [Google Scholar] [CrossRef]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.-S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 305. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Applegate, T. Zearalenone (ZEN) in Livestock and Poultry: Dose, Toxicokinetics, Toxicity and Estrogenicity. Toxins 2020, 12, 377. [Google Scholar] [CrossRef]
- Čulig, B.; Bevardi, M.; Bošnir, J.; Serdar, S.; Lasić, D.; Racz, A.; Galić, A.; Kuharić, Ž. Presence of Citrinin in Grains and Its Possible Health Effects. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Kasahara, K.; Miyamoto, T.; Fujimoto, T.; Oguri, H.; Tokiwano, T.; Oikawa, H.; Ebizuka, Y.; Fujii, I. Solanapyrone Synthase, a Possible Diels-Alderase and Iterative Type I Polyketide Synthase Encoded in a Biosynthetic Gene Cluster from Alternaria solani. ChemBioChem 2010, 11, 1245–1252. [Google Scholar] [CrossRef]
- Klausmeyer, P.; McCloud, T.G.; Tucker, K.D.; Cardellina, J.H.; Shoemaker, R.H. Aspirochlorine Class Compounds from Aspergillus flavus Inhibit Azole-Resistant Candida albicans. J. Nat. Prod. 2005, 68, 1300–1302. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, R.E.; Bhatnagar, D.; Ganley, R.J.; Gillman, C.J.; Monahan, B.J.; Seconi, J.M. Dothistroma pini, a Forest Pathogen, Contains Homologs of Aflatoxin Biosynthetic Pathway Genes. Appl. Environ. Microbiol. 2002, 68, 2885–2892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidy, P.; Baldwin, R.; Greasham, R.; Keith, C.; Mcmullen, J. Zearalenone and Some Derivatives: Production and Biological Activities. In Advances in Applied Microbiology; Perlman, D., Ed.; Academic Press: Cambridge, MA, USA, 1977; Volume 22, pp. 59–82. [Google Scholar]
- Harvey, A.M. Mode of Action of Dothistromin. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 1974. [Google Scholar]
- Doughari, J.H. The Occurrence, Properties and Significance of Citrinin Mycotoxin. J. Plant Pathol. Microbiol. 2015, 6, 2. [Google Scholar]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonah, H.; Deshmukh, R.; Bélanger, R.R. Computational Prediction of Effector Proteins in Fungi: Opportunities and Challenges. Front. Plant Sci. 2016, 7, 126. [Google Scholar] [CrossRef] [Green Version]
- Quarantin, A.; Hadeler, B.; Kröger, C.; Schäfer, W.; Favaron, F.; Sella, L.; Martínez-Rocha, A.L. Different Hydrophobins of Fusarium graminearum Are Involved in Hyphal Growth, Attachment, Water-Air Interface Penetration and Plant Infection. Front. Microbiol. 2019, 10, 751. [Google Scholar] [CrossRef] [Green Version]
- Casarrubia, S.; Daghino, S.; Kohler, A.; Morin, E.; Khouja, H.-R.; Daguerre, Y.; Veneault-Fourrey, C.; Martin, F.M.; Perotto, S.; Martino, E. The Hydrophobin-Like OmSSP1 May Be an Effector in the Ericoid Mycorrhizal Symbiosis. Front. Plant Sci. 2018, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Guzmán-Guzmán, P.; Alemán-Duarte, M.I.; Delaye, L.; Herrera-Estrella, A.; Olmedo-Monfil, V. Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 2017, 18, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, J.M.; Espadas, J.; Luque-Garcia, J.; Reynolds, T.; Casadevall, A. Lipid Biosynthetic Genes Affect Candida albicans Extracellular Vesicle Morphology, Cargo, and Immunostimulatory Properties. Eukaryot. Cell 2015, 14, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, S.-H.; Guo, Y.; Wang, Y.-Z.; Zhang, D.; Xu, L.; Tang, W.-H. A cytoplasmic Cu-Zn superoxide dismutase SOD1 contributes to hyphal growth and virulence of Fusarium graminearum. Fungal Genet. Biol. 2016, 91, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Mondola, P.; Ruggiero, G.; Serù, R.; Damiano, S.; Grimaldi, S.; Garbi, C.; Monda, M.; Greco, D.; Santillo, M. The Cu, Zn superoxide dismutase in neuroblastoma SK-N-BE cells is exported by a microvesicles dependent pathway. Mol. Brain Res. 2003, 110, 45–51. [Google Scholar] [CrossRef]
