Abstract
Effective management and treatment of fungal diseases is hampered by poor diagnosis, limited options for antifungal therapy, and the emergence of antifungal drug resistance. An understanding of molecular mechanisms contributing to resistance is essential to optimize the efficacy of currently available antifungals. In this perspective, one of the oldest antifungals, 5-fluorocytosine (5-FC), has been the focus of recent studies applying advanced genomic and transcriptomic techniques to decipher the order of events at the molecular level that lead to resistance. These studies have highlighted the complexity of resistance and provided new insights that are reviewed in the present paper.
1. Introduction
Significant advances in antifungal therapy have been achieved during the last few decades with the introduction of new generations of antifungal agents, improved fungal diagnostics, and standardization of in vitro susceptibility testing. Nevertheless, management and appropriate treatment of fungal infections remains a challenge that is aggravated by the emergence of antifungal resistance [1,2]. Antifungal drug resistance is associated with increased probability of treatment failure and mortality [3]. Elucidation of the molecular mechanisms of antifungal resistance may help to develop better diagnostic approaches and new therapeutic strategies that can overcome resistance and optimize the treatment efficacity.
5-Fluorocytosine (5-FC) is one of the earliest antifungal agents, initially synthesized in 1957 as an anticancer drug. Unlike 5-fluorouracil (5-FU), a closely related fluorinated pyrimidine, 5-FC does not exhibit antineoplastic activity, but was found to possess antifungal activity. The compound has been used since 1968 to treat human cryptococcosis and candidiasis [4]. 5-FC is active against ascomycete and basidiomycete yeasts, and some hyaline and melanized filamentous fungi [4]. The drug is particularly significant for treatment of fungal infections at body sites with limited drug penetration of other antifungal agents, such as infections of the urinary tract, brain and eyes, or heart valves [5]. Effective therapy of fungal infections of these sanctuary sites is challenging, as other drug classes, such as the lipophilic triazoles, echinocandins, and polyenes, show very limited penetration. The use of 5-FC as a monotherapy, however, is restricted by the rapid emergence of resistance, and is only recommended for some cases of chromoblastomycosis and uncomplicated lower urinary tract infection due to Candida species that are resistant to fluconazole [6]. When administered in combination with amphotericin B, 5-FC has become the gold standard for treating cryptococcal meningitis and complex Candida infections [7,8]. Although resistance to 5-FC is well known, the genetic basis of resistance regulation has remained enigmatic. An understanding of these mechanisms may help to discover novel ways to reduce or reverse resistance and broaden the clinical use of this drug, benefiting from its excellent tissue distribution. In recent years, several studies have revisited this classical drug to elucidate the molecular mechanisms of resistance in different fungi. In the present review, we discuss the mode of action of 5-FC and give an overview of the current resistance mechanisms that have been reported in the literature.
2. Spectrum of Activity
5-FC exhibits a broad spectrum of activity against common pathogenic yeasts. For Candida species, using the standardized National Committee for Clinical Laboratory Standards (NCCLS) antifungal susceptibility testing method, the MIC90 ranges from 0.12 to 1 µg/mL and the susceptibility rate is 95 to 100% for the following species: C. dubliniensis, C. glabrata, C. kefyr, C. lusitaniae, C. glabrata, and C. albicans. However, it is less effective against C. krusei (MIC90 32 µg/mL and 5% sensitive/susceptible) [9]. 5-FC demonstrates in vitro activity against Cryptococcus species; the MIC values, determined according to the recommendations proposed by the European Committee for Antibiotic Susceptibility Testing of fermentative yeasts (AFST-EUCAST, definitive document 7.1), are higher for C. neoformans isolates than for C. gattii (MIC50 of 4 and 1 µg/mL, respectively) [10]. 5-FC is also active in vitro against some uncommon pathogenic fungi, including species of Cyberlindnera, Debaryomyces, Diutina, Rhodotorula, and Saccharomyces, while other yeasts exhibit systematically elevated MICs, for example Cutaneotrichosporon mucoides, Dipodascus geotrichum, Trichosporon asahii, Yarrowia lipolytica, and species of Naganishia and Pichia [11]. Aspergillus and some agents of chromoblastomycosis in vitro are less susceptible to 5-FC [4].
3. Indication
5-FC is clinically licensed for the treatment of cryptococcosis, chromoblastomycosis, and aspergillosis. 5-FC is a first-line therapy for the treatment of cryptococcal meningitis. It is administered in combination with amphotericin B as induction therapy [7]. Adding 5-FC to amphotericin B resulted in faster sterilization of the cerebrospinal fluid (CSF) [12].
This latter effect is also obtained with the combination of 5-FC with fluconazole as compared with fluconazole monotherapy. Furthermore, the association of 5-FC with fluconazole has shown a noninferiority with two weeks of amphotericin B-based therapy [13]. Thus, is also recommended in areas where amphotericin B is not available [14]. It has the advantages of being well tolerated, administered orally, and prevented treatment failure as it suppresses the amplification of hetero-resistance to fluconazole [15].
5-FC is also used as adjunctive with amphotericin B or fluconazole in treating complex systemic fungal infections, including septicemia, endocarditis, meningitis, or pulmonary infections caused by susceptible strains of Candida or Cryptococcus [6,8]. 5-FC monotherapy is limited only for the treatment of Candida cystitis in cases of fluconazole resistance given the high urinary concentrations of 5-FC and treatment of chromoblastomycosis caused by various melanized fungi [4,6].
4. Mode of Action
The mode of action of 5-FC is unique among antifungal agents, as it targets DNA, RNA, and protein synthesis. 5-FC is a prodrug, i.e., for its activation the compound needs to be metabolized via the pyrimidine salvage pathway, where it acts as a subversive substrate with the subsequent production of toxic nucleotides and disruption of DNA and protein synthesis (Figure 1). After being actively transported into the fungal cell by membrane permeases (cytosine permease, encoded by the FCY2 gene and other homologs encoded by FCY21, FCY22), 5-FC is converted via 5-FU to 5-fluoro-uridylate (synonymous with 5-fluoro-UMP (5-FUMP)) under the action of the enzymes cytosine deaminase, encoded by FCY1, and uracil phosphoribosyltransferase (UPRT), which is encoded by the FUR1 gene, respectively. The 5-FUMP is in turn phosphorylated by two specific kinases to 5-fluoro-UTP, which is incorporated into the RNA. 5-FUMP is also reduced to 5-fluoro-2′-deoxyuridylate, which inhibits the enzyme thymidylate synthetase and thus DNA synthesis by decreasing the available nucleotide pool. Mammalian cells lack the enzyme cytosine deaminase. Therefore, 5-FC is not converted to 5-FU and consequently these cells are not directly subject to the toxic effects of 5-FC [3].
