Arginine Methyltransferase PeRmtC Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Media and Culture Conditions
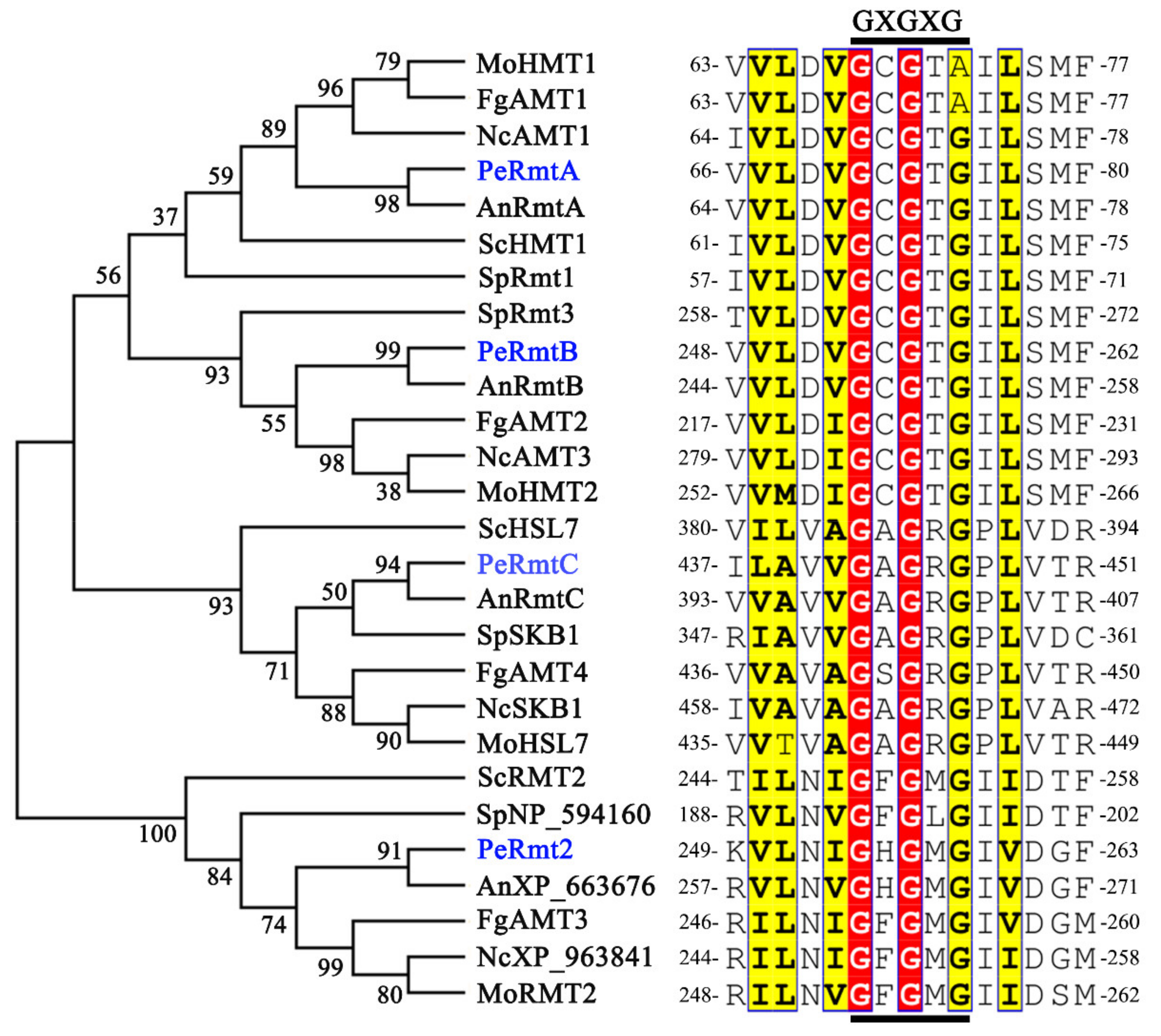
2.2. Conserved Motif and Phylogenetic Analysis
2.3. Construction of Gene Deletion and Complementation Strains
2.4. Phenotype Assay
2.5. Stress Tolerance Tests
2.6. Gene Relative Expression Analysis
2.7. Protein Extraction and Western Blot Analysis
2.8. Subcellular Localization Analysis
2.9. Statistical Analysis
3. Results
3.1. Identification of PRMTs Homologs in P. expansum
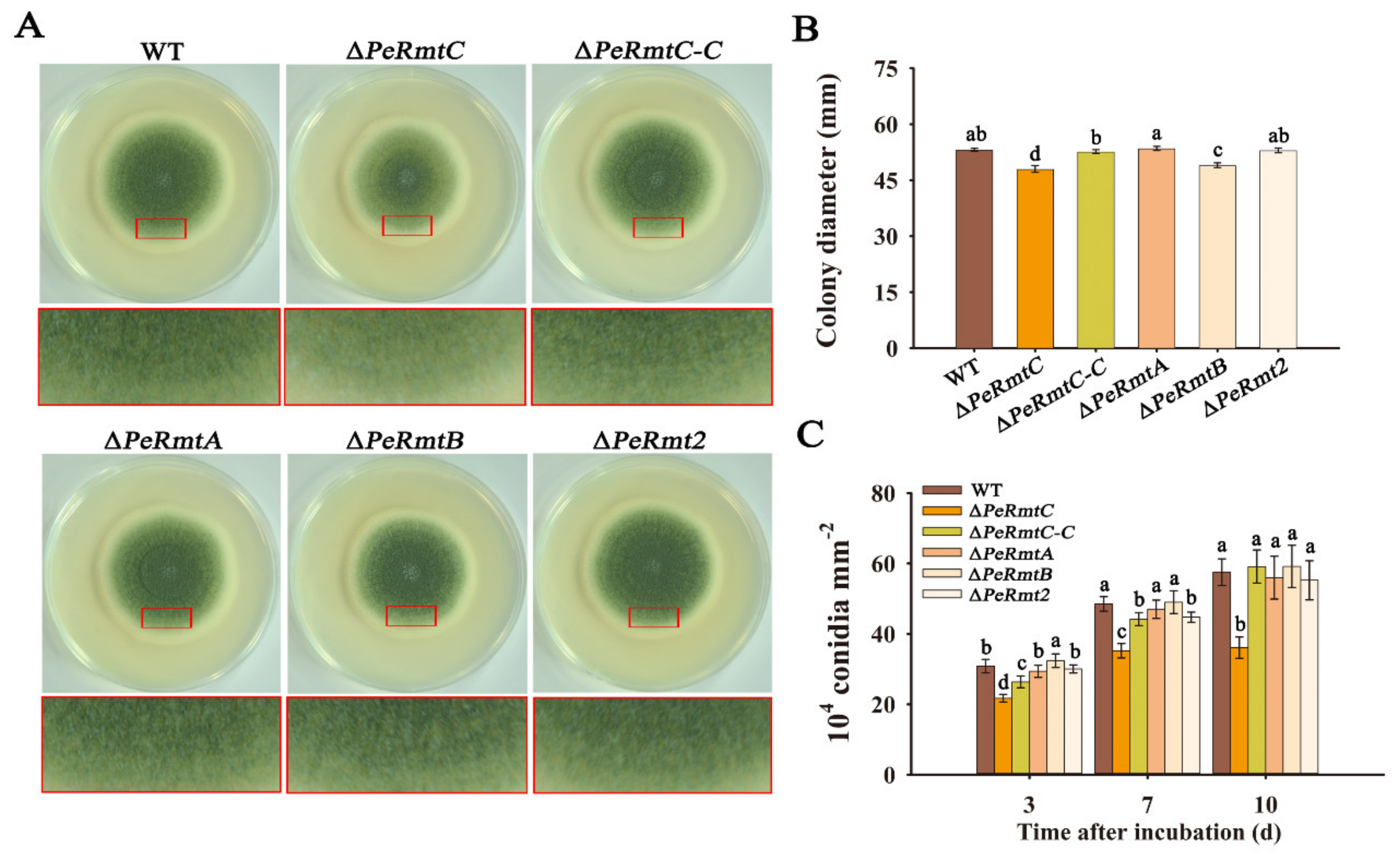
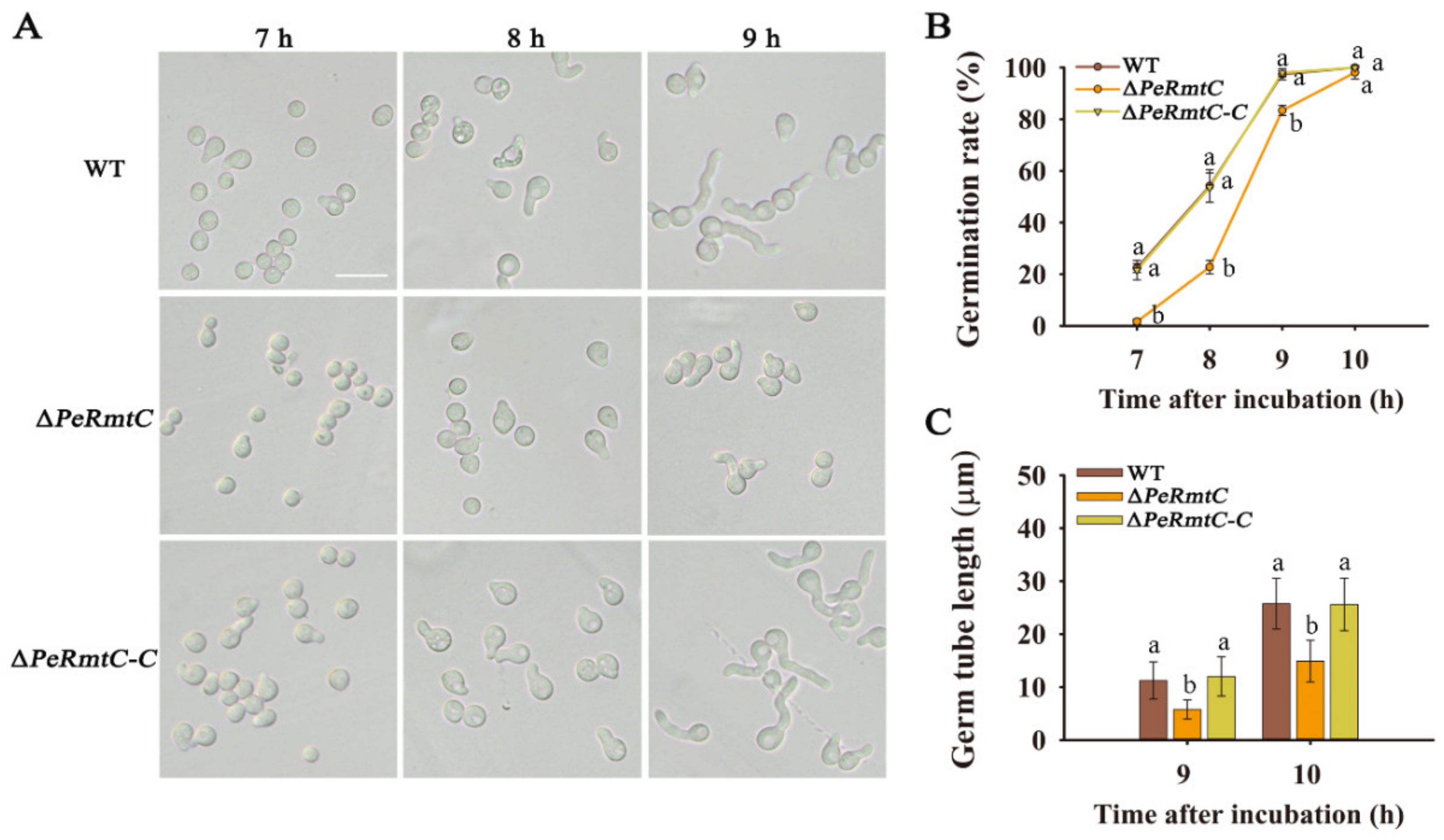
3.2. PeRmtC Is Important for Conidiation and Conidial Germination
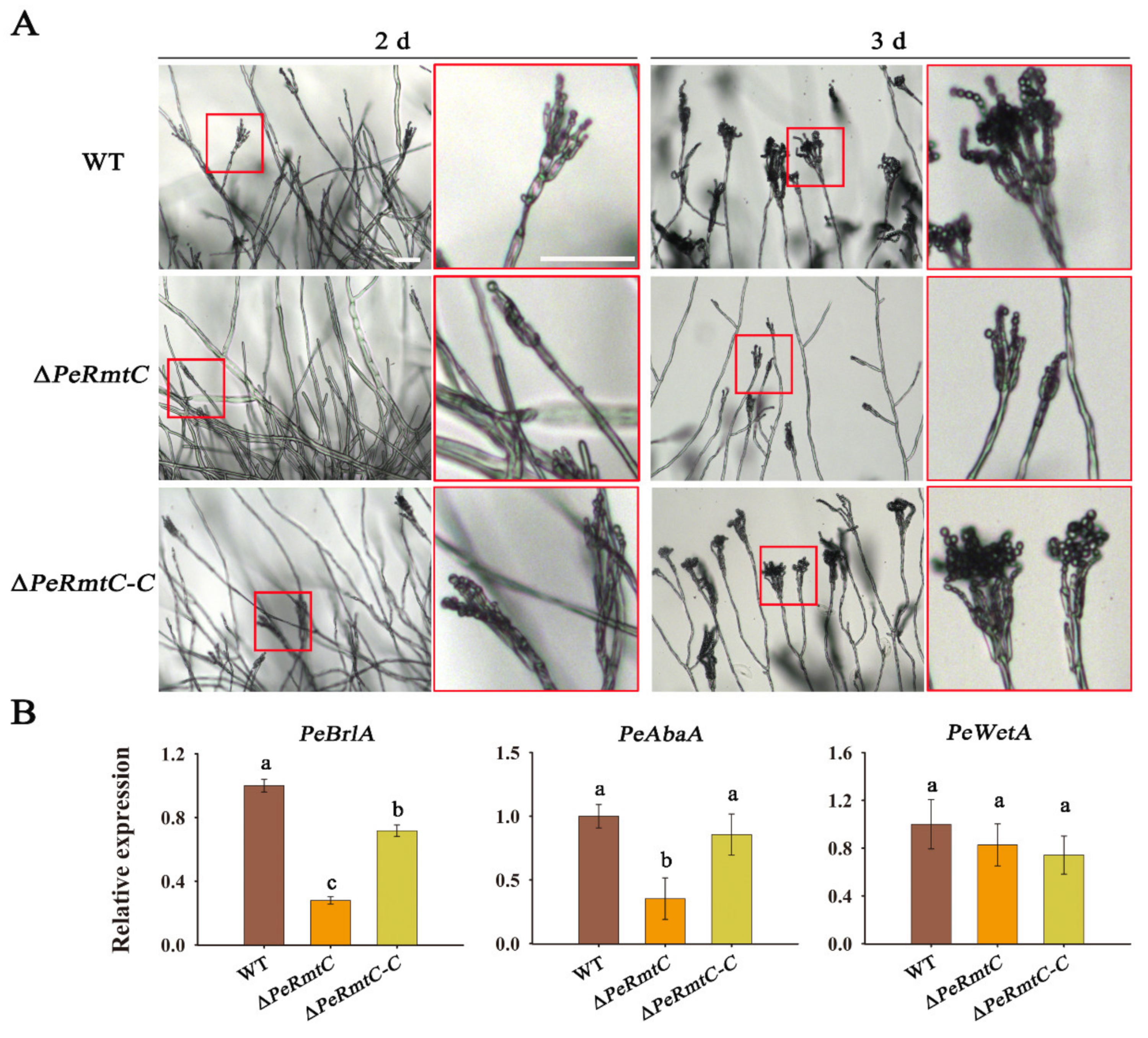
3.3. PeRmtC Is Involved in Conidiophore Development
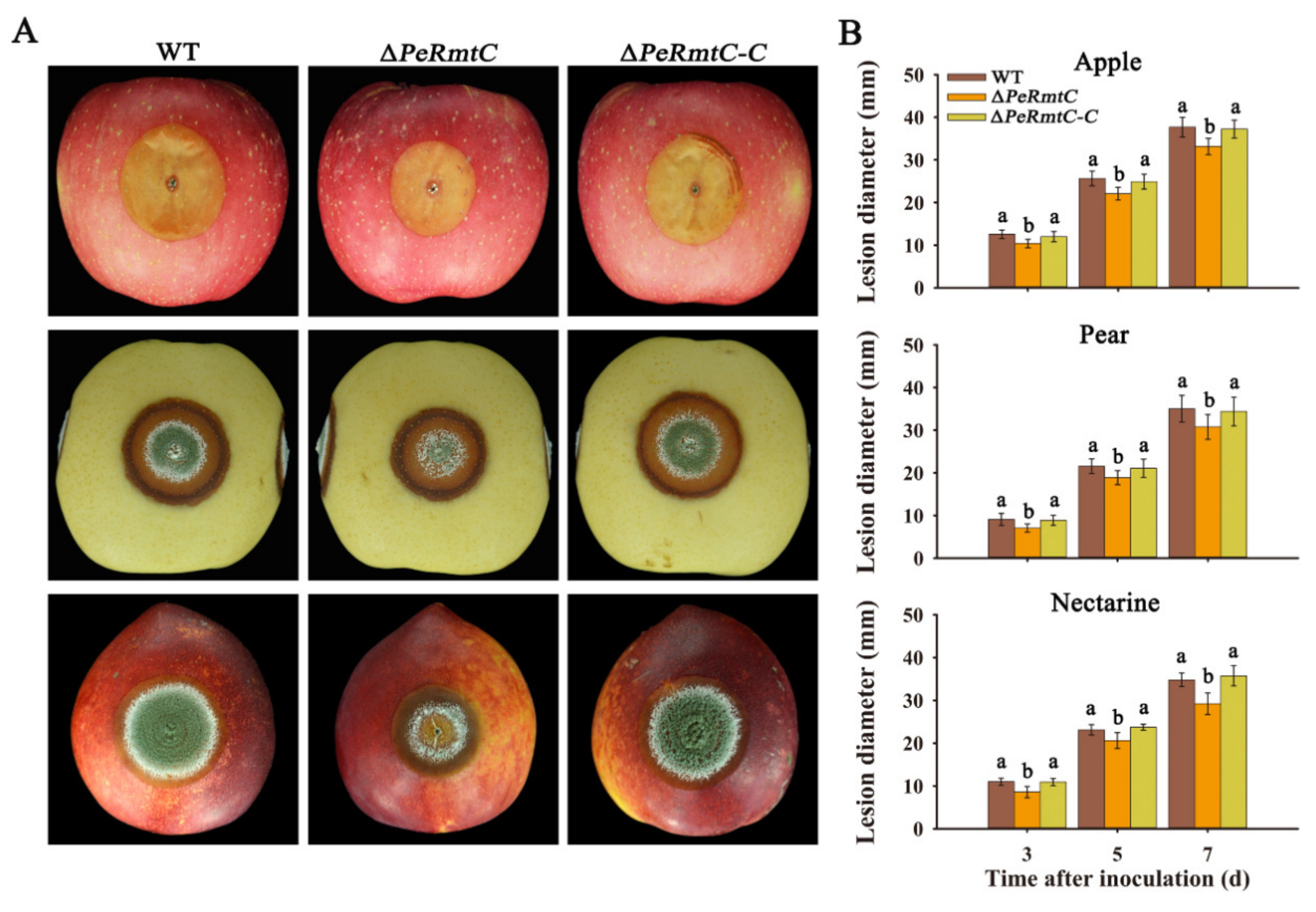
3.4. PeRmtC Affects Pathogenicity of P. expansum
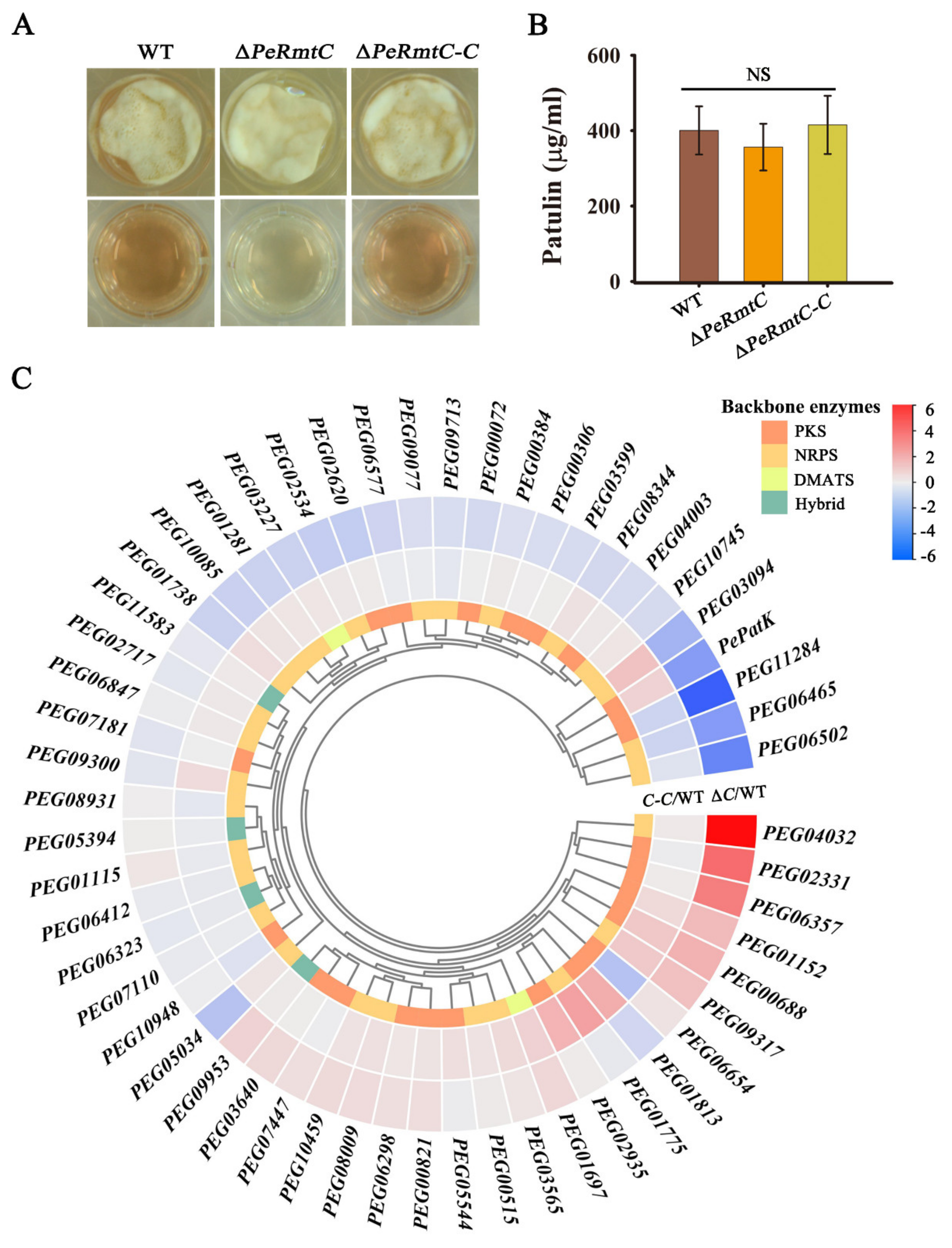
3.5. PeRmtC Plays Roles in Secondary Metabolism
3.6. PeRmtC Is Involved in Stress Responses of P. expansum
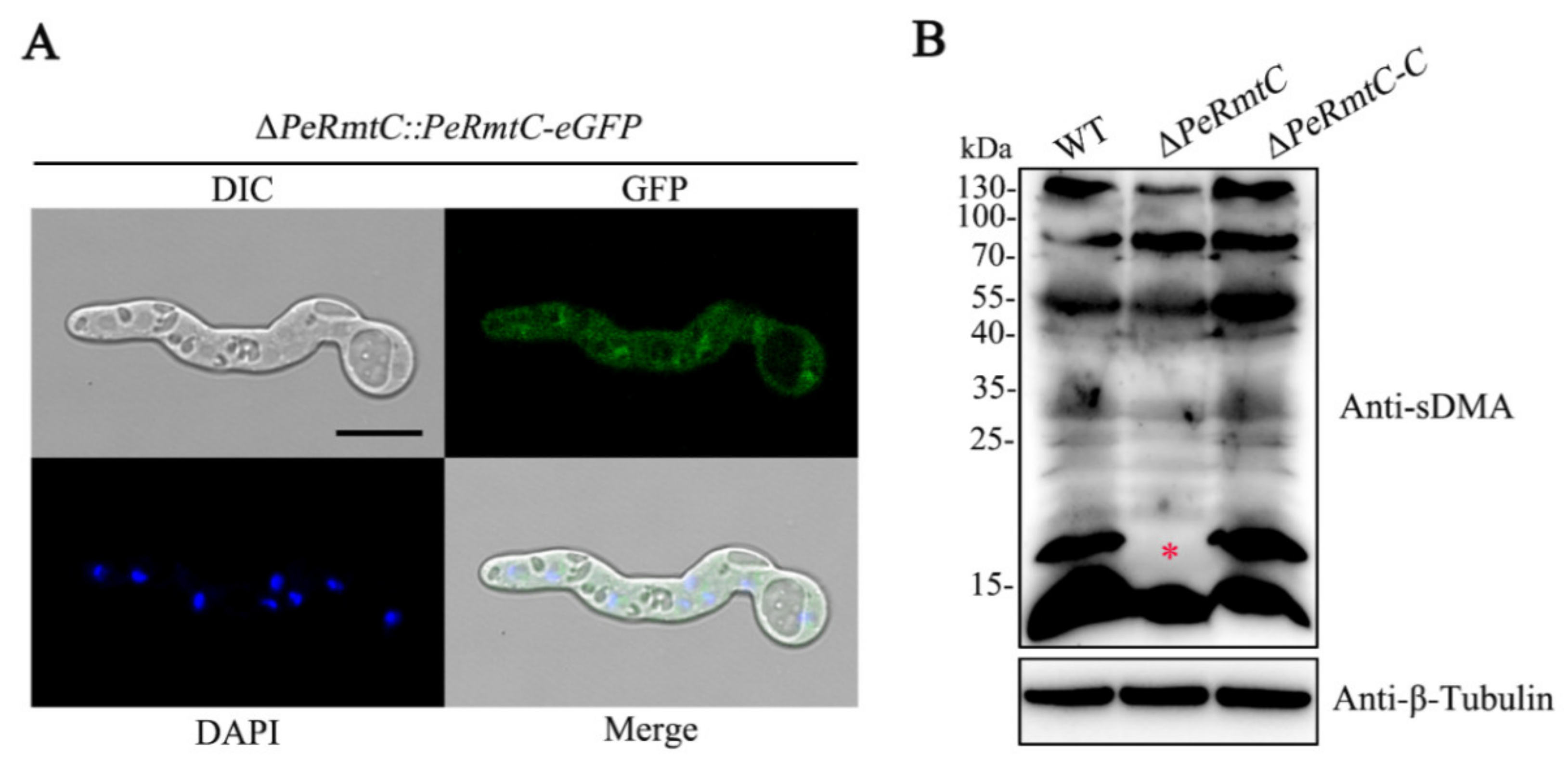
3.7. PeRmtC Affects sDMA Levels in P. expansum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, G.; Tian, S.; Chan, Z.; Li, B. Crucial role of antioxidant proteins and hydrolytic enzymes in pathogenicity of Penicillium expansum: Analysis based on proteomics approach. Mol. Cell. Proteomics 