- Tian, L.; Li, J.; Huang, C.; Zhang, D.; Xu, Y.; Yang, X.; Song, J.; Wang, D.; Qiu, N.; Short, D.P.G.; et al. Cu/Zn superoxide dismutase (VdSOD1) mediates reactive oxygen species detoxification and modulates virulence in Verticillium dahliae. Mol. Plant Pathol. 2021, 22, 1092–1108. [Google Scholar] [CrossRef]
- Cruz-Garcia, D.; Brouwers, N.; Duran, J.M.; Mora, G.; Curwin, A.J.; Malhotra, V. A diacidic motif determines unconventional secretion of wild-type and ALS-linked mutant SOD1. J. Cell Biol. 2017, 216, 2691–2700. [Google Scholar] [CrossRef] [Green Version]
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant Copper-Zinc Superoxide Dismutase (SOD1) Induces Protein Secretion Pathway Alterations and Exosome Release in Astrocytes. J. Biol. Chem. 2013, 288, 15699–15711. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, J.; Chaze, T.; Miranda, K.; Roberson, R.W.; Gorgette, O.; Nimrichter, L.; Matondo, M.; Latgé, J.-P.; Beauvais, A.; Rodrigues, M.L. Characterization of Extracellular Vesicles Produced by Aspergillus fumigatus Protoplasts. mSphere 2020, 5, 00476-20. [Google Scholar] [CrossRef]
- Wolf, J.M.; Espadas-Moreno, J.; Luque-Garcia, J.L.; Casadevall, A. Interaction of Cryptococcus neoformans Extracellular Vesicles with the Cell Wall. Eukaryot. Cell 2014, 13, 1484–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Wang, Z.; Zhang, S.; Peng, Q.; Liu, X. Characterization and proteome analysis of the extracellular vesicles of Phytophthora capsici. J. Proteom. 2021, 238, 104137. [Google Scholar] [CrossRef]
- Romero, D.; Thon, M.; de Vicente, A. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar] [CrossRef]
- Burg, H.A.V.D.; Harrison, S.J.; Joosten, M.H.A.J.; Vervoort, J.; de Wit, P.J.G.M. Cladosporium fulvum Avr4 Protects Fungal Cell Walls Against Hydrolysis by Plant Chitinases Accumulating During Infection. Mol. Plant-Microbe Interact. 2006, 19, 1420–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorin, G.L.; Sanchéz-Vallet, A.; Thomazella, D.P.D.T.; Prado, P.F.V.D.; Nascimento, L.C.D.; Figueira, A.; Thomma, B.P.; Pereira, G.A.G.; Teixeira, P.J.P.L. Suppression of Plant Immunity by Fungal Chitinase-like Effectors. Curr. Biol. 2018, 28, 3023–3030.e5. [Google Scholar] [CrossRef] [Green Version]
- Hasper, A.A.; Dekkers, E.; van Mil, M.; van de Vondervoort, P.J.I.; de Graaff, L.H. EglC, a New Endoglucanase from Aspergillus niger with Major Activity towards Xyloglucan. Appl. Environ. Microbiol. 2002, 68, 1556–1560. [Google Scholar] [CrossRef] [Green Version]
- Rouxel, T.; Grandaubert, J.; James, K.H.; Claire, H.; Angela, P.V.D.W.; Arnaud, C.; Victoria, D.; Véronique, A.; Pascal, B.; Salim, B.; et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2011, 2, 202. [Google Scholar] [CrossRef] [Green Version]
- Bolton, M.D.; van Esse, H.P.; Vossen, J.H.; de Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.E.; Berg, G.C.M.V.D.; Borrás-Hidalgo, O.; Dekker, H.L.; de Koster, C.G.; et al. The novel Cladosporium fulvumlysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Qiu, D.; Zeng, H.; Guo, L.; Yang, X. BcGs1, a glycoprotein from Botrytis cinerea, elicits defence response and improves disease resistance in host plants. Biochem. Biophys. Res. Commun. 2015, 457, 627–634. [Google Scholar] [CrossRef]