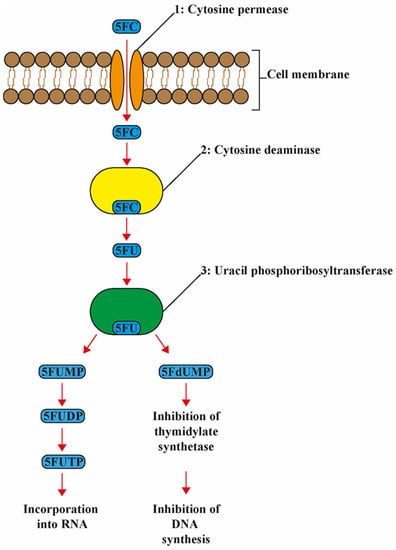
Figure 1.
Schematic and simplified presentation of the mode of action of 5-flucytosine. 5-Flucytosine (5-FC) is transported into the cell by the enzyme 1: cytosine permease. Here 5-FC is deaminated to 5-fluoraucil (5-FU) by the enzyme 2: cytosine deaminase. Then, 5FU is converted to 5-fluorouridine monophosphate (FUMP) by the enzyme 3: uridine monophosphate pyrophosphorylase, then into 5-fluorouridine diphosphate (FUDP) and eventually into 5-fluorouridine triphosphate (FUTP). FUTP is incorporated into RNA altering the aminoacylation of tRNA and inhibiting protein synthesis. Furthermore, 5FU is converted to 5-fluorodeoxyuridine monophosphate (FdUMP) by the enzyme 3: uridine monophosphate pyrophosphorylase. FdUMP inhibits DNA biosynthesis via inhibition of thymidylate and incorporation into DNA.
5. Pharmacokinetics
5-FC is available as an oral capsule and as an intravenous formulation [16], but is only available in a limited number of countries [17]. 5-FC is generally administrated at 100 mg/kg/day in 2–4 divided dosages. The oral bioavailability is approximately 75–90% in healthy patients [18,19]. A lower oral bioavailability of approximately 45% was found in late-stage HIV-infected patients from Thailand [16]. It is eliminated by the kidneys and either dose or frequency or both should be reduced in patients with impaired renal function [20,21,22]. 5-FC distributes well into various body sites and compartments. In a cohort of patients treated for cryptococcal meningitis, CSF 5-FC concentrations were 84% of the corresponding plasma concentrations [16].
6. Toxicity
5-FC is associated with gastrointestinal side effects (in 6% of patients treated with 5-FC) such as nausea, diarrhea, vomiting, and diffuse abdominal pain, but also more severe side effects such as hepatotoxicity and bone-marrow depression [23]. 5-FC exposure is limited by toxicity and therapeutic drug monitoring is needed to avoid toxic concentrations. 5-FC serum concentrations of 100 mg/L and above are associated with increased incidence of bone marrow depression and hepatotoxicity [23,24,25,26]. Furthermore, higher concentrations of 5-FU are detectable in plasma from patients that received oral 5-FC in comparison to intravenous 5-FC, possibly due to conversion of 5-FC to 5-FU by the intestinal microflora [16]. However, no differences in toxicity or treatment efficacy were observed [16]. Flucytosine is contraindicated in pregnancy [27].
7. 5-FC Resistance
The clinical use of 5-FC is limited by the common occurrence of primary (in subpopulations of C. albicans and C. dubliniensis) and acquired resistance, particularly when used as a monotherapy. Most of the studies on resistance mechanisms have focused on S. cerevisiae and C. albicans, and the detection of mutations in important enzymes of the pyrimidine salvage pathways. Results of these studies were reviewed previously [5,28,29]. It was shown that 5-FC resistance may result from loss or mutation of any of the enzymes involved in the activation of cytosine permease, cytosine deaminase, or uracil phosphoribosyltransferase or from the increased production of pyrimidines (Table 1). However, the absence of mutations in these enzymes in many clinically 5-FC-resistant isolates led to hypotheses of the existence of other mechanisms that might regulate the response of fungal cells, other than the genes involved in the pyrimidine salvage pathways.

Table 1.
Mechanisms of resistance to 5-FC.
To date, no studies have evaluated the mutant prevention concentration of 5-FC. Furthermore, it is not known whether resistance developed more often in sanctuary sites, where drug levels may be lower due to reduced 5-FC penetration. While resistance to 5-FC is a single-step process, 5-FC resistance may develop rapidly regardless of the dose or the site of infection when used as a single agent.
7.1. Saccharomyces cerevisiae
Mutations in three key enzymes of the pyrimidine salvage pathways, i.e., FCY1, FCY2, and FUR1, have been shown to cause 5-FC resistance [45,46]. While disruption of cytosine deaminase is associated with a strong dose-independent resistance, disruption of cytosine permease leads to a low but dose-dependent level of toxicity indicating involvement of two other cytosine permeases, FCY21 and FCY22. In addition, two other FCY2 homologues, TPN1 and FUR4, and one encoded by yOR071c, were found to contribute significantly to 5-FC toxicity, thus revealing alternative 5-FC entry routes via other cytosine adenine permease homologues [47]. Kern et al. [30] correlated phenotypic 5-FC resistance to a single point mutation, Arg134Ser in the FUR1 gene.
Additional resistance mechanisms have been reported that include upregulation of pyrimidines in de novo synthesis and a loss of function or disturbance in uridine monophosphate pyrophosphorylase [7]. Zhang et al. [48] applied genome-wide yeast DNA microarray and identified 96 responsive genes, of which 57 showed over two-fold upregulation and 40 were more than two-fold downregulated. Most of the upregulated genes, involved in DNA repair, synthesis, and replication, represented the highest proportion of induced genes that corresponded with the mechanism of action of 5-FC. Among the upregulated genes were UBC5 and UBC14, both involved in the ubiquitin-dependent pathway, which is known to be related to DNA repair and selective removal of damaged or obsolete proteins. UBC5 and UBC14 are thought to be responsive to the faulty protein synthesis caused by 5-FC. The observed cellular resistance to 5-FC might also be enhanced by increased DNA repair in the cell. Among the downregulated genes, the pleiotropic drug resistance (PDR) genes may play a significant role. PDR15, which encodes a multidrug transporter protein, was downregulated by a factor of 2.3, indicating that 5-FC resistance may not be ascribed to extrusion by drug pumps in the cell membrane. Two enzyme-encoding genes, CTS1 and EGT2, which function in the release of daughter cells from their mother cells, were downregulated by a factor of 3.7 and 10.2, respectively, indicating that 5-FC may also inhibit the separation of fungal cells. Other genes with significant repression were five genes related to protein synthesis; eight ADE genes that encode the enzymes of the purine nucleotide biosynthetic pathway; INO1, related to phospholipid synthesis; three genes, PHO3, PHO5, and PHO11, that contribute to increased phosphate levels; and the genes ARE2 and FAS2, related to lipid metabolism.