2007, 6, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Luciano-Rosario, D.; Keller, N.P.; Jurick, W.M. Penicillium expansum: Biology, omics, and management tools for a global post-harvest pathogen causing blue mould of pome fruit. Mol. Plant Pathol. 2020, 21, 1391–1404. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular basis and regulation of pathogenicity and patulin biosynthesis in Penicillium expansum. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3416–3438. [Google Scholar] [CrossRef]
- Andersen, B.; Smedsgaard, J.; Frisvad, J. Penicillium expansum: Consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J. Agric. Food Chem. 2004, 52, 2421–2428. [Google Scholar] [CrossRef]
- Zong, Y.; Li, B.; Tian, S. Effects of carbon, nitrogen and ambient pH on patulin production and related gene expression in Penicillium expansum. Int. J. Food Microbiol. 2015, 206, 102–108. [Google Scholar] [CrossRef]
- Lorton, B.M.; Shechter, D. Cellular consequences of arginine methylation. Cell. Mol. Life Sci. 2019, 76, 2933–2956. [Google Scholar] [CrossRef]
- Wolf, S.S. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 2009, 66, 2109–2121. [Google Scholar] [CrossRef]
- Paik, W.K.; Paik, D.C.; Kim, S. Historical review: The field of protein methylation. Trends Biochem. Sci. 2007, 32, 146–152. [Google Scholar] [CrossRef]