- Soyer, J.L.; El Ghalid, M.; Glaser, N.; Ollivier, B.; Linglin, J.; Grandaubert, J.; Balesdent, M.-H.; Connolly, L.R.; Freitag, M.; Rouxel, T.; et al. Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus Leptosphaeria maculans. PLoS Genet. 2014, 10, e1004227. [Google Scholar] [CrossRef]
- Phan, H.T.; Rybak, K.; Furuki, E.; Breen, S.; Solomon, P.; Oliver, R.P.; Tan, K. Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 2016, 87, 343–354. [Google Scholar] [CrossRef]
- Nafisi, M.; Stranne, M.; Zhang, L.; van Kan, J.; Sakuragi, Y. The Endo-Arabinanase BcAra1 Is a Novel Host-Specific Virulence Factor of the Necrotic Fungal Phytopathogen Botrytis cinerea. Mol. Plant-Microbe Interact. 2014, 27, 781–792. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krombach, S.; Reissmann, S.; Kreibich, S.; Bochen, F.; Kahmann, R. Virulence function of the Ustilago maydissterol carrier protein 2. New Phytol. 2018, 220, 553–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Jensen, J.D.; Svensson, B.; Jørgensen, H.J.L.; Collinge, D.B.; Finnie, C. Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat. Mol. Plant Pathol. 2011, 13, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.H.; Solomon, P.S. Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici. Fungal Biol. Biotechnol. 2020, 7, 13. [Google Scholar] [CrossRef] [PubMed]


| Uniprot ID | Protein Name | log2FC | GO Terms |
|---|---|---|---|
| I1S3S6 1 | Putative subtilisin-like serine protease (E-value: 0.0) | 7.17 | C: cell wall |
| I1RJE2 | Polyol transporter 5 | 5.99 | P: transmembrane transport |
| A0A1C3YMP0 1 | Peptide hydrolase | 5.76 | F: aminopeptidase activity |
| I1RQZ5 1 | AB hydrolase-1 domain-containing protein | 5.39 | |
| A0A098DKT1 1 | Carboxylic ester hydrolase | 5.14 | F: hydrolase activity |
| I1RY25 | Niemann–Pick type C-related protein 1 (E-value: 0.0) | 5.07 | C: integral component of membrane |
| I1RUM2 1 | Extracellular protein (E-value: 4.5 × 10−164) | 4.96 | |
| A0A1C3YIM6 1 | Peptidase_M14 domain-containing protein | 4.79 | F: metallocarboxypeptidase activity |
| I1S050 | Casein kinase I isoform gamma 2 | 4.70 | F: protein serine/threonine kinase |
| A0A1C3YJM7 1 | Amine oxidase | 4.67 | P: oxidation-reduction process |
| I1S2H9 | Magnesium and cobalt transporter | 4.49 | C: integral component of membrane |
| I1RP91 | Siderophore iron transporter 1 | 4.31 | P: transmembrane transport |
| A0A1C3YNA9 1 | Putative serine carboxypeptidase | 4.30 | F: serine-type carboxypeptidase |
| A0A098DS79 1 | Gamma-glutamyltransferase (E-value: 0.0) | 4.28 | F: glutathione hydrolase activity |
| V6R949 | K(+)/H(+) antiporter 1 | 4.28 | F: solute:proton antiporter activity |
| A0A098E0Z5 | H(+)/Cl(−) exchange transporter 5 | 4.27 | F: voltage-gated Cl channel activity |
| I1RJ42 1 | Alpha-amylase (E-value: 0.0) | 4.26 | F: alpha-amylase activity |
| I1RDK3 | Flotillin-like protein 1 | 4.20 | |
| I1RMG9 1 | Iron transport multicopper oxidase FET3 precursor | 4.11 | F: oxidoreductase activity |
| I1RF73 1 | Beta-fructofuranosidase (E-value: 0.0) | 4.07 | P: carbohydrate metabolic process |
| Uniprot ID | Protein Name | GO Terms |
|---|---|---|
| Host-pathogen interactions | ||
| A0A1C3YLT0 | Allergen Asp f 9-like | F: hydrolase; P: cell wall organization |