7.2. Candida
Primary 5-FC resistance occurs in <5% of all Candida species, except for C. krusei, where resistance is detected in up to 35% of isolates [9,49]. Many factors have been shown to influence the development of the resistance in Candida, which varies with species, clade, or genotype, and also with ploidy and the presence of a phenotype mutator.
7.2.1. Candida albicans
5-FC resistance in C. albicans has been reported to vary from 0 to 3% [50]. A relationship between 5-FC resistance and a decreased susceptibility to azoles was reported by Cuena-Estrella et al. [49]. Dodgson et al. [31] showed that both decreased susceptibility and increased resistance mostly correlate with a single change from cytosine to thymine at position 301 of the FUR1 gene. This change results in an amino acid substitution from arginine to cysteine at position 101 in the FUR1 protein. Strains homozygous (C/C) at position 301 were susceptible to 5-FC, whereas heterozygous counterparts (C/T) were less susceptible, and (T/T) homozygotes were fully 5-FC resistant. This mutation was restricted to Clade I strains. Hope et al. [32] identified the same mutation and found that the substitution C101R disturbs the quaternary structure of the enzyme, which was postulated to compromise its optimal activity. Additionally, another homozygous polymorphism in FCA1 was described in this study that resulted in a glycine to aspartate substitution at position 28 in the cytosine deaminase.
Resistance was more frequent in C. albicans serotype B than in serotype A [51,52,53]. However, since the two serotypes have been shown to be interconvertible within a single strain [54], the association with 5-FC resistance that was found did not necessarily have a genomic background. Using rRNA biotyping, Mercure et al. [55] demonstrated that DNA type 1A (3.7 kbp) was mostly resistant to 5-FC, while DNA type IB comprised almost exclusively susceptible strains. Using DNA fingerprinting with the complex probe Ca3, Pujol et al. [56] demonstrated that all naturally resistant isolates of C. albicans were members of group 1, and 72% of the isolates of this group had 5-FC MICs >0.5 µg/mL, compared to 2% of non-group 1 isolates.
7.2.2. Candida dubliniensis
C. dubliniensis was first described in 1995 as a sibling of the predominant human pathogen C. albicans. Three well-defined intraspecific clades can be distinguished, the species being significantly less diverse than C. albicans [57]. Resistance to 5-FC in C. dubliniensis is exclusively found in isolates of Clade 3 and was correlated to a homozygous S29L mutation in the cytosine deaminase gene [33,34,35]. McManus et al. [35] proved, using genetic transformation methods, that this mutation is responsible for the resistance to this drug. This radical substitution results in the replacement of a hydrophilic polar amino acid (serine) with a hydrophobic nonpolar residue (leucine) in the β1 strand of the cytosine deaminase enzyme and is closely linked to an active-site residue. The substitution may lead to disruption of the quaternary structure of the enzyme, distorting the active site and inhibiting the conversion of the 5-FC prodrug to its toxic form, 5-FU [35].
7.2.3. Candida lusitaniae
C. lusitaniae has a remarkable ability to develop antifungal resistance, mainly to amphotericin B but also to azoles, echinocandins, and 5-FC. Many studies have aimed to decipher the molecular mechanisms of 5-FC resistance and its cross resistance with fluconazole. It was shown that inactivation of the genes FCY2, FCY1, and FUR1 in C. lusitaniae produced two patterns of 5-FC resistance. The mutant FUR1 also demonstrated resistance to 5-FU, whereas mutants FCY1 and FCY2 demonstrated fluconazole resistance in the presence of subinhibitory 5-FC concentrations, and extracellular 5-FC would behave as a competitive inhibitor of fluconazole uptake [58,59]. Florent et al. [36] demonstrated, by using yeast transformations, that two genetic events are responsible for both 5-FC and 5-FC/fluconazole resistance in clinical isolates of C. lusitaniae: either the nonsense mutation C505T in FCY2 and its putative nonfunctional truncated purine cytosine permease of 168 amino acids unable to transport 5-FC, or the missense mutation T26C in the FCY1 gene, result in an amino acid replacement M9T in cytosine deaminase.
Kannan et al. [37] used comparative genomics to elucidate the mechanism of multidrug resistance in C. lusitaniae and described a new mechanism of 5-FC resistance with a mutation in a putative transcriptional activator (MRR1). The V668G substitution was responsible for the upregulation of the MFS7, a multidrug transporter that mediated the resistance to 5-FC and 5-FC/fluconazole cross-resistance.
7.2.4. Candida glabrata
C. glabrata has emerged in recent years as the second most common agent of mucosal and invasive candidiasis [60]. The yeast is characterized by a high frequency of acquired resistance to multiple classes of antifungals drugs. This may be related to the particular genetic background of this species, as no sexual reproduction is known. In contrast to C. albicans, C. glabrata possesses a haploid genome and therefore a single mutation in an appropriate gene may result in acquired antifungal resistance phenotype. Recently, genetic defects in the mismatch repair (MMR), the DNA repair pathway due to the presence of MSH2 mutations, were described to enhance the development of resistance [61]. Contrary to expectations, resistance of C. glabrata to 5-FC is reported to be 1%, with resistant mutants occurring at a relatively low frequency of 2 × 10−7 [9]. Normak and Schonebek [62] analyzed pyrimidine incorporation and UPRT activity of five C. glabrata clinical isolates and demonstrated a decreased UPRT activity in one of them. Fasoli et al. [63] investigated the metabolism of fluoropyrimidines in a selected mutant of C. glabrata by nuclear magnetic resonance, which revealed a partial loss of cytosine permease activity. Chapeland-Leclerc et al. [38] provided indirect evidence without formal proof that a defective FUR1 protein bearing a G190D substitution was responsible for the fluoropyrimidine resistant phenotype, suggesting that the mechanism of resistance to 5-FC is due to mutations that result in enzyme deficiency. Multiple mutations were also described in in vitro 5-FC resistant mutants of C. glabrata in the main enzymes of the pyrimidine salvage pathway. Mutations in FUR1, i.e., I83K and D193G, were responsible for the replacement of charged with uncharged residues. The ΔG73-V81 mutation involving a 9-amino-acid deletion was found in isolates with 5-FC/5-FU cross-resistance, while strains with mutations in FCY1 (A15D, G11D, and W148R), and FCY2 (G246S and I384F) were not cross-resistant to 5-FU [39,40]. Edlinde et al. [39] confirmed, by isolating the mutants in URA3 strains and then testing their ability to assimilate the cytosine, that the mutants are null. They also found some of these mutations (FUR1 G210D and L136R, FCY1 T84L, and FCY2J I384F) in clinical isolates. 5-FC-azole antagonism was described in C. glabrata as a result of upregulation of the Pdr1-dependent CDR1 induced by 5-FC [64].
