Bacterial Quorum-Quenching Lactonase Hydrolyzes Fungal Mycotoxin and Reduces Pathogenicity of Penicillium expansum—Suggesting a Mechanism of Bacterial Antagonism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Growth Conditions
2.2. Recombinant Expression and Purification of Lactonases
2.3. Enzyme Kinetics Analysis
2.4. Addition of Purified Lactonase to P. expansum Liquid Culture
2.5. Effect of Purified Lactonase on Spores Germination and Colony Growth
2.6. Pathogenicity Assay of P. expansum in Apples
2.7. RNA Isolation and Quantitative Real-Time PCR (qPCR)
2.8. Sequence Identification, Alignment of Putative Lactonase, and Structure Modeling
2.9. Putative Lactonase from P. expansum Characterization
3. Results
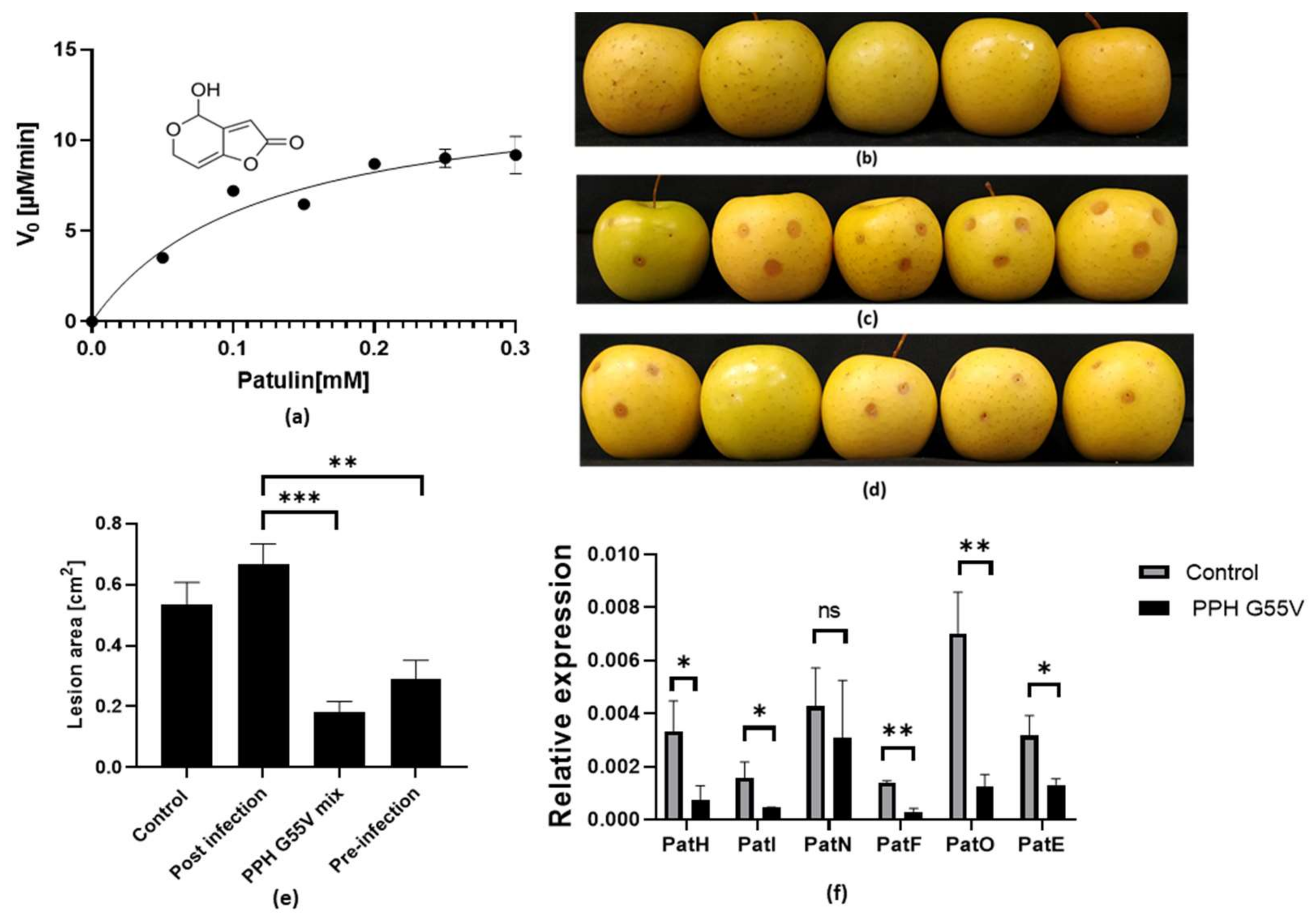
3.1. Bacterial QQ Lactonase Degrades Patulin, Inhibits Apples Infection, and Modulates Gene Expression in P. expansum during Infection
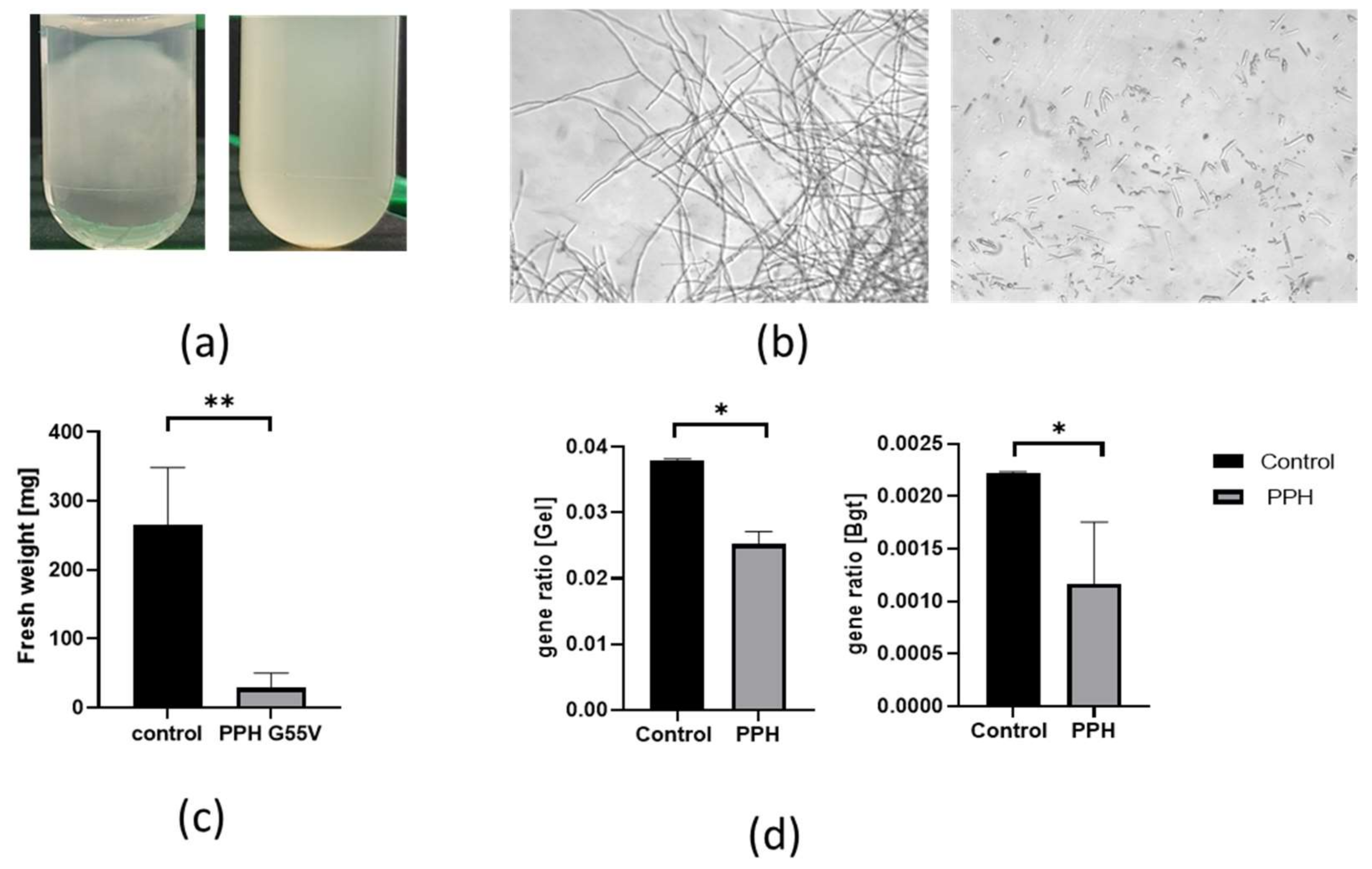
3.2. Bacterial Lactonase Modulate Fungal Growth of P. expansum and Gene Expression in Culture
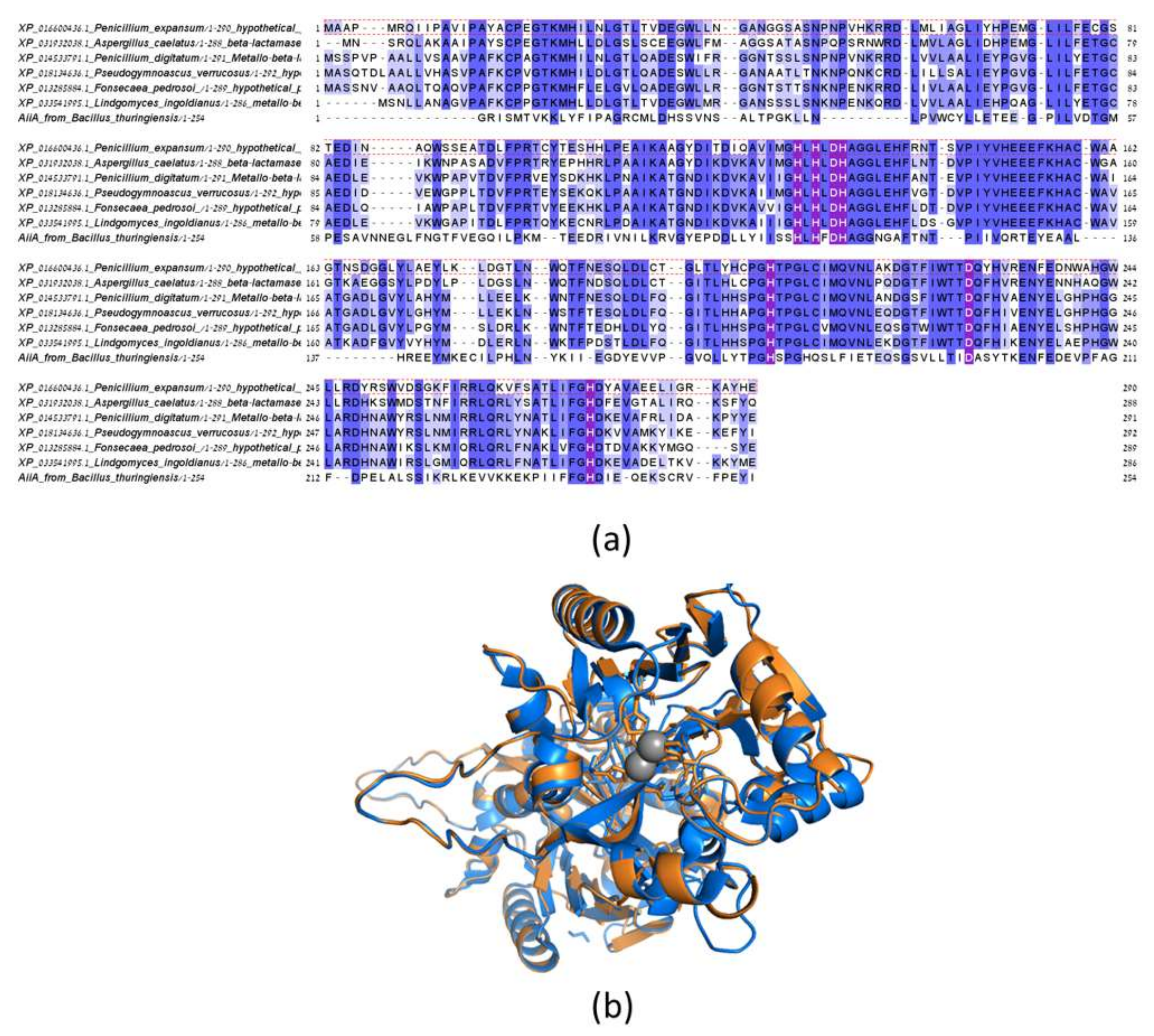
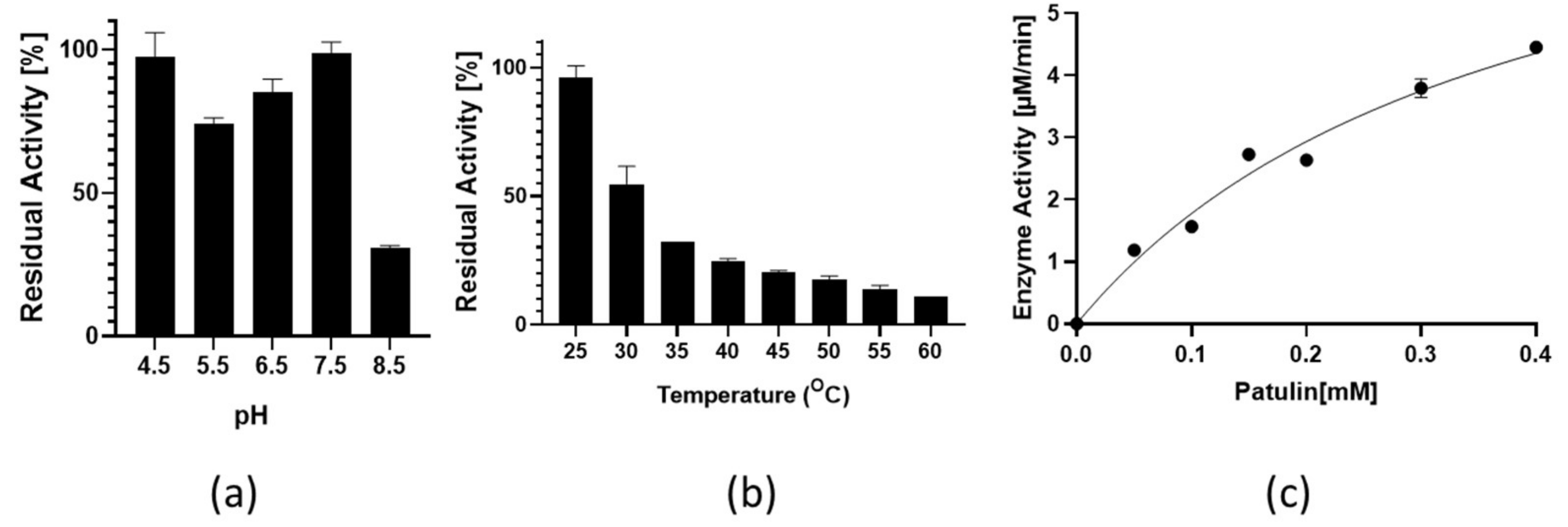
3.3. Identification of a Putative Lactonases in Fungal Species and Verification of Activity with Patulin for the Homolog from P. expansum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global analysis of the apple fruit microbiome: Are all apples the same? Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Köberl, M.; Rybakova, D.; Müller, H.; Grosch, R.; Smalla, K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Walsh, T.J.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 2004, 10 (Suppl. S1), 48–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egbuta, M.A.; Mwanza, M.; Babalola, O.O. Health risks associated with exposure to filamentous fungi. Int. J. Environ. Res. Public Health 2017, 14, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luciano-Rosario, D.; Keller, N.P.; Jurick, W.M. Penicillium expansum: Biology, omics, and management tools for a global postharvest pathogen causing blue mould of pome fruit. Mol. Plant Pathol. 2020, 21, 1391–1404. [Google Scholar] [CrossRef]
