A Systematic Survey of Characteristic Features of Yeast Cell Death Triggered by External Factors
Abstract
1. Introduction
2. Methods
3. Methodological Aspects of the Literature Studying Cell Death
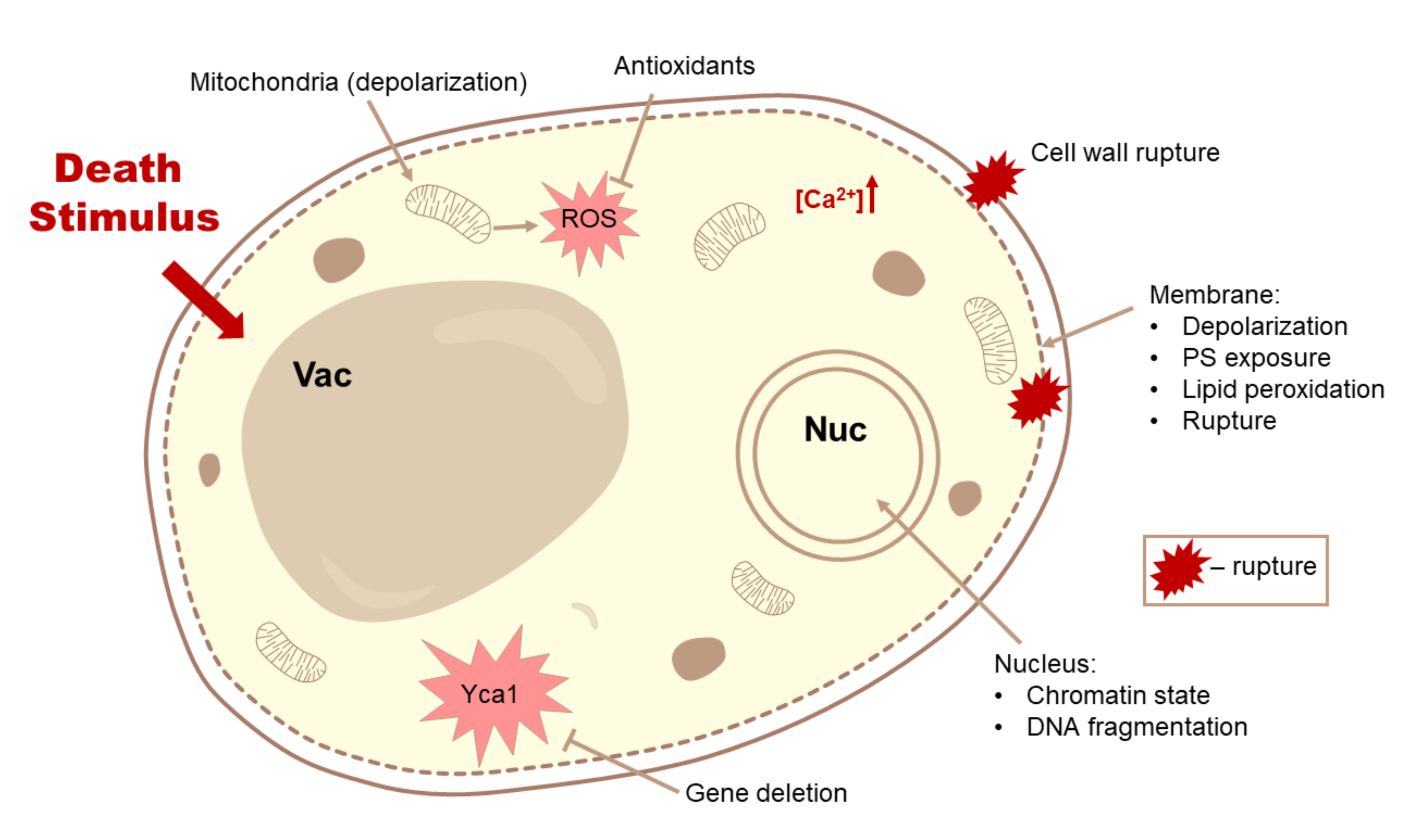
3.1. Methods for Detection of Cell Death
3.2. Differential Hallmarks of Cell Death
3.2.1. Detection of PS Exposure
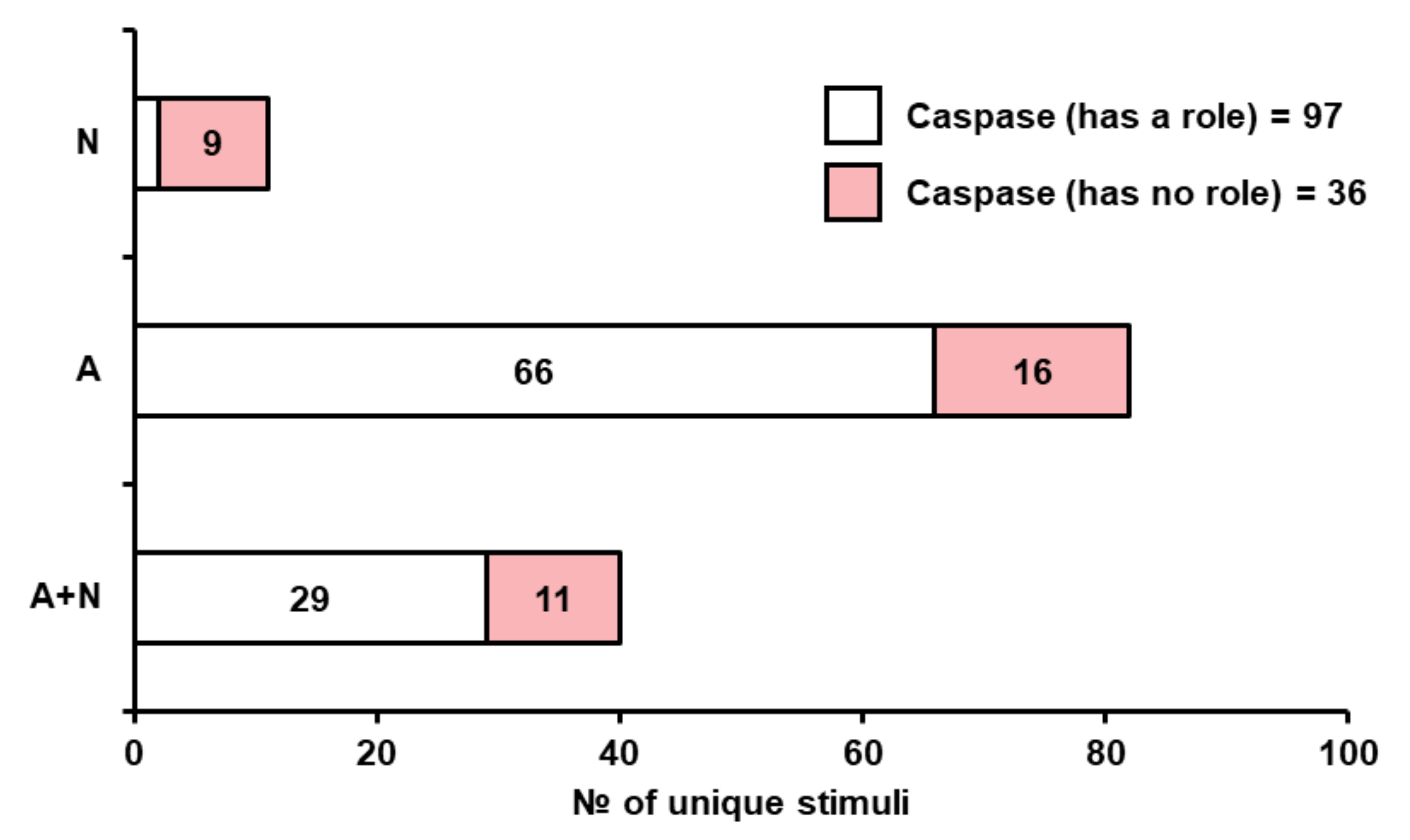
3.2.2. Yeast Metacaspase
3.2.3. Reduction of Cell Death by Cycloheximide Treatment
3.2.4. Lysosome- and Autophagy-Mediated Cell Death
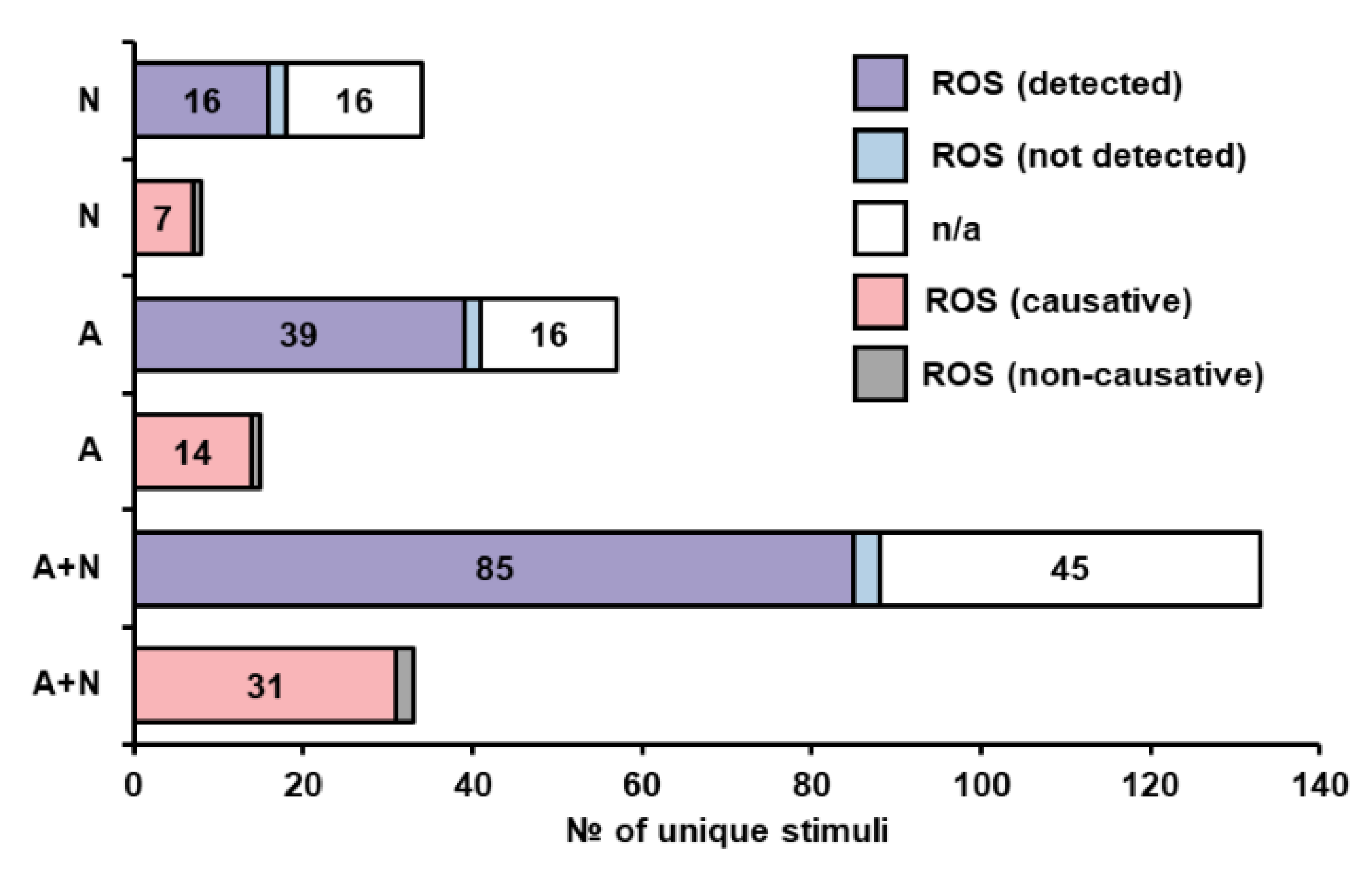
3.2.5. Oxidative Stress and Reactive Oxygen Species
3.2.6. Calcium Signaling
3.2.7. Changes in the Functionality and Morphology of Organelles
3.2.8. Role of Treatment Severity in Determining Cell Death Type
3.3. Death-Inducing Perturbations in Yeast
3.3.1. Yeast Life Cycle Related Stimuli
Replicative Aging
Chronological Aging
Colony Aging
Sporulation
Mating Associated Death
Interspecies Death Induction
3.3.2. Shocks of Physical Nature
3.3.3. Nutrient Imbalance and Depletion
3.3.4. Oxidants
3.3.5. Acids
3.3.6. Alcohols
3.3.7. Antimicrobial Peptides
3.3.8. Additional Compounds Used in Medicine and Agriculture
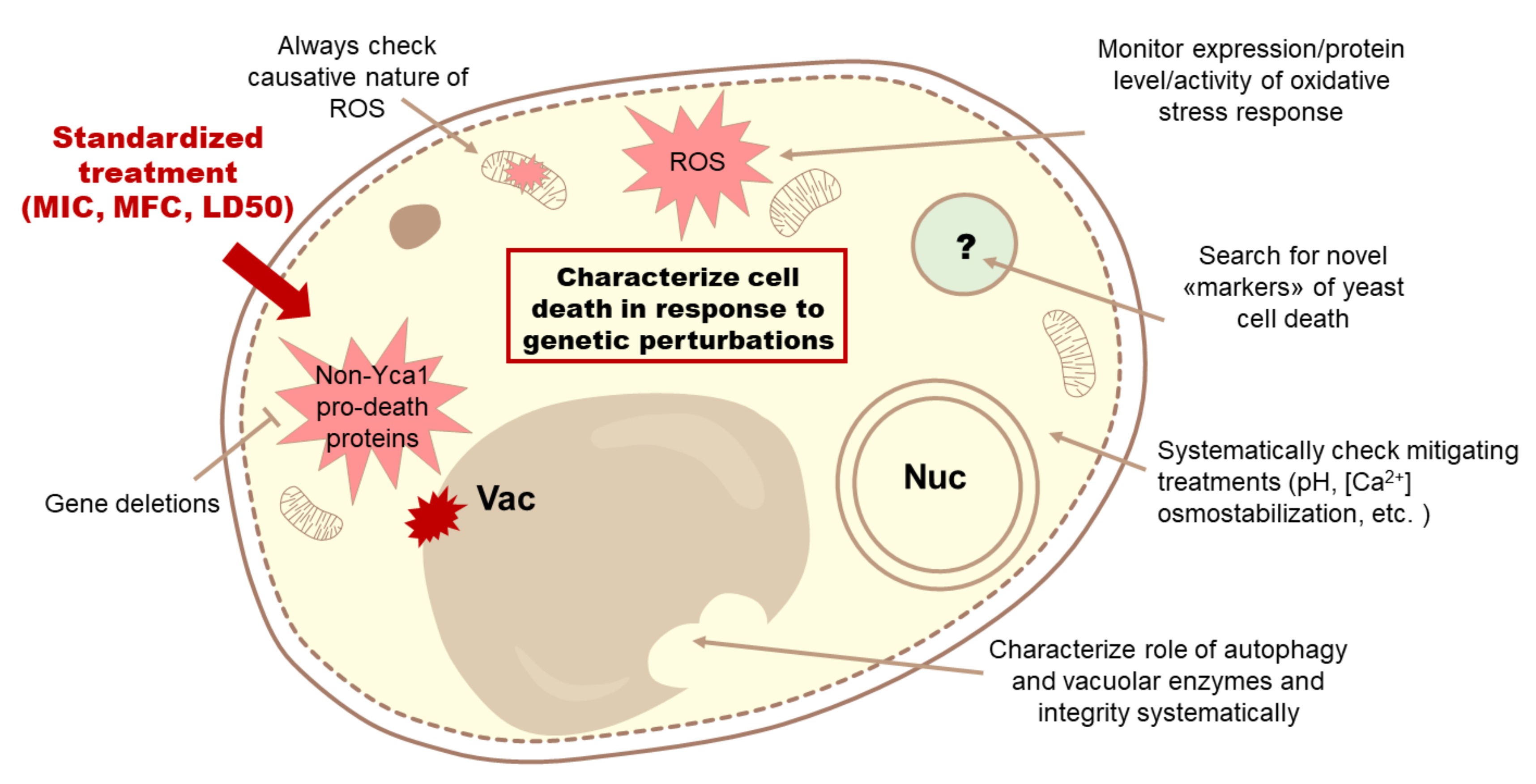
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Yuan, J. Expansion and evolution of cell death programmes. Nat. Rev. Mol. Cell Biol. 2008, 9, 378–390. [Google Scholar] [CrossRef]
- Durand, P.M. On the molecular evolution of the Plasmodium falciparum genome: Origin and evolution of a parasite’s genome. Heal. San Fr. 2010, 9, 367–393. [Google Scholar] [CrossRef]
- Syntichaki, P.; Tavernarakis, N. Death by necrosis. Uncontrollable catastrophe, or is there order behind the chaos? EMBO Rep. 2002, 3, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Festjens, N.; Vanden Berghe, T.; Vandenabeele, P. Necrosis, a well-orchestrated form of cell demise: Signalling cascades, important mediators and concomitant immune response. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 1371–1387. [Google Scholar] [CrossRef] [PubMed]
- Golstein, P.; Kroemer, G. Cell death by necrosis: Towards a molecular definition. Trends Biochem. Sci. 2007, 32, 37–43. [Google Scholar] [CrossRef]
- Madeo, F.; Fröhlich, E.; Fröhlich, K.-U. A Yeast Mutant Showing Diagnostic Markers of Early and Late Apoptosis. J. Cell Biol. 1997, 139, 729–734. [Google Scholar] [CrossRef]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lächelt, S.; Herlan, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A Caspase-Related Protease Regulates Apoptosis in Yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef]
- Klim, J.; Gładki, A.; Kucharczyk, R.; Zielenkiewicz, U.; Kaczanowski, S. Ancestral State Reconstruction of the Apoptosis Machinery in the Common Ancestor of Eukaryotes. G3 Genes Genomes Genet. 2018, 8, 2121–2134. [Google Scholar] [CrossRef]
- Kulkarni, M.; Stolp, Z.D.; Hardwick, J.M. Targeting intrinsic cell death pathways to control fungal pathogens. Biochem. Pharmacol. 2019, 162, 71–78. [Google Scholar] [CrossRef]
- Lohr, J.N.; Galimov, E.R.; Gems, D. Does senescence promote fitness in Caenorhabditis elegans by causing death? Ageing Res. Rev. 2019, 50, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Palková, Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 2005, 169, 711–717. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA 2012, 109, 1595–1600. [Google Scholar] [CrossRef]
- Eisenberg, T.; Carmona-Gutierrez, D.; Büttner, S.; Tavernarakis, N.; Madeo, F. Necrosis in yeast. Apoptosis 2010, 15, 257–268. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Eisenberg, T.; Büttner, S.; Meisinger, C.; Kroemer, G.; Madeo, F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010, 17, 763–773. [Google Scholar] [CrossRef]
- Hardwick, J.M. Do fungi undergo apoptosis-like programmed cell death? MBio 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef]
- Trutneva, K.A.; Shleeva, M.O.; Demina, G.R.; Vostroknutova, G.N.; Kaprelyans, A.S. One-Year Old Dormant, “Non-culturable” Mycobacterium tuberculosis Preserves Significantly Diverse Protein Profile. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Yue, Q.; Zhou, X.; Leng, Q.; Zhang, L.; Cheng, B.; Zhang, X. 7-ketocholesterol-induced caspase-mediated apoptosis in Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Weis, R.; Luiten, R.; Skranc, W.; Schwab, H.; Wubbolts, M.; Glieder, A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004, 5, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Pankey, G.A.; Sabath, L.D. Clinical Relevance of Bacteriostatic versus Bactericidal Mechanisms of Action in the Treatment of Gram-Positive Bacterial Infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef]
- Kirchhoff, C.; Cypionka, H. Propidium ion enters viable cells with high membrane potential during live-dead staining. J. Microbiol. Methods 2017, 142, 79–82. [Google Scholar] [CrossRef]
- Dirmeier, R.; O’Brien, K.M.; Engle, M.; Dodd, A.; Spears, E.; Poyton, R.O. Exposure of Yeast Cells to Anoxia Induces Transient Oxidative Stress. J. Biol. Chem. 2002, 277, 34773–34784. [Google Scholar] [CrossRef]
- Shi, L.; Günther, S.; Hübschmann, T.; Wick, L.Y.; Harms, H.; Müller, S. Limits of propidium iodide as a cell viability indicator for environmental bacteria. Cytom. Part A 2007, 71, 592–598. [Google Scholar] [CrossRef]
