Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host
Abstract
1. Introduction
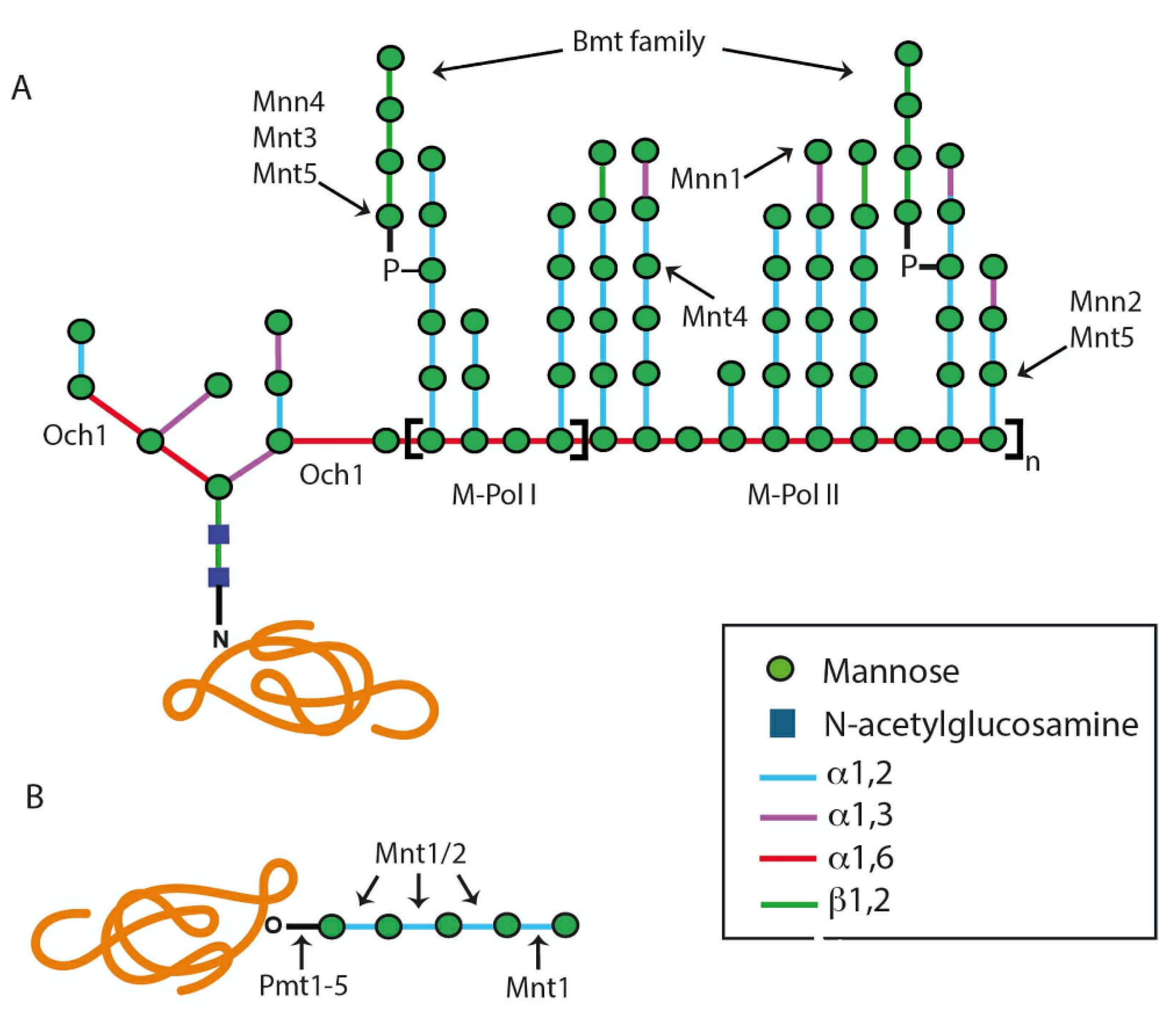
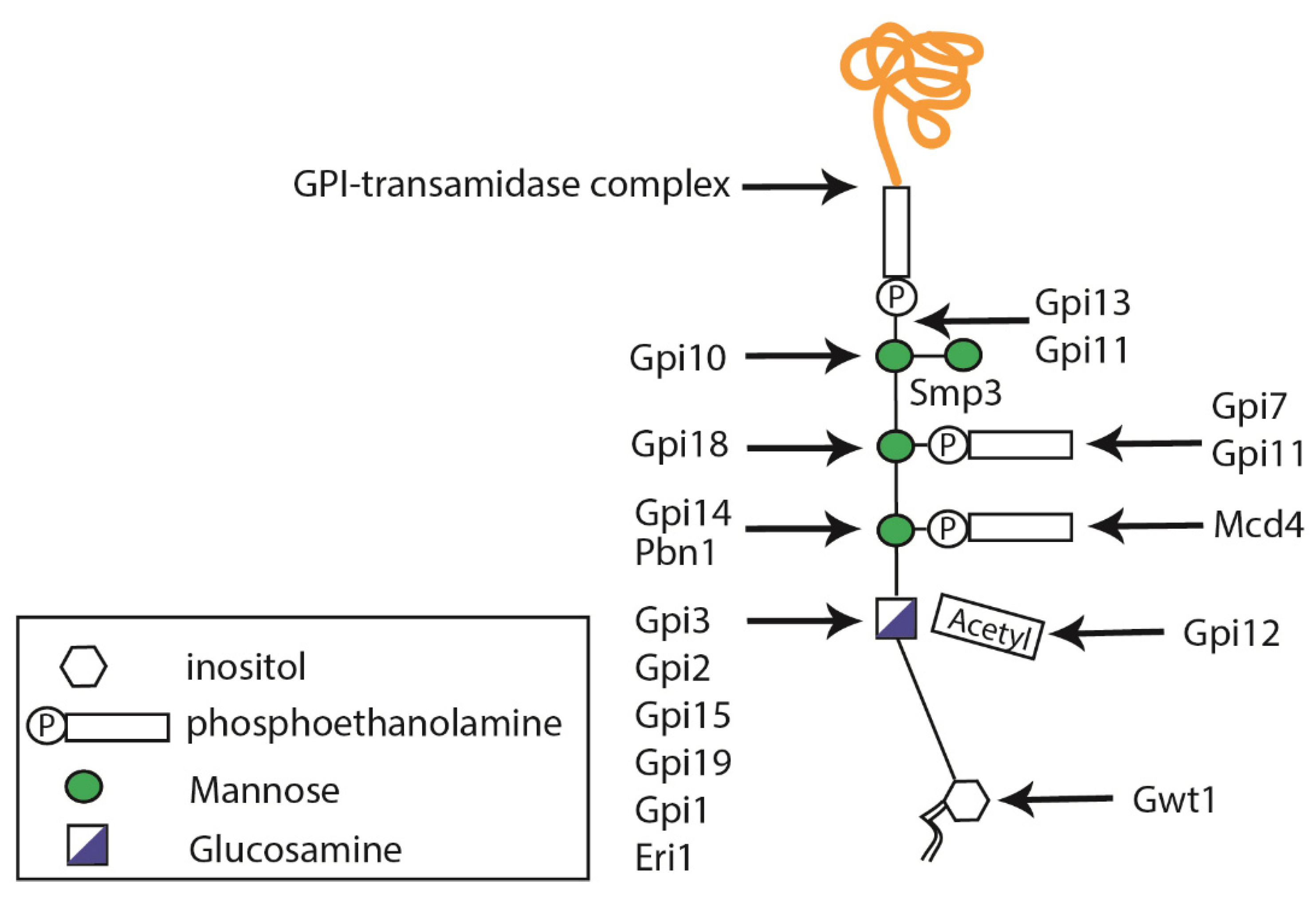
2. The Candida albicans Protein Glycosylation Pathways
2.1. The N-Linked Glycosylation Pathway
2.1.1. Assembly of Lipid-Linked Oligosaccharide
2.1.2. Modification of the N-Linked Glycan Core by Glycosidases and Transferases
2.2. The O-Linked Glycosylation Pathway
2.3. Glycosylphosphatidylinositol Anchor Synthesis
3. Protein Glycosylation in Other Medically Relevant Fungal Pathogens
4. Relevance of Fungal Protein Glycosylations during Host–Pathogen Interaction
4.1. Relevance of N-Linked Glycosylations during Host–Pathogen Interaction
4.2. Relevance of O-Linked Glycosylation during Host–Pathogen Interaction
5. Relevance of Glycosylphosphatidylinositol Anchors during Host–Pathogen Interaction
6. Relevance of Non-Conventional Glycosylation Processes during Host–Pathogen Interaction
7. Protein Glycosylation as Potential Targets of Antifungal Drugs
8. Protein Glycosylation as a Target for the Development of Antifungal Vaccines
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, B.; Qing, X.; Liao, J.; Zhuo, K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9, 1022. [Google Scholar] [CrossRef]
- Kornfeld, R.; Kornfeld, S. Assembly of Asparagine-Linked Oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef]
- Herscovics, A.; Orlean, P. Glycoprotein biosynthesis in yeast. FASEB J. 1993, 7, 540–550. [Google Scholar] [CrossRef]
- Gavel, Y.; von Heijne, G.; Creaser, E.; Murali, C.; Britt, K. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. Des. Sel. 1990, 3, 433–442. [Google Scholar] [CrossRef]
- Tanner, W.; Lehle, L. Protein glycosylation in yeast. Biochim. Biophys. Acta Rev. Biomembr. 1987, 906, 81–99. [Google Scholar] [CrossRef]
- Yadav, U.; Khan, M.A. Targeting the GPI biosynthetic pathway. Pathog. Glob. Health. 2018, 112, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.-Y.; Chang, C.-F.; Lan, C.-Y. The interaction between Carbohydrates and the Antimicrobial Peptide P-113Tri is Involved in the Killing of Candida albicans. Microorganisms 2020, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.F.; Pérez-García, L.A.; Martínez-Álvarez, J.A.; Mora-Montes, H.M. Role of the Fungal Cell Wall in Pathogenesis and Antifungal Resistance. Curr. Fungal Infect. Rep. 2012, 6, 275–282. [Google Scholar] [CrossRef]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P. Tasting the fungal cell wall. Cell. Microbiol. 2010, 12, 863–872. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Martinez-Duncker, I.; Díaz-Jímenez, D.F.; Mora-Montes, H.M. Comparative Analysis of Protein Glycosylation Pathways in Humans and the Fungal Pathogen Candida albicans. Int. J. Microbiol. 2014, 2014, 267497. [Google Scholar] [CrossRef]
- Parodi, A.J. N-Glycosylation in trypanosomatid protozoa. Glycobiology 1993, 3, 193–199. [Google Scholar] [CrossRef]
- Burda, P.; Aebi, M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta Gen. Subj. 1999, 1426, 239–257. [Google Scholar] [CrossRef]
- Lehle, L.; Strahl, S.; Tanner, W. Protein Glycosylation, Conserved from Yeast to Man: A Model Organism Helps Elucidate Congenital Human Diseases. Angew. Chem. Int. Ed. 2006, 45, 6802–6818. [Google Scholar] [CrossRef]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta Bioenerg. 2013, 1833, 2430–2437. [Google Scholar] [CrossRef]
- Dean, N. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta Gen. Subj. 1999, 1426, 309–322. [Google Scholar] [CrossRef]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, T.; Dallner, G. The biological role of dolichol. Biochem. J. 1988, 251, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Juchimiuk, M.; Kruszewska, J.; Palamarczyk, G. Dolichol phosphate mannose synthase from the pathogenic yeast Candida albicans is a multimeric enzyme. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 2265–2275. [Google Scholar] [CrossRef]
- Samuelson, J.; Banerjee, S.; Magnelli, P.; Cui, J.; Kelleher, D.J.; Gilmore, R.; Robbins, P.W. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA 2005, 102, 1548–1553. [Google Scholar] [CrossRef] [PubMed]
- Niewiadomska, M.; Janik, A.; Perlińska-Lenart, U.; Piłsyk, S.; Palamarczyk, G.; Kruszewska, J.S. The role of Alg13 N-acetylglucosaminyl transferase in the expression of pathogenic features of Candida albicans. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Ponce-Noyola, P.; Villagómez-Castro, J.C.; Gow, N.A.; Flores-Carreón, A.; López-Romero, E. Protein glycosylation in Candida. Futur. Microbiol. 2009, 4, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Lennarz, W.J. Oligosaccharyltransferase: A Complex Multisubunit Enzyme of the Endoplasmic Reticulum. Biochem. Biophys. Res. Commun. 1999, 266, 684–689. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Díaz-Jiménez, D.F.; López-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.P.; Odds, F.C.; et al. Endoplasmic Reticulum α-Glycosidases of Candida albicans Are Required for N Glycosylation, Cell Wall Integrity, and Normal Host-Fungus Interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef]
- Nishikawa, A.; Poster, J.B.; Jigami, Y.; Dean, N. Molecular and Phenotypic Analysis of CaVRG4, Encoding an Essential Golgi Apparatus GDP-Mannose Transporter. J. Bacteriol. 2002, 184, 29–42. [Google Scholar] [CrossRef]
- Bates, S.; Hughes, H.B.; Munro, C.; Thomas, W.P.; MacCallum, D.; Bertram, G.; Atrih, A.; Ferguson, M.; Brown, A.J.; Odds, F.C.; et al. Outer Chain N-Glycans Are Required for Cell Wall Integrity and Virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef]
- Mille, C.; Bobrowicz, P.; Trinel, P.-A.; Li, H.; Maes, E.; Guerardel, Y.; Fradin, C.; Martínez-Esparza, M.; Davidson, R.C.; Janbon, G.; et al. Identification of a New Family of Genes Involved in β-1,2-Mannosylation of Glycans in Pichia pastoris and Candida albicans. J. Biol. Chem. 2008, 283, 9724–9736. [Google Scholar] [CrossRef]