7.2.5. Candida tropicalis
In a study by Cuenca-Estrella et al. [49], 5-FC showed potent in vitro activity against all 117 tested isolates of C. tropicalis. However, Desnos-Olivier et al. [41] found 5-FC resistance in 45 of 130 (35%) isolates and suggested that resistance to 5-FC in C. tropicalis could be clonal and represent a subgroup of C. tropicalis. The 5-FC resistant isolates did not carry any mutation in the coding sequences of FCY1, FCY2, or FUR1, but a K177E mutation was suggested without formal proof in the URA3 gene. The URA3 enzyme (orotidine 5′-phosphate decarboxylase, ODCase) is involved in the metabolic pathway of uridyl-monophosphate (UMP), which is a substrate of thymidylate synthetase and UMP kinase, both involved in nucleic acid synthesis. The ODCase is not known to interfere directly with 5-FC activity. However, one of the resistance mechanisms against 5-FC concerns increased transcription of all genes involved in the de novo pyrimidine biosynthetic pathway (including URA3) leading to an overabundance of UMP [41].
Chen et al. [42] found that 22 out of 30 5-FC susceptible isolates with resistant progeny after exposure to the drug had a heterozygous G/T at the 145th position of FCY2 encoding purine–cytosine permease, whereas their progeny recovered from within the inhibitory ellipses had homozygous T/T. They proved, using transformation approaches, that this mutation resulted in null alleles for both copies of the gene and produced only truncated proteins, leading to 5-FC resistance. The authors concluded that C. tropicalis strains containing heterozygous alleles may develop 5-FC resistance readily, whereas patients carrying strains with homozygous G/G wild-type alleles can be treated with 5-FC.
7.2.6. Candida auris
Since its first description in 2009, C. auris has emerged as a multidrug resistant pathogen. It is associated with high mortality and therapeutic failures because of its ability of colonization and its resistance to a broad spectrum of antifungals, especially to fluconazole and amphotericin B [65].
5FC is active against C. auris; the rate of resistance varies from 0 to 47% [66]. The molecular mechanisms of resistance to 5FC in this pathogen are still unknown. Rhodes et al. [43] reported in one 5-FC resistant isolate a mutation in the FUR1 gene that causes an amino acid substitution from isoleucine to phenylalanine at position 211. Bhattacharya et al. [67] demonstrated that transient gene duplication can occur in C. auris during replicative aging, responsible for tolerance to 5-FC and other antifungal drugs in older C. auris cells (10 generations) compared to younger (0–3 generations cells).
7.3. Cryptococcus
The molecular mechanism of 5-FC resistance of C. neoformans is not well understood but has been suggested to be related to mutation in pyrimidine salvage enzymes, similar to C. albicans [28], despite large phylogenetic distances between asco- and basidiomycetous yeasts. First studies indicated that 5-FC resistance was not drug-induced, was stable, and was nearly always accompanied by 5-FU resistance. 5-FC mutation rates ranged from 1.2 to 4.8 × 10−7 per cell division [68]. Whelan et al. [28] ascribed this resistance to a reduction in FUR1 activity, based on a high frequency of cross-resistance to 5-FU. In C. gattii, deletions of FCY2 conferred resistance to 5-FC [69]. However, recent studies of 5-FC resistant C. gattii isolates presented no mutations in FCY1, FUR1, or any of three putative FCY2 paralogs that should explain drug resistance [70]. Using comparative transcriptomics, Song et al. [71] identified 177 responsive genes to 5-FC in C. neoformans. Among these, an APES-like transcription factor Mbs1 was present, which has pleiotropic functions in C. neoformans, including regulation of susceptibility to diverse antifungal drugs such as 5-FC, azoles, and polyene drugs. The gene further played a role in ergosterol biosynthesis, cell membrane stability, genotoxic, oxidative, and salt stress responses, in vitro and in vivo morphological differentiation, and virulence. Jung et al. [72] showed that 27 transcription factors (TFs) differentially regulate 5-FC resistance and described some TF mutants with increased resistance (HLH3, RIM101, GAT204, HOB3, FZC50, ZNF2, RDS2, FZC31) as well as TF mutants with increased susceptibility (NRG1, ZFC2, YAP1, MBS1, FZC6, YAP2, BZP3, JJJ1, HLH1, PIP2, APN2, FZC46, HAP2, FZC51, BZP5, HCM1, FZC19, BZP2, FZC44).
Billmyre et al. [44] showed that resistance to 5-FC in C. deuterogattii is acquired more frequently, greater than 15-fold in msh2Δ mutants with defects in DNA mismatch repair, which consequently confers an elevated mutation rate. All 29 resistant isolates used in this study were 5-FC/5-FU resistant. It was expected as previously described that these isolates harbor mutations in FUR1. However, this was confirmed in only 12 isolates; the FUR1 mutations occurred through multiple mechanisms, including regional deletions, homopolymer tract length changes that introduced frameshift mutations, and a splice site acceptor point mutation. Furthermore, the authors described a new mechanism of resistance, i.e., a mutation in the UDP-glucuronate decarboxylase 1 (UXS1) gene may confer resistance to both 5-FC and 5-FU. The UXS1 gene encodes the enzyme that converts UDP-glucuronic acid to UDP-xylose. This pathway is critical for the formation of the capsule, a core virulence trait of Cryptococcus, and for synthesis of other glycoconjugates. In this study, four loss of function mutations in UXS1 were identified: a single base deletion in a 3xT homopolymer, a single base insertion in a 7xC homopolymer, and two missense mutations, Y217C and D306G. The latter mutation is identified in the only isolate with FCY2 mutation (W167Stop). Mutations in UXS1 lead to the accumulation of UDP-glucuronic acid and alterations in the nucleotide metabolism, which appear to suppress the toxicity of both 5-FC and its toxic derivative 5-FU. The role of the mutation in UXS1 and some regional deletion including FUR1, which are associated with loss of virulence, is still unclear in the resistance to 5-FC in patients. Additionally, the authors found no mutation in FCY1, FCY2, FUR1, or UXS1, in eight remaining isolates that could explain the resistance to 5-FC; the existence of other unknown mechanisms may be presumed [44].