- Shen, L.; Li, Y.; Jiang, L.; Wang, X. Response of Saccharomyces cerevisiae to the stimulation of lipopolysaccharide. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Pedrini, M.R.; Dupont, S.; De Anchieta Câmara, A.; Beney, L.; Gervais, P. Osmoporation: A simple way to internalize hydrophilic molecules into yeast. Appl. Microbiol. Biotechnol. 2014, 98, 1271–1280. [Google Scholar] [CrossRef]
- Davey, H.M.; Hexley, P. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ. Microbiol. 2011, 13, 163–171. [Google Scholar] [CrossRef]
- Ahn, S.H.; Diaz, R.L.; Grunstein, M.; Allis, C.D. Histone H2B Deacetylation at Lysine 11 Is Required for Yeast Apoptosis Induced by Phosphorylation of H2B at Serine 10. Mol. Cell 2006, 24, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Valiakhmetov, A.Y.; Kuchin, A.V.; Suzina, N.E.; Zvonarev, A.N.; Shepelyakovskaya, A.O. Glucose causes primary necrosis in exponentially grown yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.M.; Hanekamp, T.; Stayton, M.M. Methylene blue: An alternative, multi-purpose stain for detection, analysis and isolation of nucleic acids. Biopolym. Cell 1997, 13, 250–253. [Google Scholar] [CrossRef]
- Krämer, C.E.M.; Wiechert, W.; Kohlheyer, D. Time-resolved, single-cell analysis of induced and programmed cell death via non-invasive propidium iodide and counterstain perfusion. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Minois, N.; Frajnt, M.; Wilson, C.; Vaupel, J.W. Advances in measuring lifespan in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2005, 102, 402–406. [Google Scholar] [CrossRef]
- Sami, M.; Ikeda, M.; Yabuuchi, S. Evaluation of the alkaline methylene blue staining method for yeast activity determination. J. Ferment. Bioeng. 1994, 78, 212–216. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. Potential role of potassium and chloride channels in regulation of silymarin-induced apoptosis in Candida albicans. IUBMB Life 2018, 70, 197–206. [Google Scholar] [CrossRef]
- Mochon, A.B.; Liu, H. The antimicrobial peptide histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog. 2008, 4. [Google Scholar] [CrossRef]
- Hsuchen, C.C.; Feingold, D.S. Two types of resistance to polyene antibiotics in Candida albicans. Nature 1974, 251, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Butts, A.; DiDone, L.; Koselny, K.; Baxter, B.K.; Chabrier-Rosello, Y.; Wellington, M.; Krysanb, D.J. A repurposing approach identifies off-patent drugs with fungicidal cryptococcal activity, a common structural chemotype, and pharmacological properties relevant to the treatment of Cryptococcosis. Eukaryot. Cell 2013, 12, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Krysan, D.D.J.; Didone, L. A high-throughput screening assay for small molecules that disrupt yeast cell integrity. J. Biomol. Screen. 2008, 13, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Cunha, D.; Cunha, R.; Côrte-Real, M.; Chaves, S.R. Cisplatin-induced cell death in Saccharomyces cerevisiae is programmed and rescued by proteasome inhibition. DNA Repair 2013, 12, 444–449. [Google Scholar] [CrossRef]
- Andrés, M.T.; Viejo-Díaz, M.; Fierro, J.F. Human lactoferrin induces apoptosis-like cell death in Candida albicans: Critical role of K+-channel-mediated K+ efflux. Antimicrob. Agents Chemother. 2008, 52, 4081–4088. [Google Scholar] [CrossRef]
- Shlomovitz, I.; Speir, M.; Gerlic, M. Flipping the dogma–phosphatidylserine in non-apoptotic cell death. Cell Commun. Signal. 2019, 17, 139. [Google Scholar] [CrossRef]
- Mankelow, T.J.; Griffiths, R.E.; Trompeter, S.; Flatt, J.F.; Cogan, N.M.; Massey, E.J.; Anstee, D.J. Autophagic vesicles on mature human reticulocytes explain phosphatidylserine-positive red cells in sickle cell disease. Blood 2015, 126, 1831–1834. [Google Scholar] [CrossRef]
- Discher, D.E.; Ney, P.A. The reason sickle reticulocytes expose PS. Blood 2015, 126, 1737–1738. [Google Scholar] [CrossRef][Green Version]
- Lee, S.-H.; Meng, X.W.; Flatten, K.S.; Loegering, D.A.; Kaufmann, S.H. Phosphatidylserine exposure during apoptosis reflects bidirectional trafficking between plasma membrane and cytoplasm. Cell Death Differ. 2013, 20, 64–76. [Google Scholar] [CrossRef]
- Wysocki, R.; Kron, S.J. Yeast cell death during DNA damage arrest is independent of caspase or reactive oxygen species. J. Cell Biol. 2004, 166, 311–316. [Google Scholar] [CrossRef]
- Váchová, L.; Palková, Z. Caspases in yeast apoptosis-like death: Facts and artefacts. FEMS Yeast Res. 2007, 7, 12–21. [Google Scholar] [CrossRef]
- Hauptmann, P.; Riel, C.; Kunz-Schughart, L.A.; Frohlich, K.-U.; Madeo, F.; Lehle, L. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 2006, 59, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Aerts, A.M.; Carmona-Gutierrez, D.; Lefevre, S.; Govaert, G.; François, I.E.J.A.; Madeo, F.; Santos, R.; Cammue, B.P.A.; Thevissen, K. The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans. FEBS Lett. 2009, 583, 2513–2516. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.E.C.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Hao, X.; Liu, B.; Nystrom, T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 2014, 344, 1389–1392. [Google Scholar] [CrossRef]
- Shrestha, A.; Brunette, S.; Stanford, W.L.; Megeney, L.A. The metacaspase Yca1 maintains proteostasis through multiple interactions with the ubiquitin system. Cell Discov. 2019, 5, 1–13. [Google Scholar] [CrossRef]