- González-Hernández, R.J.; Jin, K.; Hernández-Chávez, M.J.; Díaz-Jiménez, D.F.; Trujillo-Esquivel, E.; Clavijo-Giraldo, D.M.; Tamez-Castrellón, A.K.; Franco, B.; Gow, N.A.R.; Mora-Montes, H.M. Phosphomannosylation and the Functional Analysis of the Extended Candida albicans MNN4-Like Gene Family. Front. Microbiol. 2017, 8, 2156. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Díaz-Jiménez, D.F.; Kullberg, B.J.; Brown, A.J.; Odds, F.C.; et al. A Multifunctional Mannosyltransferase Family in Candida albicans Determines Cell Wall Mannan Structure and Host-Fungus Interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.F.; Mora-Montes, H.M.; Hernández-Cervantes, A.; Luna-Arias, J.P.; Gow, N.A.; Flores-Carreón, A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 2012, 419, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Goto, M. ProteinO-Glycosylation in Fungi: Diverse Structures and Multiple Functions. Biosci. Biotechnol. Biochem. 2007, 71, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.; Bates, S.; Buurman, E.T.; Hughes, H.B.; MacCallum, D.; Bertram, G.; Atrih, A.; Ferguson, M.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans Are Partially Redundant α-1,2-Mannosyltransferases That Participate in O-Linked Mannosylation and Are Required for Adhesion and Virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 405–420. [Google Scholar] [CrossRef]
- Newman, H.A.; Romeo, M.J.; Lewis, S.E.; Yan, B.C.; Orlean, P.; Levin, D.E. Gpi19, the Saccharomyces cerevisiae Homologue of Mammalian PIG-P, Is a Subunit of the Initial Enzyme for Glycosylphosphatidylinositol Anchor Biosynthesis. Eukaryot. Cell 2005, 4, 1801–1807. [Google Scholar] [CrossRef]
- Grimme, S.J.; Colussi, P.A.; Taron, C.H.; Orlean, P. Deficiencies in the essential Smp3 mannosyltransferase block glycosylphosphatidylinositol assembly and lead to defects in growth and cell wall biogenesis in Candida albicans. Microbiology 2004, 150, 3115–3128. [Google Scholar] [CrossRef][Green Version]
- Peter, O.; Menon, A. Thematic review series: Lipid Posttranslational Modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar] [CrossRef]
- Fraering, P.; Imhof, I.; Meyer, U.; Strub, J.-M.; Van Dorsselaer, A.; Vionnet, C.; Conzelmann, A. The GPI Transamidase Complex of Saccharomyces cerevisiae Contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 2001, 12, 3295–3306. [Google Scholar] [CrossRef]
- Kudoh, A.; Okawa, Y.; Shibata, N. Significant structural change in both O- and N-linked carbohydrate moieties of the antigenic galactomannan from Aspergillus fumigatus grown under different culture conditions. Glycobiology 2014, 25, 74–87. [Google Scholar] [CrossRef]
- Latgé, J.P.; Mouyna, I.; Tekaia, F.; Beauvais, A.; Debeaupuis, J.P.; Nierman, W. Specific molecular features in the organization and biosynthesis of the cell wall ofAspergillus fumigatus. Med. Mycol. 2005, 43, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Tefsen, B.; Ram, A.F.J.; Van Die, I.; Routier, F. Galactofuranose in eukaryotes: Aspects of biosynthesis and functional impact. Glycobiology 2011, 22, 456–469. [Google Scholar] [CrossRef]
- Katafuchi, Y.; Li, Q.; Tanaka, Y.; Shinozuka, S.; Kawamitsu, Y.; Izumi, M.; Ekino, K.; Mizuki, K.; Takegawa, K.; Shibata, N.; et al. GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 2017, 27, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. gfsAencodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen ofO-glycan inAspergillus nidulansandAspergillus fumigatus. Mol. Microbiol. 2013, 90, 1054–1073. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Schmalhorst, P.; Dörk, T.; Ferrieres, V.; Routier, F.H. A Single UDP-galactofuranose Transporter Is Required for Galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 2009, 284, 33859–33868. [Google Scholar] [CrossRef]
- Henry, C.; Li, J.; Danion, F.; Alcazar-Fuoli, L.; Mellado, E.; Beau, R.; Jouvion, G.; Latgé, J.-P.; Fontaine, T. Two KTR Mannosyltransferases Are Responsible for the Biosynthesis of Cell Wall Mannans and Control Polarized Growth in Aspergillus fumigatus. mBio 2019, 10, e02647-18. [Google Scholar] [CrossRef]
- Onoue, T.; Tanaka, Y.; Hagiwara, D.; Ekino, K.; Watanabe, A.; Ohta, K.; Kamei, K.; Shibata, N.; Goto, M.; Oka, T. Identification of Two Mannosyltransferases Contributing to Biosynthesis of the Fungal-type Galactomannan α-Core-Mannan Structure in Aspergillus fumigatus. Sci. Rep. 2018, 8, 16918. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P.; Beauvais, A.; Chamilos, G. The Cell Wall of the Human Fungal PathogenAspergillus fumigatus: Biosynthesis, Organization, Immune Response, and Virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef]
- Henry, C.; Fontaine, T.; Heddergott, C.; Robinet, P.; Aimanianda, V.; Beau, R.; Beauvais, A.; Mouyna, I.; Prevost, M.-C.; Fekkar, A.; et al. Biosynthesis of cell wall mannan in the conidium and the mycelium ofAspergillusfumigatus. Cell. Microbiol. 2016, 18, 1881–1891. [Google Scholar] [CrossRef]
- Klutts, J.S.; Yoneda, A.; Reilly, M.C.; Bose, I.; Doering, T.L. Glycosyltransferases and their products: Cryptococcal variations on fungal themes. FEMS Yeast Res. 2006, 6, 499–512. [Google Scholar] [CrossRef]
- Rodrigues, M.; Dobroff, A.S.S.; Couceiro, J.N.D.S.S.; Alviano, C.S.; Schauer, R.; Travassos, L.R. Sialylglycoconjugates and sialyltransferase activity in the fungus Cryptococcus neoformans. Glycoconj. J. 2002, 19, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Doering, T.L. How Sweet it is! Cell Wall Biogenesis and Polysaccharide Capsule Formation inCryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Schutzbach, J.; Ankel, H.; Brockhausen, I. Synthesis of cell envelope glycoproteins of Cryptococcus laurentii. Carbohydr. Res. 2007, 342, 881–893. [Google Scholar] [CrossRef][Green Version]