7.4. Aspergillus
5-FC is rarely used in the treatment of Aspergillus diseases, due to an apparent lack of activity of 5-FC in vitro. The compound is not recommended for therapy of any form of aspergillosis by the Infectious Diseases Society of America [73]. However, it would be more accurate to state that 5-FC has limited activity against most Aspergillus isolates in vitro, with MICs ranging from 0.25 to >256 µg/mL using NCCLS [74]. This contradicts evidence showing that 5-FC treatment is able to significantly improve survival in a murine model of infection [75,76]. One explanation for the lack of in vitro/in vivo concordance is the variable activity of 5-FC at pH 5 versus pH 7. Lower pH has been shown to increase 5-FC activity against A. fumigatus up to 4000-fold [77]. MICs determined at low pH better reflect results of in vitro models of infection for A. fumigatus. At neutral pH, two transcriptional regulators, PacC and the CCAAT binding complex (CBC), orchestrate 5-FC resistance via negative regulation of the gene encoding, the purine cytosine transporter, and FCY2 orthologue FCYB [77]. The authors also demonstrated that FCYB is critical for 5-FC activity, and the reduced expression of FCYB at pH 7 is the major mechanism conferring intrinsic 5-FC resistance in A. fumigatus [77]. However, Birštonas et al. [78] found that the ΔfcyB mutant strain was not able to grow at pH 5 and showed severe growth inhibition at pH 7, which indicated the existence of other uptake mechanisms. In contrast to the ΔfcyB mutant, the ΔfcyA and Δuprt mutants displayed full resistance to 5-FC up to 100 μg/mL, regardless of the pH. Furthermore, 100 μg/mL of 5-FU blocked growth of wt, ΔfcyB, and ΔfcyA strains at pH 5 as well as pH 7, while the Δuprt mutant was resistant. Furthermore, the inactivation of these genes caused no adverse effects with respect to nutrient requirements, stress resistance, or virulence in A. fumigatus, which make these genes a potential novel genetic toolbox for targeted genomic insertions of DNAs of interest (DOIs) in the genetic engineering of fungal species.
7.5. Dermatophytes
5-FC has no activity against dermatophytes; high MIC values were previously described [79,80]. Zhao et al. [81] performed cDNA microarray analysis to determine the response of Trichophyton rubrum to 5-FC and found that 474 genes were differentially expressed, of which 196 showed increased and 278 decreased expressions. Marked downregulation of genes involved in nucleotide metabolism (such as CDC21), transcription (such as E2F1), and RNA processing (such as SGN1, RIM4, and NOP1) was observed. Other genes which could be also affected by 5-FC have functions in signal transduction, chaperones, inorganic ion transport, secondary metabolite biosynthesis, amino acid transport, and lipid transport.
8. Conclusions
Although 5-FC resistance has been a well-known issue for a long time, the literature review indicates that our current knowledge regarding molecular mechanisms controlling 5-FC resistance is still limited. Resistance cannot be explained only by the alteration of the main target of this drug; it is more complex and implicates the involvement of different pathways depending on the fungal species. More work is needed, especially now that advanced molecular techniques have become available, allowing more in-depth exploration of the global response of fungi to antifungals. The use of these molecular techniques should be more oriented towards the analysis of resistant isolates that emerge in vivo in treated patients and can be compared with drug susceptible parent strains. This will allow all possible targets responsible for the development of 5-FC resistance to be identified. Furthermore, the availability of gene editing technology will provide important proof of the contribution of these mutations in 5-FC resistance. These studies will further increase our knowledge of the genetic basis of resistance to 5-FC and help to identify new potential diagnostic and treatment targets that could prevent the emergence of the resistance and may lead to a more appropriate use of this antifungal agent.
Author Contributions
Conceptualization, J.B.B. and F.Z.D.; methodology, F.Z.D.; resources, F.Z.D. and J.B.B.; data curation, F.Z.D.; writing—original draft preparation, F.Z.D.; writing—review and editing, F.Z.D., J.B.B., A.M.S.A.-H., R.J.M.B., W.J.G.M., S.d.H. and P.E.V.; visualization, J.B.B.; supervision, P.E.V. and J.B.B.; project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Scorzoni, L.; de Paula, E.S.A.C.; Marcos, C.M.; Assato, P.A.; de Melo, W.C.; de Oliveira, H.C.; Costa-Orlandi, C.B.; Mendes-Giannini, M.J.; Fusco-Almeida, A.M. Antifungal Therapy: New Advances in the Understanding and Treatment of Mycosis. Front. Microbiol. 2017, 8, 36. [Google Scholar] [CrossRef]
- Kanafani, Z.A.; Perfect, J.R. Antimicrobial resistance: Resistance to antifungal agents: Mechanisms and clinical impact. Clin. Infect. Dis. 2008, 46, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Lestrade, P.P.; Bentvelsen, R.G.; Schauwvlieghe, A.; Schalekamp, S.; van der Velden, W.; Kuiper, E.J.; van Paassen, J.; van der Hoven, B.; van der Lee, H.A.; Melchers, W.J.G.; et al. Voriconazole Resistance and Mortality in Invasive Aspergillosis: A Multicenter Retrospective Cohort Study. Clin. Infect. Dis. 2019, 68, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Vermes, A.; Guchelaar, H.J.; Dankert, J. Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 2000, 46, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Ghannoum, M.A. Flucytosine Treatment and Resistance Mechanisms. In Antimicrobial Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 407–413. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.H.; et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin. Infect. Dis. 2010, 50, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Mourad, A.; Perfect, J.R. Present and Future Therapy of Cryptococcus Infections. J. Fungi 2018, 4, 79. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Boyken, L.; Huynh, H.; Hollis, R.J.; Diekema, D.J. In vitro activities of 5-fluorocytosine against 8803 clinical isolates of Candida spp.: Global assessment of primary resistance using National Committee for Clinical Laboratory Standards susceptibility testing methods. Antimicrob. Agents Chemother. 2002, 46, 3518–3521. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lopez, A.; Zaragoza, O.; Dos Anjos Martins, M.; Melhem, M.C.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. In vitro susceptibility of Cryptococcus gattii clinical isolates. Clin. Microbiol. Infect. 2008, 14, 727–730. [Google Scholar] [CrossRef]