- Lee, R.E.C.; Puente, L.G.; Kærn, M.; Megeney, L.A. A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. PLoS ONE 2008, 3, e2956. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, A.S.; Alexandrov, A.I.; Makarova, N.E.; Gladyshev, V.N.; Dmitriev, S.E. Protein synthesis and quality control in aging. Aging 2018, 10, 4269–4288. [Google Scholar] [CrossRef] [PubMed]
- Rikhvanov, E.G.; Fedoseeva, I.V.; Varakina, N.N.; Rusaleva, T.M.; Fedyaeva, A.V. Mechanism of Saccharomyces cerevisiae yeast cell death induced by heat shock. Effect of cycloheximide on thermotolerance. Biochemistry 2014, 79, 16–24. [Google Scholar] [CrossRef]
- Sanchez, Y.; Lindquist, S.L. HSP104 required for induced thermotolerance. Science 1990, 248, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Dziedzic, S.A.; Caplan, A.B. Identification of autophagy genes participating in zinc-induced necrotic cell death in Saccharomyces cerevisiae. Autophagy 2011, 7, 490–500. [Google Scholar] [CrossRef]
- Sheibani, S.; Richard, V.; Beach, A.; Leonov, A.; Feldman, R.; Mattie, S.; Khelghatybana, L.; Piano, A.; Greenwood, M.; Vali, H.; et al. Macromitophagy, neutral lipids synthesis, and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell Cycle 2014, 13, 138–147. [Google Scholar] [CrossRef]
- Eastwood, M.D.; Meneghini, M.D. Developmental coordination of gamete differentiation with programmed cell death in sporulating yeast. Eukaryot. Cell 2015, 14, 858–867. [Google Scholar] [CrossRef]
- Perrone, G.G.; Tan, S.X.; Dawes, I.W. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1354–1368. [Google Scholar] [CrossRef]
- Kalyanaraman, B.; Darley-Usmar, V.; Davies, K.J.A.; Dennery, P.A.; Forman, H.J.; Grisham, M.B.; Mann, G.E.; Moore, K.; Roberts, L.J.; Ischiropoulos, H. Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic. Biol. Med. 2012, 52, 1–6. [Google Scholar] [CrossRef]
- Kiššová, I.; Deffieu, M.; Samokhvalov, V.; Velours, G.; Bessoule, J.J.; Manon, S.; Camougrand, N. Lipid oxidation and autophagy in yeast. Free Radic. Biol. Med. 2006, 41, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Knorre, D.A.; Smirnova, E.A.; Markova, O.V.; Sorokin, M.I.; Severin, F.F. Prooxidants prevent yeast cell death induced by genotoxic stress. Cell Biol. Int. 2011, 35, 431–435. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, X.; Yu, L.; Xu, H. Calcium signaling in membrane repair. Semin. Cell Dev. Biol. 2015, 45, 24–31. [Google Scholar] [CrossRef]
- Ghibelli, L.; Cerella, C.; Diederich, M. The dual role of calcium as messenger and stressor in cell damage, death, and survival. Int. J. Cell Biol. 2010, 2010. [Google Scholar] [CrossRef]
- Wong, A.H.H.; Yan, C.; Shi, Y. Crystal structure of the yeast metacaspase Yca1. J. Biol. Chem. 2012, 287, 29251–29259. [Google Scholar] [CrossRef]
- Yamagata, M.; Obara, K.; Kihara, A. Unperverted synthesis of complex sphingolipids is essential for cell survival under nitrogen starvation. Genes Cells 2013, 18, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Chandel, A.; Das, K.K.; Bachhawat, A.K. Glutathione depletion activates the yeast vacuolar transient receptor potential channel, Yvc1p, by reversible glutathionylation of specific Cysteines. Mol. Biol. Cell 2016, 27, 3913–3925. [Google Scholar] [CrossRef] [PubMed]
- Van der Pal, R.H.M.; Belde, P.J.M.; Theuvenet, A.P.R.; Peters, P.H.J.; Borst-Pauwels, G.W.F.H. Effect of ruthenium red upon Ca2+ and Mn2+ uptake in Saccharomyces cerevisiae. Comparison with the effect of La3+. Biochim. Biophys. Acta Biomembr. 1987, 902, 19–23. [Google Scholar] [CrossRef]
- Yun, J.E.; Lee, D.G. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans. IUBMB Life 2016, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Woo, E.R.; Lee, D.G. Effect of apigenin isolated from Aster yomena against Candida albicans: Apigenin-triggered apoptotic pathway regulated by mitochondrial calcium signaling. J. Ethnopharmacol. 2019, 231, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lupetti, A.; Brouwer, C.P.J.M.; Dogterom-Ballering, H.E.C.; Senesi, S.; Campa, M.; Van Dissel, J.T.; Nibbering, P.H. Release of calcium from intracellular stores and subsequent uptake by mitochondria are essential for the candidacidal activity of an N-terminal peptide of human lactoferrin. J. Antimicrob. Chemother. 2004, 54, 603–608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iida, H.; Yagawa, Y.; Anraku, Y. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. A study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J. Biol. Chem. 1990, 265, 13391–13399. [Google Scholar] [CrossRef]
- Zhang, N.N.; Dudgeon, D.D.; Paliwal, S.; Levchenko, A.; Grote, E.; Cunningham, K.W. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell 2006, 17, 3409–3422. [Google Scholar] [CrossRef] [PubMed]
- Alby, K.; Schaefer, D.; Sherwood, R.K.; Jones, S.K.; Bennett, R.J. Identification of a cell death pathway in Candida albicans during the response to pheromone. Eukaryot. Cell 2010, 9, 1690–1701. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Ischer, F.; Marchetti, O.; Entenza, J.; Bille, J. Calcineurin A of Candida albicans: Involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003, 48, 959–976. [Google Scholar] [CrossRef]