| I1RF56 | Rubrofusarin-specific efflux pump aurT | P: transmembrane transport |
| I1RFS2 | Secreted effector NIS1-like | |
| I1RGY5 | Allergen Asp f 9-like | F: hydrolase; P: cell wall organization |
| I1RIM4 1 | Allergen Asp f 34-like | |
| Transport | ||
| A0A1C3YHZ2 | GTP-binding protein RHO3-like | F: GTPase activity; F: GTP binding |
| A0A1C3YJH3 | Multidrug resistance protein FNX1 | P: transmembrane transport |
| A0A1C3YK53 | VPS74 | F: phosphatidylinositol-4-phosphate binding |
| I1RAS9 1 | VPS10-like | P: protein transport |
| I1RFK0 | GTP-binding protein RHY1 | F: GTPase activity; F: GTP binding |
| I1RG99 | VPS35 | P: endosome to Golgi transport |
| I1RN81 | CDC42Sp-like | F: GTPase activity; F: GTP binding |
| I1RQD6 | SEC17 homolog | P: vesicle-mediated transport |
| I1S278 | Syntaxin PEP12 | P: vesicle-mediated transport |
| I1SAM5 | v-SNARE protein VTI1 | P: vesicle-mediated transport |
| Hydrolysis | ||
| A0A098DV80 1 | Podosporapepsin-like | F: aspartic-type endopeptidase activity |
| A0A0E0RMK7 1 | N-acetyl-beta-glucosaminidase 1-like | P: carbohydrate metabolic process |
| A0A1C3YMS8 1 | Mannanase B | P: carbohydrate metabolic process |
| I1REC8 1 | Probable secreted lipase ARB_02369 | F: hydrolase activity |
| I1RF87 1 | Chitinase 1-like | P: carbohydrate metabolic process |
| I1RHG0 1 | Chitinase 1-like | P: carbohydrate metabolic process |
| I1RHW3 | Ribonuclease Trv | F: RNA binding |
| I1RJF8 1 | Oryzapsin B-like | F: aspartic-type endopeptidase activity |
| I1RLG1 1 | Aspartic proteinase yapsin-6-like | F: aspartic-type endopeptidase activity |
| I1RMU2 1 | Laminarinase eglC-like | P: carbohydrate metabolic process |
| I1RR60 1 | Subtilisin protease 6-like | C: cell wall |
| I1RRY4 1 | Endo-1,3(4)-beta-glucanase-like | P: carbohydrate metabolic process |
| I1RXM5 1 | Lipase 4-like | F: hydrolase activity |
| I1S2W9 1 | Carboxypeptidase MCPB-like | F: metallocarboxypeptidase activity |
| I1S3J9 1 | Secreted lipase ARB07186/07185-like | |
| I1S3S2 1 | Endo-1,5-alpha-L-arabinanase B-like | P: xylan catabolic process |
| V6R5G9 | Exo-1,3-beta-glucanase-like | P: carbohydrate metabolic process |
| V6R5Q6 1 | Man(9)-alpha-mannosidase 1b-like | F: mannosyl hydrolysis; C: membrane |
| Biosynthesis | ||
| A0A098DAH0 | Yanuthone D synthesis protein D | |
| A0A098DV37 | Pestheic acid biosynthesis cluster protein K-like | P: oxidation-reduction process |
| A0A098DVT4 | Sesquiterpene synthase BOT2 | C: membrane; F: lyase activity |
| A0A1C3YLJ5 | Anditomin synthesis protein L-like | C: integral component of membrane |
| A0A1C3YLR9 | Leucinostatins biosynthesis cluster protein R-like | F: phospholipase D activity |
| A0A1C3YMY7 | Aspirochlorine biosynthesis protein Q-like | |
| I1R9G1 1 | Solanapyrone biosynthesis protein 5-like | F: oxidoreductase; F: FAD binding |
| I1RII9 1 | Citrinin synthesis protein MPL7-like | F: oxidoreductase activity |
| I1RS87 | Dothistromin biosynthesis protein epoA-like | F: cis-stilbene-oxide hydrolase activity |
| I1RT88 | Pestheic acid biosynthesis cluster protein L-like | F: oxidoreductase activity |
| I1RUE8 1 | Zearalenone biosynthesis protein 1-like | F: oxidoreductase; F: FAD binding |
| I1RXR7 1 | Terrein biosynthesis cluster protein terF-like | |
| I1S011 | Himeic acid A biosynthesis cluster protein E-like | C: integral component of membrane |
| I1S1K2 | Tropolone synthesis protein G | |
| I1S6B9 1 | Prenyl xanthone synthesis protein C-like | F: oxidoreductase activity |