- Lee, D.-J.; Bahn, Y.-S.; Kim, H.-J.; Chung, S.-Y.; Kang, H.A. Unraveling the Novel Structure and Biosynthetic Pathway of O-Linked Glycans in the Golgi Apparatus of the Human Pathogenic Yeast Cryptococcus neoformans. J. Biol. Chem. 2015, 290, 1861–1873. [Google Scholar] [CrossRef]
- Reilly, M.; Aoki, K.; Wang, Z.A.; Skowyra, M.; Williams, M.; Tiemeyer, M.; Doering, T.L. A Xylosylphosphotransferase of Cryptococcus neoformans Acts in Protein O-Glycan Synthesis. J. Biol. Chem. 2011, 286, 26888–26899. [Google Scholar] [CrossRef]
- Bezerra, L.L.L.-B.L. Sporothrix schenckii Cell Wall Peptidorhamnomannans. Front. Microbiol. 2011, 2, 243. [Google Scholar] [CrossRef]
- Tamez-Castrellón, A.K.; van der Beek, S.L.; López-Ramírez, L.A.; Martínez-Duncker, I.; Lozoya-Pérez, N.E.; van Sorge, N.M.; Mora-Montes, H.M. Disruption of protein rhamnosylation affects the Sporothrix schenckii-host interaction. Cell Surf. 2021, 7, 100058. [Google Scholar] [CrossRef]
- Lopes-Bezerra, L.M.; Mora-Montes, H.M.; Zhang, Y.; Nino-Vega, G.; Rodrigues, A.M.; De Camargo, Z.P.; De Hoog, S. Sporotrichosis between 1898 and 2017: The evolution of knowledge on a changeable disease and on emerging etiological agents. Med. Mycol. 2018, 56, S126–S143. [Google Scholar] [CrossRef]
- Cutler, J.E. N-glycosylation of yeast, with emphasis on Candida albicans. Med Mycol. 2001, 39 (Suppl. S1), 75–86. [Google Scholar] [CrossRef]
- Ernst, J.F.; Prill, S.K.-H. O-Glycosylation. Med. Mycol. 2001, 39, 67–74. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, H.; Ouyang, H.; Li, Y.; Jin, C. Afcwh41is required for cell wall synthesis, conidiation, and polarity inAspergillus fumigatus. FEMS Microbiol. Lett. 2008, 289, 155–165. [Google Scholar] [CrossRef][Green Version]
- Brunke, S.; Quintin, J.; Kasper, L.; Jacobsen, I.D.; Richter, M.E.; Hiller, E.; Schwarzmüller, T.; d’Enfert, C.; Kuchler, K.; Rupp, S.; et al. Of mice, flies--and men? Comparing fungal infection models for large-scale screening efforts. Dis. Model Mech. 2015, 8, 473–486. [Google Scholar] [CrossRef]
- Mukaremera, L.; Lee, K.K.; Mora-Montes, H.M.; Gow, N.A.R. Candida albicans Yeast, Pseudohyphal, and Hyphal Morphogenesis Differentially Affects Immune Recognition. Front. Immunol. 2017, 8, 629. [Google Scholar] [CrossRef]
- Lozoya-Pérez, N.E.; Casas-Flores, S.; de Almeida, J.R.F.; Martínez-Álvarez, J.A.; López-Ramírez, L.A.; Jannuzzi, G.P.; Trujillo-Esquivel, E.; Estrada-Mata, E.; Almeida, S.R.; Franco, B.; et al. Silencing of OCH1 unveils the role of Sporothrix schenckii N-linked glycans during the host–fungus interaction. Infect. Drug Resist. 2018, 12, 67–85. [Google Scholar] [CrossRef]
- Perez-Garcia, L.A.; Csonka, K.; Flores-Carreon, A.; Estrada-Mata, E.; Mellado-Mojica, E.; Nemeth, T.; Lopez-Ramirez, L.A.; Toth, R.; Lopez, M.G.; Vizler, C.; et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front. Microbiol. 2016, 7, 306. [Google Scholar] [CrossRef]
- Csonka, K.; Vadovics, M.; Marton, A.; Vágvölgyi, C.; Zajta, E.; Tóth, A.; Tóth, R.; Vizler, C.; Tiszlavicz, L.; Mora-Montes, H.M.; et al. Investigation of OCH1 in the Virulence of Candida parapsilosis Using a New Neonatal Mouse Model. Front. Microbiol. 2017, 8, 1197. [Google Scholar] [CrossRef] [PubMed]
- Lozoya-Pérez, N.E.; García-Carnero, L.C.; Martínez-Álvarez, J.A.; Martínez-Duncker, I.; Mora-Montes, H.M. Tenebrio molitor as an Alternative Model to Analyze the Sporothrix Species Virulence. Infect. Drug Resist. 2021, 14, 2059–2072. [Google Scholar] [CrossRef] [PubMed]
- Kotz, A.; Wagener, J.; Engel, J.; Routier, F.H.; Echtenacher, B.; Jacobsen, I.; Heesemann, J.; Ebel, F. Approaching the Secrets of N-Glycosylation in Aspergillus fumigatus: Characterization of the AfOch1 Protein. PLoS ONE 2010, 5, e15729. [Google Scholar] [CrossRef] [PubMed]
- Lambou, K.; Perkhofer, S.; Fontaine, T.; Latge, J.-P. Comparative functional analysis of the OCH1 mannosyltransferase families in Aspergillus fumigatus and Saccharomyces cerevisiae. Yeast 2010, 27, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-N.; Lee, D.-J.; Kwon, O.; Oh, D.-B.; Bahn, Y.-S.; Kang, H.A. Unraveling Unique Structure and Biosynthesis Pathway of N-Linked Glycans in Human Fungal Pathogen Cryptococcus neoformans by Glycomics Analysis. J. Biol. Chem. 2012, 287, 19501–19515. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hall, R.A.; Cheetham, J.; Netea, M.G.; Maccallum, D.M.; Brown, A.J.; Odds, F.C.; Gow, N.A. Role of the Candida albicans MNN1 gene family in cell wall structure and virulence. BMC Res. Notes 2013, 6, 294. [Google Scholar] [CrossRef]
- Hall, R.; Bates, S.; Lenardon, M.D.; MacCallum, D.; Wagener, J.; Lowman, D.W.; Kruppa, M.D.; Williams, D.L.; Odds, F.C.; Brown, A.J.; et al. The Mnn2 Mannosyltransferase Family Modulates Mannoprotein Fibril Length, Immune Recognition and Virulence of Candida albicans. PLoS Pathog. 2013, 9, e1003276. [Google Scholar] [CrossRef]
- Hernández-Chávez, M.J.; Franco, B.; Clavijo-Giraldo, D.M.; Hernández, N.V.; Estrada-Mata, E.; Mora-Montes, H.M. Role of protein phosphomannosylation in the Candida tropicalis–macrophage interaction. FEMS Yeast Res. 2018, 18, 18. [Google Scholar] [CrossRef]
- Hobson, R.; Munro, C.; Bates, S.; MacCallum, D.; Cutler, J.E.; Heinsbroek, S.E.M.; Brown, G.; Odds, F.C.; Gow, N. Loss of Cell Wall Mannosylphosphate in Candida albicans Does Not Influence Macrophage Recognition. J. Biol. Chem. 2004, 279, 39628–39635. [Google Scholar] [CrossRef]
- Bai, C.; Xu, X.; Chan, F.-Y.; Lee, R.T.H.; Wang, Y. MNN5 Encodes an Iron-Regulated α-1,2-Mannosyltransferase Important for Protein Glycosylation, Cell Wall Integrity, Morphogenesis, and Virulence in Candida albicans. Eukaryot. Cell 2006, 5, 238–247. [Google Scholar] [CrossRef]