- Borman, A.M.; Muller, J.; Walsh-Quantick, J.; Szekely, A.; Patterson, Z.; Palmer, M.D.; Fraser, M.; Johnson, E.M. MIC distributions for amphotericin B, fluconazole, itraconazole, voriconazole, flucytosine and anidulafungin and 35 uncommon pathogenic yeast species from the UK determined using the CLSI broth microdilution method. J. Antimicrob. Chemother. 2020, 75, 1194–1205. [Google Scholar] [CrossRef]
- Johnson, M.D.; Perfect, J.R. Use of Antifungal Combination Therapy: Agents, Order, and Timing. Curr. Fungal Infect. Rep. 2010, 4, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Molloy, S.F.; Kanyama, C.; Heyderman, R.S.; Loyse, A.; Kouanfack, C.; Chanda, D.; Mfinanga, S.; Temfack, E.; Lakhi, S.; Lesikari, S. Antifungal combinations for treatment of cryptococcal meningitis in Africa. N. Engl. J. Med. 2018, 378, 1004–1017. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for the Diagnosis, Prevention, and Management of Cryptococcal Disease in HIV-Infected Adults, Adolescents and Children, March 2018: Supplement to the 2016 Consolidated Guidelines of the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Stone, N.R.; Rhodes, J.; Fisher, M.C.; Mfinanga, S.; Kivuyo, S.; Rugemalila, J.; Segal, E.S.; Needleman, L.; Molloy, S.F.; Kwon-Chung, J. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Investig. 2019, 129, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, A.E.; van Kan, H.J.; Johnson, E.; Rajanuwong, A.; Teparrukkul, P.; Wuthiekanun, V.; Chierakul, W.; Day, N.; Harrison, T.S. Oral versus intravenous flucytosine in patients with human immunodeficiency virus-associated cryptococcal meningitis. Antimicrob. Agents Chemother. 2007, 51, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Kneale, M.; Bartholomew, J.S.; Davies, E.; Denning, D.W. Global access to antifungal therapy and its variable cost. J. Antimicrob. Chemother. 2016, 71, 3599–3606. [Google Scholar] [CrossRef] [PubMed]
- Cutler, R.E.; Blair, A.D.; Kelly, M.R. Flucytosine kinetics in subjects with normal and impaired renal function. Clin. Pharmacol. Ther. 1978, 24, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Block, E.R.; Bennett, J.E. Pharmacological studies with 5-fluorocytosine. Antimicrob. Agents Chemother. 1972, 1, 476–482. [Google Scholar] [CrossRef]
- Daneshmend, T.K.; Warnock, D.W. Clinical pharmacokinetics of systemic antifungal drugs. Clin. Pharmacokinet. 1983, 8, 17–42. [Google Scholar] [CrossRef]
- Schonebeck, J.; Polak, A.; Fernex, M.; Scholer, H.J. Pharmacokinetic studies on the oral antimycotic agent 5-fluorocytosine in individuals with normal and impaired kidney function. Chemotherapy 1973, 18, 321–336. [Google Scholar] [CrossRef]
- Block, E.R.; Bennett, J.E.; Livoti, L.G.; Klein, W.J., Jr.; MacGregor, R.R.; Henderson, L. Flucytosine and amphotericin B: Hemodialysis effects on the plasma concentration and clearance. Studies in man. Ann. Intern. Med. 1974, 80, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Vermes, A.; van Der Sijs, H.; Guchelaar, H.J. Flucytosine: Correlation between toxicity and pharmacokinetic parameters. Chemotherapy 2000, 46, 86–94. [Google Scholar] [CrossRef]
- Stamm, A.M.; Diasio, R.B.; Dismukes, W.E.; Shadomy, S.; Cloud, G.A.; Bowles, C.A.; Karam, G.H.; Espinel-Ingroff, A. Toxicity of amphotericin B plus flucytosine in 194 patients with cryptococcal meningitis. Am. J. Med. 1987, 83, 236–242. [Google Scholar] [CrossRef]
- Bennett, J.E.; Dismukes, W.E.; Duma, R.J.; Medoff, G.; Sande, M.A.; Gallis, H.; Leonard, J.; Fields, B.T.; Bradshaw, M.; Haywood, H.; et al. A comparison of amphotericin B alone and combined with flucytosine in the treatment of cryptoccal meningitis. N. Engl. J. Med. 1979, 301, 126–131. [Google Scholar] [CrossRef]
- Kauffman, C.A.; Frame, P.T. Bone marrow toxicity associated with 5-fluorocytosine therapy. Antimicrob. Agents Chemother. 1977, 11, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Andes, D.R. Antifungal agents: Spectrum of activity, pharmacology, and clinical indications. Infect. Dis. Clin. 2016, 30, 51–83. [Google Scholar] [CrossRef]
- Whelan, W.L. The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit. Rev. Microbiol. 1987, 15, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of Antifungal Drug Resistance. Cold Spring Harb. Perspect. Med. 2014, 5, a019752. [Google Scholar] [CrossRef] [PubMed]
- Kern, L.; de Montigny, J.; Lacroute, F.; Jund, R. Regulation of the pyrimidine salvage pathway by the FUR1 gene product of Saccharomyces cerevisiae. Curr. Genet. 1991, 19, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Dodgson, A.R.; Dodgson, K.J.; Pujol, C.; Pfaller, M.A.; Soll, D.R. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 2223–2227. [Google Scholar] [CrossRef]
- Hope, W.W.; Tabernero, L.; Denning, D.W.; Anderson, M.J. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 4377–4386. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Khan, Z.U.; Joseph, L.; Asadzadeh, M.; Theyyathel, A. Genotypic heterogeneity and molecular basis of 5-flucytosine resistance among Candida dubliniensis isolates recovered from clinical specimens in Kuwait. Med. Mycol. 2012, 50, 244–251. [Google Scholar] [CrossRef]