- Barad, S.; Horowitz, S.B.; Moskovitch, O.; Lichter, A.; Sherman, A.; Prusky, D. A penicillium expansum glucose oxidase-encoding gene, GOX2, is essential for gluconic acid production and acidification during colonization of deciduous fruit. Mol. Plant-Microbe Interact. 2012, 25, 779–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Tannous, J.; Sionov, E.; Keller, N.; Prusky, D. Apple intrinsic factors modulating the global regulator, LaeA, the patulin gene cluster and patulin accumulation during fruit colonization by Penicillium expansum. Front. Plant Sci. 2018, 9, 1094. [Google Scholar] [CrossRef] [Green Version]
- Madsen, A.M.; Frederiksen, M.W.; Jacobsen, M.H.; Tendal, K. Towards a risk evaluation of workers’ exposure to handborne and airborne microbial species as exemplified with waste collection workers. Environ. Res. 2020. [Google Scholar] [CrossRef]
- Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and toxicological effects of patulin. Toxins 2010, 2, 613–631. [Google Scholar] [CrossRef] [Green Version]
- Ianiri, G.; Idnurm, A.; Wright, S.A.I.; Durán-Patrón, R.; Mannina, L.; Ferracane, R.; Ritieni, A.; Castoria, R. Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Appl. Environ. Microbiol. 2013, 79, 3101–3115. [Google Scholar] [CrossRef] [Green Version]
- Waters, C.M.; Bassler, B.L. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI Family of Quorum-Sensing Transcriptional Regulators. Annu. Rev. Microbiol 1996, 50, 727–751. [Google Scholar] [CrossRef]
- Aframian, N.; Eldar, A. A Bacterial Tower of Babel: Quorum-Sensing Signaling Diversity and Its Evolution. Annu. Rev. Microbiol. 2020, 74, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Poonguzhali, S.; Madhaiyan, M.; Sa, T. Production of acyl-homoserine lactone quorum-sensing signals is wide-spread in gram-negative Methylobacterium. J. Microbiol. Biotechnol. 2007, 17, 226–233. [Google Scholar]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The Yin and Yang of bacterial communication. ChemBioChem 2009, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar] [CrossRef]