| Uniprot ID | Protein Name (Gene Symbol) | Length (a.a.) | Enrichment | EffectorP2 | Effector Function |
|---|---|---|---|---|---|
| I1RFS2 | Effector NIS1-like (FGSG_02560) | 140 | Sec up | non-effector | cell death [35] |
| I1S341 | SnodProt1-like (FGSG_11205) | 140 | Not sig | unlikely effector | required for virulence [33,34] |
| I1RPD9 | Extracellular lipase (FGSG_05906) 1 | 349 | EV up | effector | inhibits innate immunity [36] |
| I1RUM2 | Hypothetical protein FGSG_07921 | 221 | Not sig | effector | unknown [32] |
| I1RIV3 | Hypothetical protein FGSG_03748 | 253 | EV up | effector | unknown [32] |
| I1RIE9 | Hypothetical protein FGSG_03581 | 198 | Not sig | effector | unknown [31] |
| I1REI8 | Hypothetical protein FGSG_02077 | 184 | EV up | non-effector | unknown [31] |
| I1RAQ3 | Hypothetical protein FGSG_00588 | 160 | Not sig | unlikely effector | unknown [31] |
| I1RW93 | Hypothetical protein FGSG_08554 | 207 | EV up | non-effector | unknown [31] |
| I1RK25 | AltA1 domain-containing protein FGSG_04213 | 166 | Sec up | effector | unknown [31] |
| I1S0H8 | Hypothetical protein FGSG_10206 | 162 | Not sig | effector | unknown [31] |
| I1S1J8 | Hypothetical protein FGSG_10603 | 158 | Not sig | effector | unknown [31] |
| Effector Candidate | Enrichment | Length (a.a.) | Effector P 2.0 | Cys % | Signal Peptide 1 | Secretome P 2.0 | PredGPI | Apoplast P 1.0 | Location Prediction |
|---|---|---|---|---|---|---|---|---|---|
| Hydrophobin 3 (I1RXJ5, FGSG_09066) | EV exclusive | 82 | effector | 9.8 | yes | NA | unlikely | yes | extracellular/ mitochondria |
| Superoxide dismutase [Cu-Zn] (A0A098DGQ1, FGSG_08721) | cell lysate | 228 | effector | 2.2 | no | 0.706 | - | no | cytoplasm/ nucleus |
| Chitinase (I1RIF9, FGSG_03591) | no difference | 417 | non effector | 0.5 | no | 0.505 | - | yes | cytoplasm |
| LysM domain-containing protein (I1RIC3, FGSG_03554) | no difference | 403 | non effector | 0.2 | no | 0.747 | - | no | cytoplasm/ nucleus |
| Glucoamylase (A0A1C3YK33, FGSG_06278) | no difference | 667 | non effector | 1.2 | no | 0.518 | - | yes | extracellular |
| Glucan endo-1,3-beta-glucosidase eglC-like (I1RMU2, FGSG_05292) | no difference | 409 | non effector | 1.2 | no | 0.703 | - | yes | extracellular/ cell membrane |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Ceron, D.; Lowe, R.G.T.; McKenna, J.A.; Brain, L.M.; Dawson, C.S.; Clark, B.; Berkowitz, O.; Faou, P.; Whelan, J.; Bleackley, M.R.; et al. Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn. J. Fungi 2021, 7, 977. https://doi.org/10.3390/jof7110977
Garcia-Ceron D, Lowe RGT, McKenna JA, Brain LM, Dawson CS, Clark B, Berkowitz O, Faou P, Whelan J, Bleackley MR, et al. Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn. Journal of Fungi. 2021; 7(11):977. https://doi.org/10.3390/jof7110977
Chicago/Turabian StyleGarcia-Ceron, Donovan, Rohan G. T. Lowe, James A. McKenna, Linda M. Brain, Charlotte S. Dawson, Bethany Clark, Oliver Berkowitz, Pierre Faou, James Whelan, Mark R. Bleackley, and et al. 2021. "Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn" Journal of Fungi 7, no. 11: 977. https://doi.org/10.3390/jof7110977
APA StyleGarcia-Ceron, D., Lowe, R. G. T., McKenna, J. A., Brain, L. M., Dawson, C. S., Clark, B., Berkowitz, O., Faou, P., Whelan, J., Bleackley, M. R., & Anderson, M. A. (2021). Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn. Journal of Fungi, 7(11), 977. https://doi.org/10.3390/jof7110977