- Southard, S.B.; Specht, C.A.; Mishra, C.; Chen-Weiner, J.; Robbins, P.W. Molecular Analysis of the Candida albicans Homolog of Saccharomyces cerevisiae MNN9, Required for Glycosylation of Cell Wall Mannoproteins. J. Bacteriol. 1999, 181, 7439–7448. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Ouyang, H.; Voglmeir, J.; Wilson, I.B.; Jin, C. Aspergillus fumigatus Mnn9 is responsible for mannan synthesis and required for covalent linkage of mannoprotein to the cell wall. Fungal Genet. Biol. 2019, 128, 20–28. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Zou, Z.; Shen, H.; Shen, S.S.; Miao, Q.; Huang, X.; Liu, W.; Li, L.P.; Chen, S.M.; Yan, L.; et al. Mnn10 Maintains Pathogenicity in Candida albicans by Extending α-1,6-Mannose Backbone to Evade Host Dectin-1 Mediated Antifungal Immunity. PLoS Pathog. 2016, 12, e1005617. [Google Scholar] [CrossRef] [PubMed]
- West, L.; Lowman, D.W.; Mora-Montes, H.M.; Grubb, S.; Murdoch, C.; Thornhill, M.H.; Gow, N.; Williams, D.; Haynes, K. Differential Virulence of Candida glabrata Glycosylation Mutants. J. Biol. Chem. 2013, 288, 22006–22018. [Google Scholar] [CrossRef]
- Bates, S.; MacCallum, D.M.; Bertram, G.; Munro, C.A.; Hughes, H.B.; Buurman, E.T.; Brown, A.J.; Odds, F.C.; Gow, N.A. Candida albicans Pmr1p, a Secretory Pathway P-type Ca2+/Mn2+-ATPase, Is Required for Glycosylation and Virulence. J. Biol. Chem. 2005, 280, 23408–23415. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Arias, M.J.; Defosse, T.A.; Dementhon, K.; Csonka, K.; Mellado-Mojica, E.; Valério, A.D.; González-Hernández, R.J.; Courdavault, V.; Clastre, M.; Hernández, N.V.; et al. Disruption of Protein Mannosylation Affects Candida guilliermondii Cell Wall, Immune Sensing, and Virulence. Front. Microbiol. 2016, 7, 1951. [Google Scholar] [CrossRef] [PubMed]
- Pinchai, N.; Juvvadi, P.; Fortwendel, J.R.; Perfect, B.Z.; Rogg, L.E.; Asfaw, Y.G.; Steinbach, W.J. The Aspergillus fumigatus P-Type Golgi Apparatus Ca 2+ /Mn 2+ ATPase PmrA Is Involved in Cation Homeostasis and Cell Wall Integrity but Is Not Essential for Pathogenesis. Eukaryot. Cell 2010, 9, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Warit, S.; Zhang, N.; Short, A.; Walmsley, R.M.; Oliver, S.G.; Stateva, L.I. Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol. Microbiol. 2000, 36, 1156–1166. [Google Scholar] [CrossRef]
- Prill, S.K.-H.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schröppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 2004, 55, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Timpel, C.; Strahl-Bolsinger, S.; Ziegelbauer, K.; Ernst, J.F. Multiple Functions of Pmt1p-mediated ProteinO-Mannosylation in the Fungal Pathogen Candida albicans. J. Biol. Chem. 1998, 273, 20837–20846. [Google Scholar] [CrossRef]
- Timpel, C.; Zink, S.; Strahl-Bolsinger, S.; SchröppelK.; Ernst, J. Morphogenesis, Adhesive Properties, and Antifungal Resistance Depend on the Pmt6 Protein Mannosyltransferase in the Fungal Pathogen Candida albicans. J. Bacteriol. 2000, 182, 3063–3071. [Google Scholar] [CrossRef]
- Mouyna, I.; Kniemeyer, O.; Jank, T.; Loussert, C.; Mellado, E.; Aimanianda, V.; Beauvais, A.; Wartenberg, D.; Sarfati, J.; Bayry, J.; et al. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol. Microbiol. 2010, 76, 1205–1221. [Google Scholar] [CrossRef]
- Willger, S.D.; Ernst, J.F.; Alspaugh, J.A.; Lengeler, K.B. Characterization of the PMT Gene Family in Cryptococcus neoformans. PLoS ONE 2009, 4, e6321. [Google Scholar] [CrossRef]
- Rouabhia, M.; Schaller, M.; Corbucci, C.; Vecchiarelli, A.; Prill, S.K.-H.; Giasson, L.; Ernst, J.F. Virulence of the Fungal Pathogen Candida albicans Requires the Five Isoforms of Protein Mannosyltransferases. Infect. Immun. 2005, 73, 4571–4580. [Google Scholar] [CrossRef]
- Buurman, E.T.; Westwater, C.; Hube, B.; Brown, A.J.P.; Odds, F.C.; Gow, N.A.R. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 1998, 95, 7670–7675. [Google Scholar] [CrossRef]
- Umemura, M.; Okamoto, M.; Nakayama, K.-I.; Sagane, K.; Tsukahara, K.; Hata, K.; Jigami, Y. GWT1 Gene Is Required for Inositol Acylation of Glycosylphosphatidylinositol Anchors in Yeast. J. Biol. Chem. 2003, 278, 23639–23647. [Google Scholar] [CrossRef]
- Watanabe, N.-A.; Miyazaki, M.; Horii, T.; Sagane, K.; Tsukahara, K.; Hata, K. E1210, a New Broad-Spectrum Antifungal, Suppresses Candida albicans Hyphal Growth through Inhibition of Glycosylphosphatidylinositol Biosynthesis. Antimicrob. Agents Chemother. 2011, 56, 960–971. [Google Scholar] [CrossRef]
- Miyazaki, M.; Horii, T.; Hata, K.; Watanabe, N.-A.; Nakamoto, K.; Tanaka, K.; Shirotori, S.; Murai, N.; Inoue, S.; Matsukura, M.; et al. In VitroActivity of E1210, a Novel Antifungal, against Clinically Important Yeasts and Molds. Antimicrob. Agents Chemother. 2011, 55, 4652–4658. [Google Scholar] [CrossRef]
- Pfaller, M.; Duncanson, F.; Messer, S.A.; Moet, G.J.; Jones, R.N.; Castanheira, M. In Vitro Activity of a Novel Broad-Spectrum Antifungal, E1210, Tested against Aspergillus spp. Determined by CLSI and EUCAST Broth Microdilution Methods. Antimicrob. Agents Chemother. 2011, 55, 5155–5158. [Google Scholar] [CrossRef] [PubMed]
- Lavie-Richard, M.; Ibata-Ombetta, S.; Dromer, F.; Bordon-Pallier, F.; Jouault, T.; Gaillardin, C. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 2002, 44, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Garai, P.; Sethi, S.C.; Naqvi, N.; Yadav, B.; Kumar, P.; Singh, S.L.; Yadav, U.; Bhatnagar, S.; Rahul; et al. Modulation of azole sensitivity and filamentation by GPI15, encoding a subunit of the first GPI biosynthetic enzyme, in Candida albicans. Sci. Rep. 2019, 9, 8508. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, H.; Luo, Y.; Ouyang, H.; Hu, H.; Jin, C. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 2007, 64, 1014–1027. [Google Scholar] [CrossRef]