- Al Mosaid, A.; Sullivan, D.J.; Polacheck, I.; Shaheen, F.A.; Soliman, O.; Al Hedaithy, S.; Al Thawad, S.; Kabadaya, M.; Coleman, D.C. Novel 5-flucytosine-resistant clade of Candida dubliniensis from Saudi Arabia and Egypt identified by Cd25 fingerprinting. J. Clin. Microbiol. 2005, 43, 4026–4036. [Google Scholar] [CrossRef][Green Version]
- McManus, B.A.; Moran, G.P.; Higgins, J.A.; Sullivan, D.J.; Coleman, D.C. A Ser29Leu substitution in the cytosine deaminase Fca1p is responsible for clade-specific flucytosine resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 2009, 53, 4678–4685. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Florent, M.; Noel, T.; Ruprich-Robert, G.; Da Silva, B.; Fitton-Ouhabi, V.; Chastin, C.; Papon, N.; Chapeland-Leclerc, F. Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob. Agents Chemother. 2009, 53, 2982–2990. [Google Scholar] [CrossRef] [PubMed]
- Kannan, A.; Asner, S.A.; Trachsel, E.; Kelly, S.; Parker, J.; Sanglard, D. Comparative Genomics for the Elucidation of Multidrug Resistance in Candida lusitaniae. mBio 2019, 10, e02512-19. [Google Scholar] [CrossRef]
- Chapeland-Leclerc, F.; Hennequin, C.; Papon, N.; Noel, T.; Girard, A.; Socie, G.; Ribaud, P.; Lacroix, C. Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob. Agents Chemother. 2010, 54, 1360–1362. [Google Scholar] [CrossRef]
- Edlind, T.D.; Katiyar, S.K. Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob. Agents Chemother. 2010, 54, 4733–4738. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, P.; Pineau, L.; Larcher, G.; Noel, T.; Brethes, D.; Chabasse, D.; Bouchara, J.P. Molecular mechanisms of resistance to 5-fluorocytosine in laboratory mutants of Candida glabrata. Mycopathologia 2011, 171, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Desnos-Ollivier, M.; Bretagne, S.; Bernede, C.; Robert, V.; Raoux, D.; Chachaty, E.; Forget, E.; Lacroix, C.; Dromer, F.; Yeasts, G. Clonal population of flucytosine-resistant Candida tropicalis from blood cultures, Paris, France. Emerg. Infect. Dis. 2008, 14, 557–565. [Google Scholar] [CrossRef]
- Chen, Y.N.; Lo, H.J.; Wu, C.C.; Ko, H.C.; Chang, T.P.; Yang, Y.L. Loss of heterozygosity of FCY2 leading to the development of flucytosine resistance in Candida tropicalis. Antimicrob. Agents Chemother. 2011, 55, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.; Abdolrasouli, A.; Farrer, R.A.; Cuomo, C.A.; Aanensen, D.M.; Armstrong-James, D.; Fisher, M.C.; Schelenz, S. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 2018, 7, 43. [Google Scholar] [CrossRef]
- Billmyre, R.B.; Applen Clancey, S.; Li, L.X.; Doering, T.L.; Heitman, J. 5-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus. Nat. Commun. 2020, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Jund, R.; Lacroute, F. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J. Bacteriol. 1970, 102, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Chevallier, M.R.; Jund, R.; Lacroute, F. Characterization of cytosine permeation in Saccharomyces cerevisiae. J. Bacteriol. 1975, 122, 629–641. [Google Scholar] [CrossRef]
- Paluszynski, J.P.; Klassen, R.; Rohe, M.; Meinhardt, F. Various cytosine/adenine permease homologues are involved in the toxicity of 5-fluorocytosine in Saccharomyces cerevisiae. Yeast 2006, 23, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Zhou, Y.; Zhao, Y.; Zhou, Y.; Cheng, J. Expression profiling of the response of Saccharomyces cerevisiae to 5-fluorocytosine using a DNA microarray. Int. J. Antimicrob. Agents 2002, 20, 444–450. [Google Scholar] [CrossRef]
- Cuenca-Estrella, M.; Diaz-Guerra, T.M.; Mellado, E.; Rodriguez-Tudela, J.L. Flucytosine primary resistance in Candida species and Cryptococcus neoformans. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 276–279. [Google Scholar] [CrossRef]
- Barchiesi, F.; Arzeni, D.; Caselli, F.; Scalise, G. Primary resistance to flucytosine among clinical isolates of Candida spp. J. Antimicrob. Chemother. 2000, 45, 408–409. [Google Scholar] [CrossRef] [PubMed]
- Auger, P.; Dumas, C.; Joly, J. A study of 666 strains of Candida albicans: Correlation between serotype and susceptibility to 5-fluorocytosine. J. Infect. Dis. 1979, 139, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Drouhet, E.; Mercier-Soucy, L.; Montplaisir, S. [Sensitivity and resistance of pathogenic yeasts to 5-fluoropyrimidines. I.—Relation between the phenotypes of resistance to 5-fluorocytosine, the serotype of Candida albicans and the ecology of various species of Candida of human origin (author’s transl.)]. Ann. Microbiol. 1975, 126B, 25–39. [Google Scholar]
- Stiller, R.L.; Bennett, J.E.; Scholer, H.J.; Wall, M.; Polak, A.; Stevens, D.A. Susceptibility to 5-fluorocytosine and prevalence of serotype in 402 Candida albicans isolates from the United States. Antimicrob. Agents Chemother. 1982, 22, 482–487. [Google Scholar] [CrossRef]
- Poulain, D.; Hopwood, V.; Vernes, A. Antigenic variability of Candida albicans. Crit. Rev. Microbiol. 1985, 12, 223–270. [Google Scholar] [CrossRef]
- Mercure, S.; Montplaisir, S.; Lemay, G. Correlation between the presence of a self-splicing intron in the 25S rDNA of C. albicans and strains susceptibility to 5-fluorocytosine. Nucleic Acids Res. 1993, 21, 6020–6027. [Google Scholar] [CrossRef] [PubMed]
- Pujol, C.; Pfaller, M.A.; Soll, D.R. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 262–266. [Google Scholar] [CrossRef] [PubMed]