- Peng, C.; Wong, C.C. The story of protein arginine methylation: Characterization, regulation, and function. Expert Rev. Proteom. 2017, 14, 157–170. [Google Scholar] [CrossRef]
- Wesche, J.; Kühn, S.; Kessler, B.M.; Salton, M.; Wolf, A. Protein arginine methylation: a prominent modification and its de-methylation. Cell. Mol. Life Sci. 2017, 74, 3305–3315. [Google Scholar] [CrossRef]
- Ahmad, A.; Cao, X. Plant PRMTs broaden the scope of arginine methylation. J. Genet. Genom. 2012, 39, 195–208. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [Green Version]
- Olsson, I.; Berrez, J.-M.; Leipus, A.; Östlund, C.; Mutvei, A. The arginine methyltransferase Rmt2 is enriched in the nucleus and co-purifies with the nuclear porins Nup49, Nup57 and Nup100. Exp. Cell Res. 2007, 313, 1778–1789. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.; Gary, J.D.; Clarke, S. δ-N-methylarginine is a novel posttranslational modification of arginine residues in yeast proteins. J. Biol. Chem. 1998, 273, 29283–29286. [Google Scholar] [CrossRef] [Green Version]
- Bauer, I.; Lechner, L.; Pidroni, A.; Petrone, A.-M.; Merschak, P.; Lindner, H.; Kremser, L.; Graessle, S.; Golderer, G.; Allipour, S.; et al. Type I and II PRMTs regulate catabolic as well as detoxifying processes in Aspergillus nidulans. Fungal Genet. Biol. 2019, 129, 86–100. [Google Scholar] [CrossRef]