- Fedoseeva, I.V.; Pjatricas, D.V.; Varakina, N.N.; Rusaleva, T.M.; Stepanov, A.V.; Rikhvanov, E.G.; Borovskii, G.B.; Voinikov, V.K. Effect of amiodarone on thermotolerance and Hsp104p synthesis in the yeast Saccharomyces cerevisiae. Biochemistry 2012, 77, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Hwang, J.S.; Lee, D.G. Scolopendin 2 leads to cellular stress response in Candida albicans. Apoptosis 2016, 21, 856–865. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. Reactive oxygen species modulate itraconazole-induced apoptosis via mitochondrial disruption in Candida albicans. Free Radic. Res. 2018, 52, 39–50. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.G. Role of calcium in reactive oxygen species-induced apoptosis in Candida albicans: An antifungal mechanism of antimicrobial peptide, PMAP-23. Free Radic. Res. 2019, 53, 8–17. [Google Scholar] [CrossRef]
- Marx, G.; Moody, A.; Bermúdez-Aguirre, D. A comparative study on the structure of Saccharomyces cerevisiae under nonthermal technologies: High hydrostatic pressure, pulsed electric fields and thermo-sonication. Int. J. Food Microbiol. 2011, 151, 327–337. [Google Scholar] [CrossRef]
- Camarillo-Márquez, O.; Córdova-Alcántara, I.M.; Hernández-Rodríguez, C.H.; García-Pérez, B.E.; Martínez-Rivera, M.A.; Rodríguez-Tovar, A.V. Antagonistic interaction of Staphylococcus aureus toward Candida glabrata during in vitro biofilm formation is caused by an apoptotic mechanism. Front. Microbiol. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Guaragnella, N.; Ždralevic, M.; Antonacci, L.; Passarella, S.; Marra, E.; Giannattasio, S. The role of mitochondria in yeast programmed cell death. Front. Oncol. 2012, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, G.; Balzan, R. Oxidative stress and programmed cell death in yeast. Front. Oncol. 2012, 2, 64. [Google Scholar] [CrossRef] [PubMed]
- Pozniakovsky, A.I.; Knorre, D.A.; Markova, O.V.; Hyman, A.A.; Skulachev, V.P.; Severin, F.F. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 2005, 168, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Severin, F.F.; Hyman, A.A. Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 2002, 12, 233–235. [Google Scholar] [CrossRef]
- Ruckenstuhl, C.; Büttner, S.; Carmona-Gutierrez, D.; Eisenberg, T.; Kroemer, G.; Sigrist, S.J.; Fröhlich, K.U.; Madeo, F. The warburg effect suppresses oxidative stress induced apoptosis yeast model for cancer. PLoS ONE 2009, 4, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Reisenbichler, A.; Heimbucher, P.; Bauer, M.A.; Braun, R.J.; Ruckenstuhl, C.; Büttner, S.; Eisenberg, T.; Rockenfeller, P.; Fröhlich, K.U.; et al. Ceramide triggers metacaspase-independent mitochondrial cell death in yeast. Cell Cycle 2011, 10, 3973–3978. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Zaldívar, M.; Andrés, M.T.; Rego, A.; Pereira, C.S.; Fierro, J.F.; Côrte-Real, M. Human lactoferrin triggers a mitochondrial- and caspase-dependent regulated cell death in Saccharomyces cerevisiae. Apoptosis 2016, 21, 163–173. [Google Scholar] [CrossRef]
- Emrick, D.; Ravichandran, A.; Gosai, J.; Lu, S.; Gordon, D.M.; Smith, L. The antifungal occidiofungin triggers an apoptotic mechanism of cell death in yeast. J. Nat. Prod. 2013, 76, 829–838. [Google Scholar] [CrossRef]
- Rockenfeller, P.; Ring, J.; Muschett, V.; Beranek, A.; Buettner, S.; Carmona-Gutierrez, D.; Eisenberg, T.; Khoury, C.; Rechberger, G.; Kohlwein, S.D.; et al. Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle 2010, 9, 2908–2914. [Google Scholar] [CrossRef]
- Klassen, R.; Meinhardt, F. Induction of DNA damage and apotosis in Saccharomyces cerevisiae by a yeast killer toxin. Cell. Microbiol. 2005, 7, 393–401. [Google Scholar] [CrossRef]
- Laun, P.; Pichova, A.; Madeo, F.; Fuchs, J.; Ellinger, A.; Kohlwein, S.; Dawes, I.; Fröhlich, K.-U.; Breitenbach, M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 2004, 39, 1166–1173. [Google Scholar] [CrossRef]
- Fehrmann, S.; Paoletti, C.; Goulev, Y.; Ungureanu, A.; Aguilaniu, H.; Charvin, G. Aging Yeast Cells Undergo a Sharp Entry into Senescence Unrelated to the Loss of Mitochondrial Membrane Potential. Cell Rep. 2013, 5, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.M.; Russell, A.E.; Schafer, B.J.; Blue, B.W.; Whalen, R.; Almazan, J.; Hong, M.G.; Nguyen, B.; Goings, J.E.; Chen, K.L.; et al. DNA damage checkpoint activation impairs chromatin homeostasis and promotes mitotic catastrophe during aging. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Herker, E.; Jungwirth, H.; Lehmann, K.A.; Maldener, C.; Fröhlich, K.-U.; Wissing, S.; Büttner, S.; Fehr, M.; Sigrist, S.; Madeo, F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004, 164, 501–507. [Google Scholar] [CrossRef]
- Allen, C.; Büttner, S.; Aragon, A.D.; Thomas, J.A.; Meirelles, O.; Jaetao, J.E.; Benn, D.; Ruby, S.W.; Veenhuis, M.; Madeo, F.; et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006, 174, 89–100. [Google Scholar] [CrossRef]
- Burtner, C.R.; Murakami, C.J.; Kennedy, B.K.; Kaeberlein, M. A molecular mechanism of chronological aging in yeast. Cell Cycle 2009, 8, 1256–1270. [Google Scholar] [CrossRef]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef]
- Eastwood, M.D.; Cheung, S.W.T.; Lee, K.Y.; Moffat, J.; Meneghini, M.D. Developmentally Programmed Nuclear Destruction during Yeast Gametogenesis. Dev. Cell 2012, 23, 35–44. [Google Scholar] [CrossRef]