- McManus, B.A.; Coleman, D.C.; Moran, G.; Pinjon, E.; Diogo, D.; Bougnoux, M.E.; Borecka-Melkusova, S.; Bujdakova, H.; Murphy, P.; d’Enfert, C.; et al. Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J. Clin. Microbiol. 2008, 46, 652–664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Noel, T.; Francois, F.; Paumard, P.; Chastin, C.; Brethes, D.; Villard, J. Flucytosine-fluconazole cross-resistance in purine-cytosine permease-deficient Candida lusitaniae clinical isolates: Indirect evidence of a fluconazole uptake transporter. Antimicrob. Agents Chemother. 2003, 47, 1275–1284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papon, N.; Noel, T.; Florent, M.; Gibot-Leclerc, S.; Jean, D.; Chastin, C.; Villard, J.; Chapeland-Leclerc, F. Molecular mechanism of flucytosine resistance in Candida lusitaniae: Contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob. Agents Chemother. 2007, 51, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.; Chew, S.Y.; Than, L.T.L. Candida glabrata: Pathogenicity and Resistance Mechanisms for Adaptation and Survival. J. Fungi 2021, 7, 667. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Zhao, Y.; Perez, W.B.; Lockhart, S.R.; Sobel, J.D.; Farmakiotis, D.; Kontoyiannis, D.P.; Sanglard, D.; Taj-Aldeen, S.J.; Alexander, B.D.; et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016, 7, 11128. [Google Scholar] [CrossRef]
- Normark, S.; Schonebeck, J. In vitro studies of 5-fluorocytosine resistance in Candida albicans and Torulopsis glabrata. Antimicrob. Agents Chemother. 1972, 2, 114–121. [Google Scholar] [CrossRef]
- Fasoli, M.; Kerridge, D. Isolation and characterization of fluoropyrimidine-resistant mutants in two Candida species. Ann. N. Y. Acad. Sci. 1988, 544, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Steier, Z.; Vermitsky, J.P.; Toner, G.; Gygax, S.E.; Edlind, T.; Katiyar, S. Flucytosine antagonism of azole activity versus Candida glabrata: Role of transcription factor Pdr1 and multidrug transporter Cdr1. Antimicrob. Agents Chemother. 2013, 57, 5543–5547. [Google Scholar] [CrossRef]
- Lamoth, F.; Kontoyiannis, D.P. The Candida auris alert: Facts and perspectives. J. Infect. Dis. 2018, 217, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Frías-De-León, M.G.; Hernández-Castro, R.; Vite-Garín, T.; Arenas, R.; Bonifaz, A.; Castañón-Olivares, L.; Acosta-Altamirano, G.; Martínez-Herrera, E. Antifungal resistance in Candida auris: Molecular determinants. Antibiotics 2020, 9, 568. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Holowka, T.; Orner, E.P.; Fries, B.C. Gene duplication associated with increased fluconazole tolerance in Candida auris cells of advanced generational age. Sci. Rep. 2019, 9, 5052. [Google Scholar] [CrossRef] [PubMed]
- Block, E.R.; Jennings, A.E.; Bennett, J.E. 5-fluorocytosine resistance in Cryptococcus neoformans. Antimicrob. Agents Chemother. 1973, 3, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Khanal Lamichhane, A.; Garraffo, H.M.; Cai, H.; Walter, P.J.; Kwon-Chung, K.J.; Chang, Y.C. A Novel Role of Fungal Type I Myosin in Regulating Membrane Properties and Its Association with d-Amino Acid Utilization in Cryptococcus gattii. mBio 2019, 10, e01867-19. [Google Scholar] [CrossRef]
- Vu, K.; Thompson, G.R., 3rd; Roe, C.C.; Sykes, J.E.; Dreibe, E.M.; Lockhart, S.R.; Meyer, W.; Engelthaler, D.M.; Gelli, A. Flucytosine resistance in Cryptococcus gattii is indirectly mediated by the FCY2-FCY1-FUR1 pathway. Med. Mycol. 2018, 56, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Song, M.H.; Lee, J.W.; Kim, M.S.; Yoon, J.K.; White, T.C.; Floyd, A.; Heitman, J.; Strain, A.K.; Nielsen, J.N.; Nielsen, K.; et al. A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryot Cell 2012, 11, 53–67. [Google Scholar] [CrossRef]
- Jung, K.W.; Yang, D.H.; Maeng, S.; Lee, K.T.; So, Y.S.; Hong, J.; Choi, J.; Byun, H.J.; Kim, H.; Bang, S.; et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 2015, 6, 6757. [Google Scholar] [CrossRef]
- Patterson, T.F.; Thompson, G.R., 3rd; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef] [PubMed]
- Te Dorsthorst, D.T.; Mouton, J.W.; van den Beukel, C.J.; van der Lee, H.A.; Meis, J.F.; Verweij, P.E. Effect of pH on the in vitro activities of amphotericin B, itraconazole, and flucytosine against Aspergillus isolates. Antimicrob. Agents Chemother. 2004, 48, 3147–3150. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Te Dorsthorst, D.T.; Verweij, P.E.; Meis, J.F.; Mouton, J.W. Efficacy and pharmacodynamics of flucytosine monotherapy in a nonneutropenic murine model of invasive aspergillosis. Antimicrob. Agents Chemother. 2005, 49, 4220–4226. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Te Dorsthorst, D.T.; Janssen, W.H.; Meis, J.F.; Mouton, J.W. In vitro activities at pH 5.0 and pH 7.0 and in vivo efficacy of flucytosine against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2008, 52, 4483–4485. [Google Scholar] [CrossRef][Green Version]
- Gsaller, F.; Furukawa, T.; Carr, P.D.; Rash, B.; Jochl, C.; Bertuzzi, M.; Bignell, E.M.; Bromley, M.J. Mechanistic Basis of pH-Dependent 5-Flucytosine Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2018, 62, e02593-17. [Google Scholar] [CrossRef]
- Birstonas, L.; Dallemulle, A.; Lopez-Berges, M.S.; Jacobsen, I.D.; Offterdinger, M.; Abt, B.; Strassburger, M.; Bauer, I.; Schmidt, O.; Sarg, B.; et al. Multiplex Genetic Engineering Exploiting Pyrimidine Salvage Pathway-Based Endogenous Counterselectable Markers. mBio 2020, 11, e00230-20. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, R.; Katoh, T.; Nishioka, K. Isolation of dermatophytes from house dust on a medium containing gentamicin and flucytosine: Zur Isolierung von Dermatophyten aus Hausstaub auf einem Gentamicin-und Flucytosin-haltigen Medium. Mycoses 1995, 38, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Ansari, S.; Ilkit, M.; Rafati, H.; Hedayati, M.T.; Taghizadeh-Armaki, M.; Nasrollahi-Omran, A.; Tolooe, A.; Zhan, P.; Liao, W.; et al. In Vitro Antifungal Susceptibility Profiles of 12 Antifungal Drugs against 55 Trichophyton schoenleinii Isolates from Tinea Capitis Favosa Patients in Iran, Turkey, and China. Antimicrob. Agents Chemother. 2017, 61, e01753-16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Bin, W.; Diao, Y.; Yang, J.; Liu, T.; Peng, J.; Jin, Q. Global transcriptional profiles of Trichophyton rubrum in response to Flucytosine. Sci. China Ser. C Life Sci. 2009, 52, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).