- Bauer, I.; Graessle, S.; Loidl, P.; Hohenstein, K.; Brosch, G. Novel insights into the functional role of three protein arginine methyltransferases in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 551–561. [Google Scholar] [CrossRef]
- Li, Y.; He, Y.; Li, X.; Fasoyin, O.E.; Hu, Y.; Liu, Y.; Yuan, J.; Zhuang, Z.; Wang, S. Histone methyltransferase aflrmtA gene is involved in the morphogenesis, mycotoxin biosynthesis, and pathogenicity of Aspergillus flavus. Toxicon 2017, 127, 112–121. [Google Scholar] [CrossRef]
- Li, Z.; Wu, L.; Wu, H.; Zhang, X.; Mei, J.; Zhou, X.; Wang, G.L.; Liu, W. Arginine methylation is required for remodelling pre-mRNA splicing and induction of autophagy in rice blast fungus. New Phytol. 2020, 225, 413–429. [Google Scholar] [CrossRef] [Green Version]
- Ben Lovely, C.; Aulakh, K.B.; Perlin, M.H. Role of Hsl7 in morphology and pathogenicity and its interaction with other signaling components in the plant pathogen Ustilago maydis. Eukaryot. Cell 2011, 10, 869–883. [Google Scholar] [CrossRef] [Green Version]
- Satterlee, T.; Cary, J.W.; Calvo, A.M. RmtA, a putative arginine methyltransferase, regulates secondary metabolism and de-velopment in Aspergillus flavus. PLOS ONE 2016, 11, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Feldman, D.; Ziv, C.; Gorovits, R.; Efrat, M.; Yarden, O. Neurospora crassa protein arginine methyl transferases are involved in growth and development and interact with the NDR kinase COT1. PLOS ONE 2013, 8, e80756. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, C.; Hou, R.; Zhou, X.; Li, G.; Zhang, S.; Xu, J.-R. The AMT1 arginine methyltransferase gene is important for plant infection and normal hyphal growth in Fusarium graminearum. PLOS ONE 2012, 7, e38324. [Google Scholar] [CrossRef]
- Zhenglin, D.; Zong, Y.; Du, Z.; Chen, Y.; Zhang, Z.; Qin, G.; Zhao, W.; Tian, S. Genomic characterization reveals insights into patulin biosynthesis and pathogenicity in Penicillium Species. Mol. Plant-Microbe Interactions 2015, 28, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, B.; Xu, X.; Zhang, Z.; Tian, S. The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 2018, 20, 4063–4078. [Google Scholar] [CrossRef]
- Prakash, P.Y.; Bhargava, K. A modified micro chamber agar spot slide culture technique for microscopic examination of fila-mentous fungi. J. Microbiol. Methods 2016, 123, 126–129. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; He, C.; Qin, G.; Tian, S. Comparative proteomics reveals the potential targets of BcNoxR, a putative regu-latory subunit of NADPH oxidase of Botrytis cinerea. Mol. Plant-Microbe Interact. 2016, 29, 990–1003. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zong, Y.; Shang, Y.; Zhang, Z.; Xu, X.; Wang, X.; Long, M.; Tian, S. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef]