- Zadrag, R.; Wojnar, L.; Bartosz, G.; Biliński, T. Does yeast shmooing mean a commitment to apoptosis? Cell Biol. Int. 2006, 30, 205–209. [Google Scholar] [CrossRef]
- Reiter, J.; Herker, E.; Madeo, F.; Schmitt, M.J. Viral killer toxins induce caspase-mediated apoptosis in yeast. J. Cell Biol. 2005, 168, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Alonso, A.; Belda, I.; Marquina, D. Cell cycle arrest and apoptosis, two alternative mechanisms for PMKT2 killer activity. Fungal Genet. Biol. 2013, 50, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Guyard, C.; Dehecq, E.; Tissier, J.P.; Polonelli, L.; Dei-Cas, E.; Cailliez, J.C.; Menozzi, F.D. Involvement of β-glucans in the wide-spectrum antimicrobial activity of Williopsis saturnus var. mrakii MUCL 41968 killer toxin. Mol. Med. 2002, 8, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Sesti, F.; Ilan, N.; Shih, T.M.; Sturley, S.L.; Goldstein, S.A. A Molecular Target for Viral Killer Toxin. Cell 1999, 99, 283–291. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Alonso, A.; Marquina, D.; Santos, A. The Biology of Pichia membranifaciens Killer Toxins. Toxins 2017, 9, 112. [Google Scholar] [CrossRef]
- Nissen, P.; Arneborg, N. Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch. Microbiol. 2003, 180, 257–263. [Google Scholar] [CrossRef]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial properties and death-inducing mechanisms of saccharomycin, a biocide secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Ikeda, R.; Sawamura, K. Bacterial and H2O2 stress-induced apoptosis-like events in Cryptococcus neoformans. Res. Microbiol. 2008, 159, 628–634. [Google Scholar] [CrossRef]
- Ikeda, R. Apoptosis-like cell death of cryptococcus neoformans mediated by staphylococcus aureus contact. Jpn. J. Med. Mycol. 2013, 54, 49–52. [Google Scholar] [CrossRef][Green Version]
- Schmitt, M.J.; Reiter, J. Viral induced yeast apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1413–1417. [Google Scholar] [CrossRef]
- Ivanovska, I.; Hardwick, J.M. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 2005, 170, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Nowlin, K.; Boseman, A.; Covell, A.; LaJeunesse, D. Adhesion-dependent rupturing of Saccharomyces cerevisiae on biological antimicrobial nanostructured surfaces. J. R. Soc. Interface 2014, 12. [Google Scholar] [CrossRef]
- Pyatrikas, D.V.; Fedoseeva, I.V.; Varakina, N.N.; Rusaleva, T.M.; Stepanov, A.V.; Fedyaeva, A.V.; Borovskii, G.B.; Rikhvanov, E.G. Relation between cell death progression, reactive oxygen species production and mitochondrial membrane potential in fermenting Saccharomyces cerevisiae cells under heat-shock conditions. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.F.; Whyte, B.; Bissinger, P.H.; Schiestl, R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5116–5121. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Skinner, C.; Easlon, E.; Lin, S.J. Deleting the 14-3-3 protein Bmh1 extends life span in Saccharomyces cerevisiae by increasing stress response. Genetics 2009, 183, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Cheng, W.-C.; Qi, B.; Yu, T.-X.; Ramachandran, K.; Boersma, M.D.; Hattier, T.; Lehmann, P.V.; Pineda, F.J.; Hardwick, J.M. Gene-dependent cell death in yeast. Cell Death Dis. 2011, 2, e188. [Google Scholar] [CrossRef] [PubMed]
- Fannjiang, Y.; Cheng, W.C.; Lee, S.J.; Qi, B.; Pevsner, J.; McCaffery, J.M.; Hill, R.B.; Basañez, G.; Hardwick, J.M. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004, 18, 2785–2797. [Google Scholar] [CrossRef]
- Simonin, H.; Beney, L.; Gervais, P. Sequence of occurring damages in yeast plasma membrane during dehydration and rehydration: Mechanisms of cell death. Biochim. Biophys. Acta Biomembr. 2007, 1768, 1600–1610. [Google Scholar] [CrossRef]
- Dupont, S.; Beney, L.; Ritt, J.F.; Lherminier, J.; Gervais, P. Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochim. Biophys. Acta Biomembr. 2010, 1798, 975–985. [Google Scholar] [CrossRef]
- Silva, R.D.; Sotoca, R.; Johansson, B.; Ludovico, P.; Sansonetty, F.; Silva, M.T.; Peinado, J.M.; Côrte-Real, M. Hyperosmotic stress induces metacaspase- and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 2005, 58, 824–834. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa, K.A.; Tyagi, M.B.; Sinha, R.P. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.; Hayes, J.; Laffey, J.; Rowan, N. Studies on the relationship between pulsed UV light irradiation and the simultaneous occurrence of molecular and cellular damage in clinically-relevant Candida albicans. J. Microbiol. Methods 2011, 84, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Del Carratore, R.; Della Croce, C.; Simili, M.; Taccini, E.; Scavuzzo, M.; Sbrana, S. Cell cycle and morphological alterations as indicative of apoptosis promoted by UV irradiation in S. cerevisiae. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2001, 513, 183–191. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jun, D.Y.; Park, J.E.; Kwon, G.H.; Kim, J.S.; Kim, Y.H. Pro-apoptotic role of the human YPEL5 gene identified by functional complementation of a yeast moh1Δ mutation. J. Microbiol. Biotechnol. 2017, 27, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.-P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Kim, J.K.; Park, J.; Ryu, T.H.; Nili, M. Effect of N-acetyl-l-cysteine on Saccharomyces cerevisiae irradiated with gamma-rays. Chemosphere 2013, 92, 512–516. [Google Scholar] [CrossRef]