- Schmalhorst, P.S.; Krappmann, S.; Vervecken, W.; Rohde, M.; Müller, M.; Braus, G.H.; Contreras, R.; Braun, A.; Bakker, H.; Routier, F.H. Contribution of Galactofuranose to the Virulence of the Opportunistic Pathogen Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 1268–1277. [Google Scholar] [CrossRef]
- Lamarre, C.; Beau, R.; Balloy, V.; Fontaine, T.; Hoi, J.W.S.; Guadagnini, S.; Berkova, N.; Chignard, M.; Beauvais, A.; Latgé, J.-P. Galactofuranose attenuates cellular adhesion ofAspergillus fumigatus. Cell. Microbiol. 2009, 11, 1612–1623. [Google Scholar] [CrossRef]
- Netea, M.G.; Gow, N.A.; Munro, C.A.; Bates, S.; Collins, C.; Ferwerda, G.; Hobson, R.P.; Bertram, G.; Hughes, H.B.; Jansen, T.; et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 2006, 116, 1642–1650. [Google Scholar] [CrossRef]
- Cambi, A.; Netea, M.G.; Mora-Montes, H.M.; Gow, N.A.; Hato, S.V.; Lowman, D.W.; Kullberg, B.-J.; Torensma, R.; Williams, D.L.; Figdor, C.G. Dendritic Cell Interaction with Candida albicans Critically Depends on N-Linked Mannan. J. Biol. Chem. 2008, 283, 20590–20599. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Mora-Montes, H.M.; Wagener, J.; Cunningham, I.; West, L.; Haynes, K.; Brown, A.J.; Gow, N.A. Differences in fungal immune recognition by monocytes and macrophages: N-mannan can be a shield or activator of immune recognition. Cell Surf. 2020, 6, 100042. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Mrsa, V.; Hagen, I.; Deutzmann, R.; Strahl, S.; Tanner, W. O-Mannosylation precedes and potentially controls the N -glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 2003, 4, 628–632. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.G.J.; Koser, U.; Lewis, L.E.; Bain, J.M.; Mora-Montes, H.M.; Barker, R.N.; Gow, N.; Erwig, L.P. Contribution of Candida albicans Cell Wall Components to Recognition by and Escape from Murine Macrophages. Infect. Immun. 2010, 78, 1650–1658. [Google Scholar] [CrossRef]
- Harris, M.; Mora-Montes, H.M.; Gow, N.; Coote, P.J. Loss of mannosylphosphate from Candida albicans cell wall proteins results in enhanced resistance to the inhibitory effect of a cationic antimicrobial peptide via reduced peptide binding to the cell surface. Microbiology 2009, 155, 1058–1070. [Google Scholar] [CrossRef]
- Juchimiuk, M.; Orłowski, J.; Gawarecka, K.; Świeżewska, E.; Ernst, J.F.; Palamarczyk, G. Candida albicans cis-prenyltransferase Rer2 is required for protein glycosylation, cell wall integrity and hypha formation. Fungal Genet. Biol. 2014, 69, 1–12. [Google Scholar] [CrossRef]
- Janik, A.; Niewiadomska, M.; Perlińska-Lenart, U.; Lenart, J.; Kołakowski, D.; Skorupińska-Tudek, K.; Swiezewska, E.; Kruszewska, J.S.; Palamarczyk, G. Inhibition of Dephosphorylation of Dolichyl Diphosphate Alters the Synthesis of Dolichol and Hinders Protein N-Glycosylation and Morphological Transitions in Candida albicans. Int. J. Mol. Sci. 2019, 20, 5067. [Google Scholar] [CrossRef]
- Zhou, H.; Hu, H.; Zhang, L.; Li, R.; Ouyang, H.; Ming, J.; Jin, C. O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) Is Crucial for Cell Wall Integrity and Conidium Morphology, Especially at an Elevated Temperature. Eukaryot. Cell 2007, 6, 2260–2268. [Google Scholar] [CrossRef]
- Olson, G.M.; Fox, D.S.; Wang, P.; Alspaugh, J.A.; Buchanan, K.L. Role of Protein O-Mannosyltransferase Pmt4 in the Morphogenesis and Virulence of Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 222–234. [Google Scholar] [CrossRef]
- Wagener, J.; Echtenacher, B.; Rohde, M.; Kotz, A.; Krappmann, S.; Heesemann, J.; Ebel, F. The Putative α-1,2-Mannosyltransferase AfMnt1 of the Opportunistic Fungal Pathogen Aspergillus fumigatus Is Required for Cell Wall Stability and Full Virulence. Eukaryot. Cell 2008, 7, 1661–1673. [Google Scholar] [CrossRef]
- Mutz, M.; Roemer, T. The GPI anchor pathway: A promising antifungal target? Futur. Med. Chem. 2016, 8, 1387–1391. [Google Scholar] [CrossRef]
- Tsukahara, K.; Hata, K.; Nakamoto, K.; Sagane, K.; Watanabe, N.-A.; Kuromitsu, J.; Kai, J.; Tsuchiya, M.; Ohba, F.; Jigami, Y.; et al. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol. Microbiol. 2003, 48, 1029–1042. [Google Scholar] [CrossRef]
- Hong, Y.; Maeda, Y.; Watanabe, R.; Ohishi, K.; Mishkind, M.; Riezman, H.; Kinoshita, T. Pig-n, a Mammalian Homologue of Yeast Mcd4p, Is Involved in Transferring Phosphoethanolamine to the First Mannose of the Glycosylphosphatidylinositol. J. Biol. Chem. 1999, 274, 35099–35106. [Google Scholar] [CrossRef]
- Benachour, A.; Sipos, G.; Flury, I.; Reggiori, F.; Canivenc-Gansel, E.; Vionnet, C.; Conzelmann, A.; Benghezal, M. Deletion of GPI7, a Yeast Gene Required for Addition of a Side Chain to the Glycosylphosphatidylinositol (GPI) Core Structure, Affects GPI Protein Transport, Remodeling, and Cell Wall Integrity. J. Biol. Chem. 1999, 274, 15251–15261. [Google Scholar] [CrossRef]
- Ouyang, H.; Du, T.; Zhou, H.; Wilson, I.B.H.; Yang, J.; Latgé, J.-P.; Jin, C. Aspergillus fumigatus phosphoethanolamine transferase gene gpi7 is required for proper transportation of the cell wall GPI-anchored proteins and polarized growth. Sci. Rep. 2019, 9, 5857. [Google Scholar] [CrossRef]
- Victoria, G.S.; Kumar, P.; Komath, S.S. The Candida albicans homologue of PIG-P, CaGpi19p: Gene dosage and role in growth and filamentation. Microbiology 2010, 156, 3041–3051. [Google Scholar] [CrossRef]
- Victoria, G.S.; Yadav, B.; Hauhnar, L.; Jain, P.; Bhatnagar, S.; Komath, S.S. Mutual co-regulation between GPI-N-acetylglucosaminyltransferase and ergosterol biosynthesis in Candida albicans. Biochem. J. 2012, 443, 619–625. [Google Scholar] [CrossRef]