- Tewary, S.K.; Zheng, Y.G.; Ho, M.-C. Protein arginine methyltransferases: Insights into the enzyme structure and mechanism at the atomic level. Cell. Mol. Life Sci. 2019, 76, 2917–2932. [Google Scholar] [CrossRef]
- Qin, Y.; Bao, L.; Gao, M.; Chen, M.; Lei, Y.; Liu, G.; Qu, Y. Penicillium decumbens BrlA extensively regulates secondary metabo-lism and functionally associates with the expression of cellulase genes. Appl. Microbiol. Biotechnol. 2013, 97, 10453–10467. [Google Scholar] [CrossRef]
- Tao, L.; Yu, J.-H. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 2011, 157, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Sun, X.; Zhu, C.; Xu, Q.; Ruan, R.; Yu, D.; Li, H. PdbrlA, PdabaA and PdwetA control distinct stages of conidiogenesis in Penicillium digitatum. Res. Microbiol. 2015, 166, 56–65. [Google Scholar] [CrossRef]
- Li, B.; Lai, T.; Qin, G.; Tian, S. Ambient pH stress inhibits spore germination of Penicillium expansum by impairing protein synthesis and folding: A proteomic-based study. J. Proteome Res. 2010, 9, 298–307. [Google Scholar] [CrossRef]