- Rémy, B.; Mion, S.; Plener, L.; Elias, M.; Chabrière, E.; Daudé, D. Interference in bacterial quorum sensing: A biopharmaceutical perspective. Front. Pharmacol. 2018, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Boon, N.; Bossier, P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010, 6, e1000989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Contreras, R.; Maeda, T.; Wood, T.K. Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 2013, 79, 6840–6846. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O.; Andersen, J.B.; Koch, B.; Larsen, T.O.; Hentzer, M.; et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2005, 151 Pt 5, 1325–1340. [Google Scholar] [CrossRef] [Green Version]
- Afonso, T.B.; Simões, L.C.; Lima, N. Effect of quorum sensing and quenching molecules on inter-kingdom biofilm formation by Penicillium expansum and bacteria. Biofouling 2020, 36, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Afonso, T.B.; Simões, L.C.; Lima, N. Methylobacterium oryzae influences isoepoxydon dehydrogenase gene expression and patulin production by penicillium expansum. Water 2021, 13, 1427. [Google Scholar] [CrossRef]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Leong Ong, S.; Renton Leadbetter, J.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar] [CrossRef]
- Afriat, L.; Roodveldt, C.; Manco, G.; Tawfik, D.S. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 2006, 45, 13677–13686. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Thomas, P.W.; Momb, J.; Hoang, Q.Q.; Petsko, G.A.; Ringe, D.; Fast, W. Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry 2007, 46, 11789–11799. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2015, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef]
- Liu, D.; Momb, J.; Thomas, P.W.; Moulin, A.; Petsko, G.A.; Fast, W.; Ringe, D. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 2008, 47, 7706–7714. [Google Scholar] [CrossRef] [Green Version]