- Yadav, B.; Bhatnagar, S.; Ahmad, M.F.; Jain, P.; Pratyusha, V.A.; Kumar, P.; Komath, S.S. First Step of Glycosylphosphatidylinositol (GPI) Biosynthesis Cross-talks with Ergosterol Biosynthesis and Ras Signaling in Candida albicans. J. Biol. Chem. 2014, 289, 3365–3382. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Du, T.; Zhao, W.; Hartmann, T.; Lu, H.; Lü, Y.; Ouyang, H.; Jiang, X.; Sun, L.; Jin, C. Transcriptome and Biochemical Analysis Reveals That Suppression of GPI-Anchor Synthesis Leads to Autophagy and Possible Necroptosis in Aspergillus fumigatus. PLoS ONE 2013, 8, e59013. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Simmer, M.I.; D’Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2004, 55, 1452–1472. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C. Molecular mechanism of Aspergillus fumigatus adherence to host constituents. Curr. Opin. Microbiol. 2011, 14, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Calugi, C.; Trabocchi, A.; Guarna, A. Novel small molecules for the treatment of infections caused by Candida albicans: A patent review (2002–2010). Expert Opin. Ther. Pat. 2011, 21, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, L.A.; Martínez-Duncker, I.; Montes, H.M.M. The Endoplasmic Reticulum Alpha-Glycosidases as Potential Targets for Virus Control. Curr. Protein Pept. Sci. 2017, 18, 1090–1097. [Google Scholar] [CrossRef]
- Orchard, M.G.; Neuss, J.C.; Galley, C.M.; Carr, A.; Porter, D.W.; Smith, P.; Scopes, D.I.; Haydon, D.; Vousden, K.; Stubberfield, C.R.; et al. Rhodanine-3-acetic acid derivatives as inhibitors of fungal protein mannosyl transferase 1 (PMT1). Bioorganic Med. Chem. Lett. 2004, 14, 3975–3978. [Google Scholar] [CrossRef]
- Bai, L.; Kovach, A.; You, Q.; Kenny, A.; Li, H. Structure of the eukaryotic protein O-mannosyltransferase Pmt1−Pmt2 complex. Nat. Struct. Mol. Biol. 2019, 26, 704–711. [Google Scholar] [CrossRef]
- Salci, T.P.; Negri, M.; Abadio, A.K.R.; Bonfim-Mendonça, P.; Capoci, I.; Caparroz-Assef, S.M.; Donatti, L.; Felipe, M.S.S.; Kioshima, E.S.; Svidzinski, T.I. A new small-molecule KRE2 inhibitor against invasive Candida parapsilosis infection. Futur. Microbiol. 2017, 12, 1283–1295. [Google Scholar] [CrossRef]
- Singh, V.; Praveen, V.; Tripathi, D.; Haque, S.; Somvanshi, P.; Katti, S.B.; Tripathi, C.K.M. Isolation, characterization and antifungal docking studies of wortmannin isolated from Penicillium radicum. Sci. Rep. 2015, 5, 11948. [Google Scholar] [CrossRef]
- Sawada, Y.; Nishio, M.; Yamamoto, H.; Hatori, M.; Miyaki, T.; Konishi, M.; Oki, T. New antifungal antibiotics, pradimicins D and E. Glycine analogs of pradimicins A and C. J. Antibiot. 1990, 43, 771–777. [Google Scholar] [CrossRef]
- Takeuchi, T.; Hara, T.; Naganawa, H.; Okada, M.; Hamada, M.; Umezawat, H.; Gomi, S.; Sezaki, M.; Kondo, S. New antifungal antibiotics, benanomicins A and B from an Actinomycete. J. Antibiot. 1988, 41, 807–811. [Google Scholar] [CrossRef]
- Ueki, T.; Numata, K.-I.; Ada, Y.S.; Nakajima, T.; Fukagawa, Y.; Oki, T. Studies on the mode of antifungal action of pradimicin antibiotics. I.Lectin-mimic binding of BMY-28864 to yeast mannan in the presence of calcium. J. Antibiot. 1993, 46, 149–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Watanabe, M.; Tohyama, H.; Hiratani, T.; Watabe, H.; Inoue, S.; Kondo, S.-I.; Takeuchi, T.; Yamaguchi, H. Mode of antifungal action of benanomicin A in Saccharomyces cerevisiae. J. Antibiot. 1997, 50, 1042–1051. [Google Scholar] [CrossRef][Green Version]
- Hiramoto, F.; Nomura, N.; Furumai, T.; Oki, T.; Igarashi, Y. Apoptosis-like cell death of Saccharomyces cerevisiae induced by a mannose-binding antifungal antibiotic, pradimicin. J. Antibiot. 2003, 56, 768–772. [Google Scholar] [CrossRef]
- Oki, T.; Tenmyo, O.; Hirano, M.; Tomatsu, K.; Kamei, H. Pradimicins A, B and C: New antifungal antibiotics. II. In vitro and in vivo biological activities. J. Antibiot. 1990, 43, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Yasuoka, A.; Oka, S.; Komuro, K.; Shimizu, H.; Kitada, K.; Nakamura, Y.; Shibahara, S.; Takeuchi, T.; Kondo, S.; Shimada, K. Successful treatment of Pneumocystis carinii Pneumonia in mice with benanomicin A (ME1451). Antimicrob. Agents Chemother. 1995, 39, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Regente, M.; Taveira, G.B.; Pinedo, M.; Elizalde, M.M.; Ticchi, A.J.; Diz, M.S.S.; Carvalho, A.O.; De La Canal, L.; Gomes, V.M. A Sunflower Lectin with Antifungal Properties and Putative Medical Mycology Applications. Curr. Microbiol. 2014, 69, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.; de la Canal, L.; Pinedo, M.; Mora-Montes, H.M.; Regente, M. Effects of the binding of a Helianthus annuus lectin to Candida albicans cell wall on biofilm development and adhesion to host cells. Phytomedicine 2019, 58, 152875. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Schell, W.A.; Covel, J.; Duboc, G.; Giamberardino, C.; Kapoor, M.; Moloney, M.; Soltow, Q.A.; Tenor, J.L.; Toffaletti, D.L.; et al. In Vitro and In Vivo Evaluation of APX001A/APX001 and Other Gwt1 Inhibitors against Cryptococcus. Antimicrob. Agents Chemother. 2018, 62, e00523-18. [Google Scholar] [CrossRef]
- Fernandes, C.M.; Dasilva, D.; Haranahalli, K.; McCarthy, J.B.; Mallamo, J.; Ojima, I.; Del Poeta, M. The Future of Antifungal Drug Therapy: Novel Compounds and Targets. Antimicrob. Agents Chemother. 2020, 65, 01719–01720. [Google Scholar] [CrossRef]
- Rauseo, A.; Coler-Reilly, A.; Larson, L.; Spec, A. Hope on the Horizon: Novel Fungal Treatments in Development. Open Forum Infect. Dis. 2020, 7, ofaa016. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Boas, D.V.; Haynes, K.; Henriques, M. The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata. Biomolecules 2018, 8, 130. [Google Scholar] [CrossRef]
- Juchimiuk, M.; Pasikowska, M.; Zatorska, E.; Laudy, A.E.; Smoleńska-Sym, G.; Palamarczyk, G. Defect in dolichol-dependent glycosylation increases sensitivity of Saccharomyces cerevisiae towards anti-fungal drugs. Yeast 2010, 27, 637–645. [Google Scholar] [CrossRef]