- Sanchez-Torres, P.; Vilanova, L.; Ballester, A.R.; López-Pérez, M.; Teixidó, N.; Viñas, I.; Usall, J.; González-Candelas, L.; Torres, R. Unravelling the contribution of the Penicillium expansum PeSte12 transcription factor to virulence during apple fruit infection. Food Microbiol. 2018, 69, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Sewall, T.C.; Mims, C.W.; Timberlake, W.E. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 1990, 2, 731–739. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.-H. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 2010, 38, 229. [Google Scholar] [CrossRef] [Green Version]
- Zetina-Serrano, C.; Rocher, O.; Naylies, C.; Lippi, Y.; Oswald, I.P.; Lorber, S.; Puel, O. The brlA gene deletion reveals that patulin biosynthesis is not related to conidiation in Penicillium expansum. Int. J. Mol. Sci. 2020, 21, 6660. [Google Scholar] [CrossRef]
- Viggiano, A.; Salo, O.; Ali, H.; Szymanski, W.; Lankhorst, P.P.; Nygård, Y.; Bovenberg, R.A.L.; Driessen, A.J.M. Pathway for the biosynthesis of the pigment chrysogine by Penicillium chrysogenum. Appl. Environ. Microbiol. 2018, 84, e02246-17. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.J.; Zhang, X.; Cao, H.; Niu, Y.J.; Li, C.; Liu, H. Chemical composition, security and bioactivity of the red pigment from Penicillium purpurogenum Li-3. Chem. Biodivers. 2018, 15, e1800300. [Google Scholar] [CrossRef]