- Bar-Rogovsky, H.; Hugenmatter, A.; Tawfik, D.S. The Evolutionary origins of detoxifying Enzymes: The mammalian serum paraoxonaes (PONs) relate to bacerial homoserine lactonases. J. Biol. Chem. 2013, 288, 23914–23927. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Ye, T.; Li, Q.; Bhatt, P.; Zhang, L.; Chen, S. Potential of a Quorum Quenching Bacteria Isolate Ochrobactrum intermedium D-2 Against Soft Rot Pathogen Pectobacterium carotovorum subsp. carotovorum. Front. Microbiol. 2020, 11, 898. [Google Scholar] [CrossRef]
- Wang, L.H.; Weng, L.X.; Dong, Y.H.; Zhang, L.H. Specificity and Enzyme Kinetics of the Quorum-quenching N-Acyl Homoserine Lactone Lactonase (AHL-lactonase). J. Biol. Chem. 2004, 279, 13645–13651. [Google Scholar] [CrossRef] [Green Version]
- Bzdrenga, J.; Daudé, D.; Rémy, B.; Jacquet, P.; Plener, L.; Elias, M.; Chabrière, E. Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact. 2017, 267, 104–115. [Google Scholar] [CrossRef]
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-Quenching Activity of the AHL-Lactonase from Bacillus licheniformis DAHB1 Inhibits Vibrio Biofilm Formation In Vitro and Reduces Shrimp Intestinal Colonisation and Mortality. Mar. Biotechnol. 2014, 16, 707–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remy, B.; Plener, L.; Elias, M.; Daude, D.; Chabriere, E. Enzymes for disrupting bacterial communication, an alternative to antibiotics? Ann. Pharm. Fr. 2016, 74, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Guendouze, A.; Plener, L.; Bzdrenga, J.; Jacquet, P.; Rémy, B.; Elias, M.; Lavigne, J.P.; Daudé, D.; Chabrière, E. Effect of quorum quenching lactonase in clinical isolates of pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 2017, 8, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Gao, Y.; Chen, X.; Yu, Z.; Li, X. Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent infection. Int. J. Mol. Sci. 2013, 14, 17477–17500. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, G.F.; Sartorio, R.; Lee, S.-H.; Rogers, C.J.; Meijler, M.M.; Moss, J.A.; Clapham, B.; Brogan, A.P.; Dickerson, T.J.; Janda, K.D. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Natl. Acad. Sci. USA 2005, 102, 309–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöckli, M.; Lin, C.; Sieber, R.; Plaza, D.F.; Ohm, R.A.; Künzler, M. Coprinopsis cinerea intracellular lactonases hydrolyze quorum sensing molecules of Gram-negative bacteria. Fungal Genet. Biol. 2017, 102, 49–62. [Google Scholar] [CrossRef]