- Reliene, R.; Pollard, J.M.; Sobol, Z.; Trouiller, B.; Gatti, R.A.; Schiestl, R.H. N-acetyl cysteine protects against ionizing radiation-induced DNA damage but not against cell killing in yeast and mammals. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2009, 665, 37–43. [Google Scholar] [CrossRef]
- Bryan, R.A.; Huang, X.; Morgenstern, A.; Bruchertseifer, F.; Casadevall, A.; Dadachova, E. Radiofungicidal Effects of External Gamma Radiation and Antibody-Targeted Beta and Alpha Radiation on Cryptococcus neoformans. Antimicrob. Agents Chemother. 2008, 52, 2232–2235. [Google Scholar] [CrossRef][Green Version]
- Kang, M.S.; Yu, S.L.; Lim, H.S.; Choi, B.; Park, C.S.; Kang, J.H.; Lee, S.K. Mitotic catastrophe induced by overexpression of budding yeast Rad2p. Yeast 2010, 27, 399–411. [Google Scholar] [CrossRef]
- Radhakrishnan, V.S.; Dwivedi, S.P.; Siddiqui, M.H.; Prasad, T. In vitro studies on oxidative stress-independent, Ag nanoparticles-induced cell toxicity of Candida albicans, an opportunistic pathogen. Int. J. Nanomed. 2018, 13, 91–96. [Google Scholar] [CrossRef][Green Version]
- Seong, M.; Lee, D.G. Reactive oxygen species-independent apoptotic pathway by gold nanoparticles in Candida albicans. Microbiol. Res. 2018, 207, 33–40. [Google Scholar] [CrossRef]
- Granot, D.; Levine, A.; Dor-Hefetz, E. Sugar-induced apoptosis in yeast cells. FEMS Yeast Res. 2003, 4, 7–13. [Google Scholar] [CrossRef]
- Lee, Y.J.; Burlet, E.; Galiano, F.; Circu, M.L.; Aw, T.Y.; Williams, B.J.; Witt, S.N. Phosphate and succinate use different mechanisms to inhibit sugar-induced cell death in yeast: Insight into the Crabtree effect. J. Biol. Chem. 2011, 286, 20267–20274. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Guan, G.; Li, X.; Gulati, M.; Tao, L.; Cao, C.; Johnson, A.D.; Nobile, C.J. N-Acetylglucosamine-Induced Cell Death in Candida albicans and Its. MBio 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Postma, E.; Verduyn, C.; Kuiper, A.; Scheffers, W.A.; Van Dijken, J.P. Substrate-accelerated death of Saccharomyces cerevisiae CBS 8066 under maltose stress. Yeast 1990, 6, 149–158. [Google Scholar] [CrossRef]
- Dziedzic, S.A.; Caplan, A.B. Autophagy proteins play cytoprotective and cytocidal roles in leucine starvation-induced cell death in Saccharomyces cerevisiae. Autophagy 2012, 8, 731–738. [Google Scholar] [CrossRef]
- Henry, S.A.; Atkinson, K.D.; Kolat, A.I.; Culbertson, M.R. Growth and metabolism of inositol starved Saccharomyces cerevisiae. J. Bacteriol. 1977, 130, 472–484. [Google Scholar] [CrossRef]
- Guérlin, R.; Beauregard, P.B.; Leroux, A.; Rokeach, L.A. Calnexin regulates apoptosis induced by inositol starvation in fission yeast. PLoS ONE 2009, 4, e6244. [Google Scholar] [CrossRef]
- Eisenberg, T.; Büttner, S. Lipids and cell death in yeast. FEMS Yeast Res. 2014, 14, 179–197. [Google Scholar] [CrossRef]
- Fakas, S.; Qiu, Y.; Dixon, J.L.; Han, G.S.; Ruggles, K.V.; Garbarino, J.; Sturley, S.L.; Carman, G.M. Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 2011, 286, 29074–29085. [Google Scholar] [CrossRef]
- Garbarino, J.; Padamsee, M.; Wilcox, L.; Oelkers, P.M.; D’Ambrosio, D.; Ruggles, K.V.; Ramsey, N.; Jabado, O.; Turkish, A.; Sturley, S.L. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 2009, 284, 30994–31005. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, T.C.; De Moraes, L.M.P.; Campos, É.G. Cell density-dependent linoleic acid toxicity to Saccharomyces cerevisiae. FEMS Yeast Res. 2011, 11, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Chen, X.; Milanova, S.; Santos, C.; Petranovic, D. PUFA-induced cell death is mediated by Yca1p-dependent and -independent pathways, and is reduced by vitamin C in yeast. FEMS Yeast Res. 2016, 16, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, K.; Muneoka, T.; Watanabe, Y.; Karashima, T.; Kitagaki, H.; Funato, K. Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol. Microbiol. 2012, 86, 1246–1261. [Google Scholar] [CrossRef]
- Rego, A.; Costa, M.; Chaves, S.R.; Matmati, N.; Pereira, H.; Sousa, M.J.; Moradas-Ferreira, P.; Hannun, Y.A.; Costa, V.; Côrte-Real, M. Modulation of Mitochondrial Outer Membrane Permeabilization and Apoptosis by Ceramide Metabolism. PLoS ONE 2012, 7, e48571. [Google Scholar] [CrossRef]
- Carmona-Gutiérrez, D.; Bauer, M.A.; Ring, J.; Knaue, H.R.; Eisenberg, T.; Büttner, S.; Ruckenstuhl, C.; Reisenbichler, A.; Magnes, C.; Rechberger, G.N.; et al. The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis. 2011, 2, e161. [Google Scholar] [CrossRef]
- Pacheco, A.; Azevedo, F.; Rego, A.; Santos, J.; Chaves, S.R.; Côrte-Real, M.; Sousa, M.J. C2-phytoceramide perturbs lipid rafts and cell integrity in Saccharomyces cerevisiae in a sterol-dependent manner. PLoS ONE 2013, 8, 1–12. [Google Scholar] [CrossRef][Green Version]
- Kavakçıoğlu, B.; Tarhan, L. Yeast caspase-dependent apoptosis in Saccharomyces cerevisiae BY4742 induced by antifungal and potential antitumor agent clotrimazole. Arch. Microbiol. 2018, 200, 97–106. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. A novel mechanism of fluconazole: Fungicidal activity through dose-dependent apoptotic responses in Candida albicans. Microbiology 2018, 164, 194–204. [Google Scholar] [CrossRef]