- Deng, X.; Gu, L.; Liu, C.; Lu, T.; Lu, F.; Lu, Z.; Cui, P.; Pei, Y.; Wang, B.; Hu, S.; et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2010, 107, 19114–19119. [Google Scholar] [CrossRef] [Green Version]
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019, 12, eaat8388. [Google Scholar] [CrossRef]
- Ryu, H.-Y.; Duan, R.; Ahn, S.H. Yeast symmetric arginine methyltransferase Hsl7 has a repressive role in transcription. Res. Microbiol. 2019, 170, 222–229. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Chen, Y.; Li, B.; Tian, S. Arginine Methyltransferase PeRmtC Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism. J. Fungi 2021, 7, 807. https://doi.org/10.3390/jof7100807
Xu X, Chen Y, Li B, Tian S. Arginine Methyltransferase PeRmtC Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism. Journal of Fungi. 2021; 7(10):807. https://doi.org/10.3390/jof7100807
Chicago/Turabian StyleXu, Xiaodi, Yong Chen, Boqiang Li, and Shiping Tian. 2021. "Arginine Methyltransferase PeRmtC Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism" Journal of Fungi 7, no. 10: 807. https://doi.org/10.3390/jof7100807
APA StyleXu, X., Chen, Y., Li, B., & Tian, S. (2021). Arginine Methyltransferase PeRmtC Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism. Journal of Fungi, 7(10), 807. https://doi.org/10.3390/jof7100807