- Uroz, S.; Heinonsalo, J. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. Proc. FEMS Microbiol. Ecol. 2008, 65, 271–278. [Google Scholar] [CrossRef] [Green Version]
- Padder, S.A.; Prasad, R.; Shah, A.H. Quorum sensing: A less known mode of communication among fungi. Microbiol. Res. 2018, 210, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum Sensing in the Dimorphic Fungus Candida albicans Is Mediated by Farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, C.F.; Černáková, L. Farnesol and tyrosol: Secondary metabolites with a crucial quorum-sensing role in candida biofilm development. Genes 2020, 11, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef]
- Venkatesh, N.; Keller, N.P. Mycotoxins in conversation with bacteria and fungi. Front. Microbiol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, S.R.; Wisniewski, M.E.; Droby, S.; Abdelfattah, A.; Freilich, S.; Mazzola, M. The Apple Microbiome: Structure, Function, and Manipulation for Improved Plant Health. In The Apple Genome, Compendium of Plant Genomes; Korban, S.S., Ed.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Angeli, D.; Sare, A.R.; Jijakli, M.H.; Pertot, I.; Massart, S. Insights gained from metagenomic shotgun sequencing of apple fruit epiphytic microbiota. Postharvest Biol. Technol. 2019, 153, 96–106. [Google Scholar] [CrossRef]
- Cui, Z.; Huntley, R.B.; Zeng, Q.; Steven, B. Temporal and spatial dynamics in the apple flower microbiome in the presence of the phytopathogen Erwinia amylovora. ISME J. 2021, 15, 318–329. [Google Scholar] [CrossRef]
- Shen, Y.; Nie, J.; Dong, Y.; Kuang, L.; Li, Y.; Zhang, J. Compositional shifts in the surface fungal communities of apple fruits during cold storage. Postharvest Biol. Technol. 2018, 144, 55–62. [Google Scholar] [CrossRef]