- Harries, E.; Carmona, L.; Muñoz, A.; Ibeas, J.I.; Read, N.D.; Gandía, M.; Marcos, J.F. Genes involved in protein glycosylation determine the activity and cell internalization of the antifungal peptide PAF26 in Saccharomyces cerevisiae. Fungal Genet. Biol. 2013, 58-59, 105–115. [Google Scholar] [CrossRef]
- Zeng, G.; Xu, X.; Gao, J.; Dantas, A.D.S.; Gow, N.A.; Wang, Y. Inactivating the mannose-ethanolamine phosphotransferase Gpi7 confers caspofungin resistance in the human fungal pathogen Candida albicans. Cell Surf. 2021, 7, 100057. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Weintraub, S.T.; Lopez-Ribot, J.; Wormley, F.L., Jr. Identification and characterization of Cryptococcus neoformans protein fractions that induce protective immune responses. Proteomics 2013, 13, 3429–3441. [Google Scholar] [CrossRef] [PubMed]
- Levitz, S.M.; Huang, H.; Ostroff, G.R.; Specht, C.A. Exploiting fungal cell wall components in vaccines. Semin. Immunopathol. 2015, 37, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Pan, B.; Liao, G.; Zhao, Q.; Gao, Y.; Chai, X.; Zhuo, X.; Wu, Q.; Jiao, B.; Pan, W.; et al. Synthesis and immunological studies of β-1,2-mannan-peptide conjugates as antifungal vaccines. Eur. J. Med. Chem. 2019, 173, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Bundle, D.R.; Paszkiewicz, E.; Elsaidi, H.R.H.; Mandal, S.S.; Sarkar, S. A Three Component Synthetic Vaccine Containing a β-Mannan T-Cell Peptide Epitope and a β-Glucan Dendritic Cell Ligand. Molecules 2018, 23, 1961. [Google Scholar] [CrossRef]
- Paulovičová, L.; Paulovičová, E.; Karelin, A.A.; Tsvetkov, Y.E.; Nifantiev, N.E.; Bystrický, S. Humoral and cell-mediated immunity following vaccination with synthetic Candida cell wall mannan derived heptamannoside–protein conjugate. Int. Immunopharmacol. 2012, 14, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Machová, E.; Neščáková, Z.; Medovarská, I.; Clemons, K.V.; Martinez, M.; Chen, V.; Bystrický, S.; Stevens, D.A. Vaccination with mannan protects mice against systemic aspergillosis. Med. Mycol. 2012, 50, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Bundle, D.R.; Nycholat, C.; Costello, C.; Rennie, R.; Lipinski, T. Design of a Candida albicans Disaccharide Conjugate Vaccine by Reverse Engineering a Protective Monoclonal Antibody. ACS Chem. Biol. 2012, 7, 1754–1763. [Google Scholar] [CrossRef]
- Han, Y.; Rhew, K.Y. Comparison of two Candida mannan vaccines: The role of complement in protection against disseminated candidiasis. Arch. Pharmacal Res. 2012, 35, 2021–2027. [Google Scholar] [CrossRef] [PubMed]
- Rosario-Colon, J.; Eberle, K.; Adams, A.; Courville, E.; Xin, H. Candida Cell-Surface-Specific Monoclonal Antibodies Protect Mice against Candida auris Invasive Infection. Int. J. Mol. Sci. 2021, 22, 6162. [Google Scholar] [CrossRef] [PubMed]
| Gene | Mutant Phenotype | Impact on Virulence | References |
|---|---|---|---|
| N-Linked Glycosylation | |||
| CWH41 | Candida albicans Cellular aggregates, defects in cell wall composition. Aspergillus fumigatus Reduction in conidia formation, changes in the cell wall composition, polar growth, and hyphal elongation. | Attenuated virulence in a murine model. None found. | [26,61] |
| ROT2 | C. albicans Cellular aggregates, defects in cell wall composition. Candida glabrata None found. | Attenuated virulence in a murine model. Decreased virulence in Drosophila melanogaster. | [26,62] |
| MNS1 | C. albicans Few phenotypical changes. C. glabrata None found. | Virulence affected. Decreased virulence in a murine model of systemic candidiasis. | [26,62,63] |
| OCH1 | C. albicans Defects in the cell wall, cellular aggregates, decreased growth rate, and reduced ability to stimulate cytokine production. Candida parapsilosis Changes in cell morphology and cell wall composition, susceptibility to cell wall perturbing agents, and reduced ability to stimulate cytokine production. Sporothrix schenckii Minimal changes in the phenotype and the cell wall. A. fumigatus: Defects in ability to sporulate, non-significant changes in phenotype. Cryptococcus neoformans None found. | Attenuated virulence in a murine model. Attenuated virulence in neonate and adult murine models. Attenuated virulence in a murine model and the invertebrates Galleria mellonella and Tenebrio molitor. None found. Slightly attenuated virulence. | [26,64,65,66,67,68,69,70] |
| ANP1 | C. glabrata None found. | Decreased virulence in both murine model of systemic candidiasis and D. melanogaster. | [62] |
| MNN14 | C. albicans Defects in cell wall composition, but normal ability to stimulate cytokine production. | Virulence attenuation in a murine model of disseminated candidiasis. | [71] |
| MNN2 | C. albicans Changes in phenotype. | Attenuated virulence in G. mellonella and a murine model. | [72] |
| MNN4 | C. albicans Changes in the C. albicans–macrophage interaction. Candida tropicalis Changes in the yeast–macrophage interaction. C. glabrata None found. | Attenuated virulence in the G. mellonella model but not in a murine model. Attenuated virulence in G. mellonella model. Attenuated virulence in a murine model of systemic candidiasis. | [30,62,73,74] |
| MNN5 | C. albicans Hypersensitivity to wall perturbing agents, inability to undergo dimorphism. | Attenuated virulence in a murine model. | [75] |
| MNN9 | C. albicans Changes in the cell wall, osmotic instability, defective hyphae, and cell agglutination. A. fumigatus Defects in the cell wall, changes in morphogenesis, sensitivity to wall perturbing agents. | None found. None found. | [76,77] |
| MNN10 | C. albicans Abnormal organization of the cell wall. C. glabrata Abnormal growth in vitro | Decreased virulence. Decreased virulence in both murine model of systemic candidiasis and D. melanogaster | [62,78] |
| MNN11 | C. glabrata None found. | Virulence is not affected. | [79] |