- Dai, B.D.; Wang, Y.; Li, D.D.; Xu, Y.; Liang, R.M.; Zhao, L.X.; Cao, Y.B.; Jia, J.H.; Jiang, Y.Y. Hsp90 Is Involved in Apoptosis of Candida albicans by Regulating the Calcineurin-Caspase Apoptotic Pathway. PLoS ONE 2012, 7, 1–11. [Google Scholar] [CrossRef][Green Version]
- Laprade, D.J.; Brown, M.S.; McCarthy, M.L.; Ritch, J.J.; Austriaco, N.P.G. Filamentation protects Candida albicans from amphotericin b-induced programmed cell death via a mechanism involving the yeast metacaspase, MCA1. Microb. Cell 2016, 3, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Djordjevic, J.T.; Oei, J.B.; Desmarini, D.; Schibeci, S.D.; Jolliffe, K.A.; Sorrell, T.C. Miltefosine induces apoptosis-like cell death in yeast via Cox9p in cytochrome c oxidase. Mol. Pharmacol. 2011, 80, 476–485. [Google Scholar] [CrossRef]
- Biswas, C.; Zuo, X.; Chen, S.C.A.; Schibeci, S.D.; Forwood, J.K.; Jolliffe, K.A.; Sorrell, T.C.; Djordjevic, J.T. Functional disruption of yeast metacaspase, Mca1, leads to miltefosine resistance and inability to mediate miltefosine-induced apoptotic effects. Fungal Genet. Biol. 2014, 67, 71–81. [Google Scholar] [CrossRef]
- De Castro Spadari, C.; Vila, T.; Rozental, S.; Ishida, K. Miltefosine has a postantifungal effect and induces apoptosis in cryptococcus yeasts. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Zhang, H.; Gajate, C.; Yu, L.P.; Fang, Y.X.; Mollinedo, F. Mitochondrial-derived ROS in edelfosine-induced apoptosis in yeasts and tumor cells. Acta Pharmacol. Sin. 2007, 28, 888–894. [Google Scholar] [CrossRef]
- Bitew, T.; Sveen, C.E.; Heyne, B.; Zaremberg, V. Vitamin E prevents lipid raft modifications induced by an anti-cancer lysophospholipid and abolishes a Yap1-mediated stress response in yeast. J. Biol. Chem. 2010, 285, 25731–25742. [Google Scholar] [CrossRef]
- Valachovic, M.; Garaiova, M.; Holic, R.; Hapala, I. Squalene is lipotoxic to yeast cells defective in lipid droplet biogenesis. Biochem. Biophys. Res. Commun. 2016, 469, 1123–1128. [Google Scholar] [CrossRef]
- Scariot, F.J.; Jahn, L.; Delamare, A.P.L.; Echeverrigaray, S. Necrotic and apoptotic cell death induced by Captan on Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 1–7. [Google Scholar] [CrossRef]
- Scariot, F.J.; Jahn, L.; Delamare, A.P.L.; Echeverrigaray, S. Necrotic cell death induced by dithianon on Saccharomyces cerevisiae. Pestic. Biochem. Physiol. 2018, 149, 137–142. [Google Scholar] [CrossRef]
- Madeo, F.; Fröhlich, E.; Ligr, M.; Grey, M.; Sigrist, S.J.; Wolf, D.H.; Fröhlich, K.U. Oxygen stress: A regulator of apoptosis in yeast. J. Cell Biol. 1999, 145, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Kwolek-Mirek, M.; Zadrąg-Tęcza, R.; Bednarska, S.; Bartosz, G. Acrolein-Induced Oxidative Stress and Cell Death Exhibiting Features of Apoptosis in the Yeast Saccharomyces cerevisiae Deficient in SOD1. Cell Biochem. Biophys. 2015, 71, 1525–1536. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Almeida, T.; Marques, M.; Mojzita, D.; Amorim, M.A.; Silva, R.D.; Almeida, B.; Rodrigues, P.; Ludovico, P.; Hohmann, S.; Moradas-Ferreira, P.; et al. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol. Biol. Cell 2008, 19, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Cao, C.L.; Liu, Y.L.; Wang, J.; Li, J.; Li, S.Y.; Deng, Y. Identification of the genetic requirements for zinc tolerance and toxicity in saccharomyces cerevisiae. G3 Genes Genomes Genet. 2020, 10, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhou, B. Copper and manganese induce yeast apoptosis via different pathways. Mol. Biol. Cell 2007, 18, 4741–4749. [Google Scholar] [CrossRef]
- Wu, L.; Yi, H.; Zhang, H. Reactive oxygen species and Ca2+ are involved in sodium arsenite-induced cell killing in yeast cells. FEMS Microbiol. Lett. 2013, 343, 57–63. [Google Scholar] [CrossRef]
- Du, L.; Yu, Y.; Chen, J.; Liu, Y.; Xia, Y.; Chen, Q.; Liu, X. Arsenic induces caspase- and mitochondria-mediated apoptosis in Saccharomyces cerevisiae. FEMS Yeast Res. 2007, 7, 860–865. [Google Scholar] [CrossRef]
- Du, L.; Yu, Y.; Li, Z.; Chen, J.; Liu, Y.; Xia, Y.; Liu, X. Tim18, a component of the mitochondrial translocator, mediates yeast cell death induced by arsenic. Biochemistry 2007, 72, 843–847. [Google Scholar] [CrossRef]
- Peyroche, G.; Saveanu, C.; Dauplais, M.; Lazard, M.; Beuneu, F.; Decourty, L.; Malabat, C.; Jacquier, A.; Blanquet, S.; Plateau, P. Sodium selenide toxicity is mediated by O2-dependent DNA breaks. PLoS ONE 2012, 7, e20592. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Bzducha-Wróbel, A.; Kot, A.M. Correction to: Effect of Selenium on Lipid and Amino Acid Metabolism in Yeast Cells (Biological Trace Element Research, (2019), 187, 1, (316-327), 10.1007/s12011-018-1342-x). Biol. Trace Elem. Res. 2019, 187, 328. [Google Scholar] [CrossRef]
- Izquierdo, A.; Casas, C.; Herrero, E. Selenite-induced cell death in Saccharomyces cerevisiae: Protective role of glutaredoxins. Microbiology 2010, 156, 2608–2620. [Google Scholar] [CrossRef]
- Ludovico, P.; Sousa, M.J.; Silva, M.T.; Leão, C.; Côrte-Real, M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 2001, 147, 2409–2415. [Google Scholar] [CrossRef]