- Wassermann, B.; Kusstatscher, P.; Berg, G. Microbiome Response to Hot Water Treatment and Potential Synergy With Biological Control on Stored Apples. Front. Microbiol. 2019, 10, 2502. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, D.; Dor, S.; Erov, M.; Dan, Y.; Moy, J.C.; Mairesse, O.; Dafny-Yelin, M.; Adler-Abramovich, L.; Afriat-Jurnou, L. Directed Enzyme Evolution and Encapsulation in Peptide Nanospheres of Quorum Quenching Lactonase as an Antibacterial Treatment against Plant Pathogen. ACS Appl. Mater. Interfaces 2021, 13, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Hadas, Y.; Goldberg, I.; Pines, O.; Prusky, D. Involvement of gluconic acid and glucose oxidase in the pathogenicity of Penicillium expansum in apples. Phytopathology 2007, 97, 384–390. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Barad, S.; Chen, Y.; Luo, X.; Tannous, J.; Dubey, A.; Glam Matana, N.; Tian, S.; Li, B.; Keller, N.; et al. LaeA regulation of secondary metabolism modulates virulence in Penicillium expansum and is mediated by sucrose. Mol. Plant Pathol. 2017, 18, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Ya’ar Bar, S.; Dor, S.; Erov, M.; Afriat-Jurnou, L. Identification and Characterization of a New Quorum-Quenching N-acyl Homoserine Lactonase in the Plant Pathogen Erwinia amylovora. J. Agric. Food Chem. 2021, 69, 5652–5662. [Google Scholar] [CrossRef]
- Roach, J.A.G.; Brause, A.R.; Eisele, T.A.; Rupp, H.S. HPLC detection of patulin in apple juice with GC/MS confirmation of patulin identity. Adv. Exp. Med. Biol. 2002, 504, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Barad, S.; Horowitz, S.B.; Kobiler, I.; Sherman, A.; Prusky, D. Accumulation of the mycotoxin patulin in the presence of gluconic acid contributes to pathogenicity of penicillium expansum. Mol. Plant-Microbe Interact. 2014, 27, 66–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. S1), S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Jami, M.S.; García-Estrada, C.; Barreiro, C.; Cuadrado, A.A.; Salehi-Najafabadi, Z.; Martín, J.F. The penicillium chrysogenum extracellular proteome. Conversion from a food-rotting strain to a versatile cell factory for white biotechnology. Mol. Cell. Proteom. 2010, 9, 2729–2744. [Google Scholar] [CrossRef] [Green Version]