| KRE2/MNT1 | C. albicans Cellular aggregates, susceptibility to cell wall perturbing agents, reduction in the ability to stimulate cytokine production. C. glabrata None found. | Attenuated virulence in a murine model. Virulence attenuation in both a murine model of candidiasis and D. melanogaster. | [31,62] |
| KTR3 | C. glabrata None found. | Virulence attenuation in D. melanogaster. | [62] |
| PMR1 | C. albicans Cellular aggregates, defects in the cell wall composition, hypersensitivity to wall perturbing agents. Candida guilliermondii Affected cell growth, morphology, cell wall, biofilm formation, and interaction with PBMC. A. fumigatus Defects in growth, changes in the cell wall composition, increased content of β-glucan and chitin. Hypersensitivity to cell wall inhibitors. | Attenuated virulence in a murine model. Attenuated virulence in a murine model and G. mellonella. None found. | [80,81,82] |
| ALG6 | C. glabrata None found. | Virulence attenuation in D. melanogaster. | [62] |
| ALG13 | C. albicans Defects in the hypha and biofilm formation, changes in the cell wall carbohydrate content. Defects in N-linked glycosylation. | None found. | [23] |
| DPM1 | C. albicans Susceptibility to wall perturbing agents, altered cell wall composition and affected growth, increased chitin levels. | None found. | [21] |
| DPM2 | C. albicans Susceptibility to wall perturbing agents and changes in the cell wall integrity. | None found. | [21] |
| DPM3 | C. albicans Growth rate attenuation and increased chitin levels. | None found. | [21] |
| SRB1 | C. albicans Changes in morphology, growth, sensitivity to antifungal agents and cell wall inhibitors, and glycosylation defects. | None found. | [83] |
| O-Linked Glycosylation | |||
| PMT1 | C. albicans Hypersensitivity to antifungal agents and changes in the cell wall components. C. glabrata None found. A. fumigatus Growth defects, changes in the cell wall, defects in cell morphology, and conidia formation. C. neoformans: None found. | Severely affected virulence in a murine model. Virulence attenuation in D. melanogaster. None found. Attenuated virulence in a macrophage death model and a murine model. | [62,84,85,86,87,88] |
| PMT3 | C. glabrata None found. | Decreased virulence in a murine model of systemic candidiasis. | [62] |
| PMT4 | C. albicans Hypersensitivity to antifungal agents and changes in the cell wall components. C. glabrata None found. A. fumigatus Growth defects, changes in the cell wall, defects in cell morphology, and conidia formation. C. neoformans: Morphological defects, alterations in the cell wall. | Severely affected virulence in a murine model. Decreased virulence in a murine model of systemic candidiasis. None found. Virulence affected. | [62,84,86,87,88,89] |
| PMT5 | C. albicans: Defects in morphogenesis and susceptibility to antifungal agents. | None found. | [84] |
| PMT6 | C. albicans: None found. | Severely affected virulence in a murine model. | [85,86] |
| MNT1 | C. albicans: Changes in cell morphology, inability to adhere to human buccal epithelial cells, hypersensitivity to wall perturbing agents. A. fumigatus: Reduction in the cell wall thickness, decreased cell growth. | Attenuated virulence in a murine model. Attenuated virulence. | [34,90] |
| MNT2 | C. albicans: Few defects in phenotype. | Attenuated virulence. | [34] |
| Glycosylphosphatidylinositol Anchors | |||
| GWT1 | Candida spp., Aspergillus spp., Fusarium spp., and Scedosporium spp Changes in the cell wall integrity, structure, growth, and dimorphism. C. albicans: Growth and morphological changes, reduced adherence, growth, and dimorphism. | Virulence affected. None found. | [6,91,92,93,94] |
| GPI7 | C. albicans Effects on chlamydospore formation, altered levels of plasma membrane GPI proteins, abnormal polarity, and growth. | None found. | [95] |
| GPI15 | C. albicans Presents hypofilamentous, ergosterol defective, and azole sensitive. | Virulence affected, less virulence in vivo. | [96] |
| GPI3 | A. fumigatus Growth rate significantly slower, sensitivity to SDS, increased levels of the chitinase ChiB. | Reduced virulence in a murine model. | [97] |
| Non-Conventional Glycosylation Processes | |||
| glfB | A. fumigatus Poor growth. | Attenuated virulence in a murine model. | [45,98] |
| umg1 | A. fumigatus Thinner cell wall, susceptibility to antifungal agents, and hyper-adherent cells. | None found. | [99] |
| ktr4 and ktr7 | A. fumigatus Severe phenotype defects, showing hyperbranched mycelium, loss of polarity during conidia germination, and hyphal elongation. | Virulence affected. | [46] |
| KTR3 | C. neoformans Sensitivity to SDS and NaCl. | Attenuated virulence in a murine model. | [54] |
| RmlD | S. schenckii Few phenotypical changes, alterations in the cell wall composition and organization. | Attenuated virulence in the G. mellonella model. | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Gaviria, M.; Vargas-Macías, A.P.; García-Carnero, L.C.; Martínez-Duncker, I.; Mora-Montes, H.M. Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host. J. Fungi 2021, 7, 875. https://doi.org/10.3390/jof7100875
Gómez-Gaviria M, Vargas-Macías AP, García-Carnero LC, Martínez-Duncker I, Mora-Montes HM. Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host. Journal of Fungi. 2021; 7(10):875. https://doi.org/10.3390/jof7100875
Chicago/Turabian StyleGómez-Gaviria, Manuela, Ana P. Vargas-Macías, Laura C. García-Carnero, Iván Martínez-Duncker, and Héctor M. Mora-Montes. 2021. "Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host" Journal of Fungi 7, no. 10: 875. https://doi.org/10.3390/jof7100875
APA StyleGómez-Gaviria, M., Vargas-Macías, A. P., García-Carnero, L. C., Martínez-Duncker, I., & Mora-Montes, H. M. (2021). Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host. Journal of Fungi, 7(10), 875. https://doi.org/10.3390/jof7100875