- Chaves, S.R.; Rego, A.; Martins, V.M.; Santos-Pereira, C.; Sousa, M.J.; Côrte-Real, M. Regulation of cell death induced by acetic acid in yeasts. Front. Cell Dev. Biol. 2021, 9, 642375. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Pereira, C.; Sousa, M.J.; Antonacci, L.; Passarella, S.; Côrte-Real, M.; Marra, E.; Giannattasio, S. YCA1 participates in the acetic acid induced yeast programmed cell death also in a manner unrelated to its caspase-like activity. FEBS Lett. 2006, 580, 6880–6884. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Passarella, S.; Marra, E.; Giannattasio, S. Knock-out of metacaspase and/or cytochrome c results in the activation of a ROS-independent acetic acid-induced programmed cell death pathway in yeast. FEBS Lett. 2010, 584, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, S.; Knorre, D.; Smirnova, E.; Markova, O.; Pozniakovsky, A.; Skulachev, V.; Severin, F. Ysp2 mediates death of yeast induced by amiodarone or intracellular acidification. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Cascio, V.; Gittings, D.; Merloni, K.; Hurton, M.; Laprade, D.; Austriaco, N. S-Adenosyl-L-Methionine protects the probiotic yeast, Saccharomyces boulardii, from acid-induced cell death. BMC Microbiol. 2013, 13, 1–11. [Google Scholar] [CrossRef]
- Lastauskiene, E.; Zinkevičiene, A.; Čitavičius, D. Ras/PKA signal transduction pathway participates in the regulation of Saccharomyces cerevisiae cell apoptosis in an acidic environment. Biotechnol. Appl. Biochem. 2014, 61, 3–10. [Google Scholar] [CrossRef]
- Giannattasio, S.; Guaragnella, N.; Corte-Real, M.; Passarella, S.; Marra, E. Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid-induced programmed cell death. Gene 2005, 354, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Stirpe, M.; Marzulli, D.; Mazzoni, C.; Giannattasio, S. Acid stress triggers resistance to acetic acid-induced regulated cell death through Hog1 activation which requires RTG2 in yeast. Oxid. Med. Cell. Longev. 2019, 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, J.M.; Contreras, M.F.; Faller, R.; Longo, M.L. Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys. J. 2012, 102, 507–516. [Google Scholar] [CrossRef]
- Kitagaki, H.; Araki, Y.; Funato, K.; Shimoi, H. Ethanol-induced death in yeast exhibits features of apoptosis mediated by mitochondrial fission pathway. FEBS Lett. 2007, 581, 2935–2942. [Google Scholar] [CrossRef] [PubMed]
- Creutz, C.E. Expression of Metazoan Annexins in Yeast Provides Protection Against Deleterious Effects of the Biofuel Isobutanol. Sci. Rep. 2019, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal Peptides as Therapeutic Agents. Front. Cell. Infect. Microbiol. 2020, 10, e02123-20. [Google Scholar] [CrossRef]
- Brauer, V.S.; Rezende, C.P.; Pessoni, A.M.; De Paula, R.G.; Rangappa, K.S.; Nayaka, S.C.; Gupta, V.K.; Almeida, F. Antifungal agents in agriculture: Friends and foes of public health. Biomolecules 2019, 9, 1–21. [Google Scholar] [CrossRef]
- Yu, Q.; Zhang, B.; Li, J.; Zhang, B.; Wang, H.; Li, M. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radic. Biol. Med. 2016, 99, 572–583. [Google Scholar] [CrossRef]
- Endo, K.; Mizuguchi, M.; Harata, A.; Itoh, G.; Tanaka, K. Nocodazole induces mitotic cell death with apoptotic-like features in Saccharomyces cerevisiae. FEBS Lett. 2010, 584, 2387–2392. [Google Scholar] [CrossRef]
- Enoiu, M.; Jiricny, J.; Schärer, O.D. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012, 40, 8953–8964. [Google Scholar] [CrossRef]
- Tanida, S.; Mizoshita, T.; Ozeki, K.; Tsukamoto, H.; Kamiya, T.; Kataoka, H.; Sakamuro, D.; Joh, T. Mechanisms of cisplatin-induced apoptosis and of cisplatin sensitivity: Potential of BIN1 to act as a potent predictor of cisplatin sensitivity in gastric cancer treatment. Int. J. Surg. Oncol. 2012, 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Mariani, D.; Castro, F.A.V.; Almeida, L.G.; Fonseca, F.L.; Pereira, M.D. Protection against cisplatin in calorie-restricted Saccharomyces cerevisiae is mediated by the nutrient-sensor proteins Ras2, Tor1, or Sch9 through its target Glutathione. FEMS Yeast Res. 2014, 14, 1147–1159. [Google Scholar] [CrossRef]
- Wei, H.; Tan, G.; Qiu, S.; Kong, G.; Yong, P.; Koh, C.; Ooi, T.H.; Lim, S.Y.; Wong, P.; Gan, S.U.; et al. One-step derivation of cardiomyocytes and mesenchymal stem cells from human pluripotent stem cells. Stem Cell Res. 2012, 9, 87–100. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grosfeld, E.V.; Bidiuk, V.A.; Mitkevich, O.V.; Ghazy, E.S.M.O.; Kushnirov, V.V.; Alexandrov, A.I. A Systematic Survey of Characteristic Features of Yeast Cell Death Triggered by External Factors. J. Fungi 2021, 7, 886. https://doi.org/10.3390/jof7110886
Grosfeld EV, Bidiuk VA, Mitkevich OV, Ghazy ESMO, Kushnirov VV, Alexandrov AI. A Systematic Survey of Characteristic Features of Yeast Cell Death Triggered by External Factors. Journal of Fungi. 2021; 7(11):886. https://doi.org/10.3390/jof7110886
Chicago/Turabian StyleGrosfeld, Erika V., Victoria A. Bidiuk, Olga V. Mitkevich, Eslam S. M. O. Ghazy, Vitaliy V. Kushnirov, and Alexander I. Alexandrov. 2021. "A Systematic Survey of Characteristic Features of Yeast Cell Death Triggered by External Factors" Journal of Fungi 7, no. 11: 886. https://doi.org/10.3390/jof7110886
APA StyleGrosfeld, E. V., Bidiuk, V. A., Mitkevich, O. V., Ghazy, E. S. M. O., Kushnirov, V. V., & Alexandrov, A. I. (2021). A Systematic Survey of Characteristic Features of Yeast Cell Death Triggered by External Factors. Journal of Fungi, 7(11), 886. https://doi.org/10.3390/jof7110886