- Levin, E.; Kishore, A.; Ballester, A.R.; Raphael, G.; Feigenberg, O.; Liu, Y.; Norelli, J.; Gonzalez-Candelas, L.; Wisniewski, M.; Droby, S. Identification of pathogenicity-related genes and the role of a subtilisin-related peptidase S8 (PePRT) in authophagy and virulence of Penicillium expansum on apples. Postharvest Biol. Technol. 2019, 149, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Bebrone, C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef]
- Jock, S.; Lksch, B.; Mansvelt, L.; Geider, K. Characterization of Bacillus strains from apple and pear trees in South Africa antagonistic to Erwinia amylovora. FEMS Microbiol. Lett. 2002, 211, 247–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelfattah, A.; Wisniewski, M.; Droby, S.; Schena, L. Spatial and compositional variation in the fungal communities of organic and conventionally grown apple fruit at the consumer point-of-purchase. Hortic. Res. 2016, 3, 16047. [Google Scholar] [CrossRef] [Green Version]
- Abdelfattah, A.; Whitehead, S.R.; Macarisin, D.; Liu, J.; Burchard, E.; Freilich, S.; Dardick, C.; Droby, S.; Wisniewski, M. Effect of washing, waxing and low-temperature storage on the postharvest microbiome of apple. Microorganisms 2020, 8, 944. [Google Scholar] [CrossRef]
- Zhang, J.W.; Xuan, C.G.; Lu, C.H.; Guo, S.; Yu, J.F.; Asif, M.; Jiang, W.J.; Zhou, Z.G.; Luo, Z.Q.; Zhang, L.Q. AidB, a novel thermostable N-Acylhomoserine lactonase from the bacterium Bosea sp. Appl. Environ. Microbiol. 2019, 85, e02065–e02119. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, X.; Li, J.; Ye, T.; Mishra, S.; Zhang, L.; Chen, S. Exploration of the Quorum-Quenching Mechanism in Pseudomonas nitroreducens W-7 and Its Potential to Attenuate the Virulence of Dickeya zeae EC1. Front. Microbiol. 2021, 12, 694161. [Google Scholar] [CrossRef]
- Bartholomew, H.P.; Bradshaw, M.; Jurick, W.M.; Fonseca, J.M. The Good, the Bad, and the Ugly: Mycotoxin Production During Postharvest Decay and Their Influence on Tritrophic Host–Pathogen–Microbe Interactions. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Snini, S.P.; Tannous, J.; Heuillard, P.; Bailly, S.; Lippi, Y.; Zehraoui, E.; Barreau, C.; Oswald, I.P.; Puel, O. Patulin is a cultivar-dependent aggressiveness factor favouring the colonization of apples by Penicillium expansum. Mol. Plant Pathol. 2016, 17, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Tannous, J.; Keller, N.P.; Atoui, A.; El Khoury, A.; Lteif, R.; Oswald, I.P.; Puel, O. Secondary metabolism in Penicillium expansum: Emphasis on recent advances in patulin research. Crit. Rev. Food Sci. Nutr. 2018, 58, 2082–2098. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, V.M.; Zara, S.; Fracchetti, F.; Capozzi, V.; Nardi, T. Microbial biocontrol as an alternative to synthetic fungicides: Boundaries between pre-and postharvest applications on vegetables and fruits. Fermentation 2021, 7, 60. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dor, S.; Prusky, D.; Afriat-Jurnou, L. Bacterial Quorum-Quenching Lactonase Hydrolyzes Fungal Mycotoxin and Reduces Pathogenicity of Penicillium expansum—Suggesting a Mechanism of Bacterial Antagonism. J. Fungi 2021, 7, 826. https://doi.org/10.3390/jof7100826
Dor S, Prusky D, Afriat-Jurnou L. Bacterial Quorum-Quenching Lactonase Hydrolyzes Fungal Mycotoxin and Reduces Pathogenicity of Penicillium expansum—Suggesting a Mechanism of Bacterial Antagonism. Journal of Fungi. 2021; 7(10):826. https://doi.org/10.3390/jof7100826
Chicago/Turabian StyleDor, Shlomit, Dov Prusky, and Livnat Afriat-Jurnou. 2021. "Bacterial Quorum-Quenching Lactonase Hydrolyzes Fungal Mycotoxin and Reduces Pathogenicity of Penicillium expansum—Suggesting a Mechanism of Bacterial Antagonism" Journal of Fungi 7, no. 10: 826. https://doi.org/10.3390/jof7100826
APA StyleDor, S., Prusky, D., & Afriat-Jurnou, L. (2021). Bacterial Quorum-Quenching Lactonase Hydrolyzes Fungal Mycotoxin and Reduces Pathogenicity of Penicillium expansum—Suggesting a Mechanism of Bacterial Antagonism. Journal of Fungi, 7(10), 826. https://doi.org/10.3390/jof7100826






