Plant Triterpenoids Regulate Endophyte Community to Promote Medicinal Plant Schisandra sphenanthera Growth and Metabolites Accumulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of S. Sphenanthera Plants in the Natural Habitat
2.2. Isolation, Purification, Identification, and Preservation of Culturable Bacteria and Fungi
2.3. Isolation of Main Triterpenoid Metabolites from S. sphenanthera
2.4. In Vitro Assay for Selective Effect of Triterpenoids
2.5. Functional Screening for Potentially Beneficial Microbes
2.6. Assessment of Dual Interactions in Culture Media
2.7. SynCom Construction and Functional Evaluation
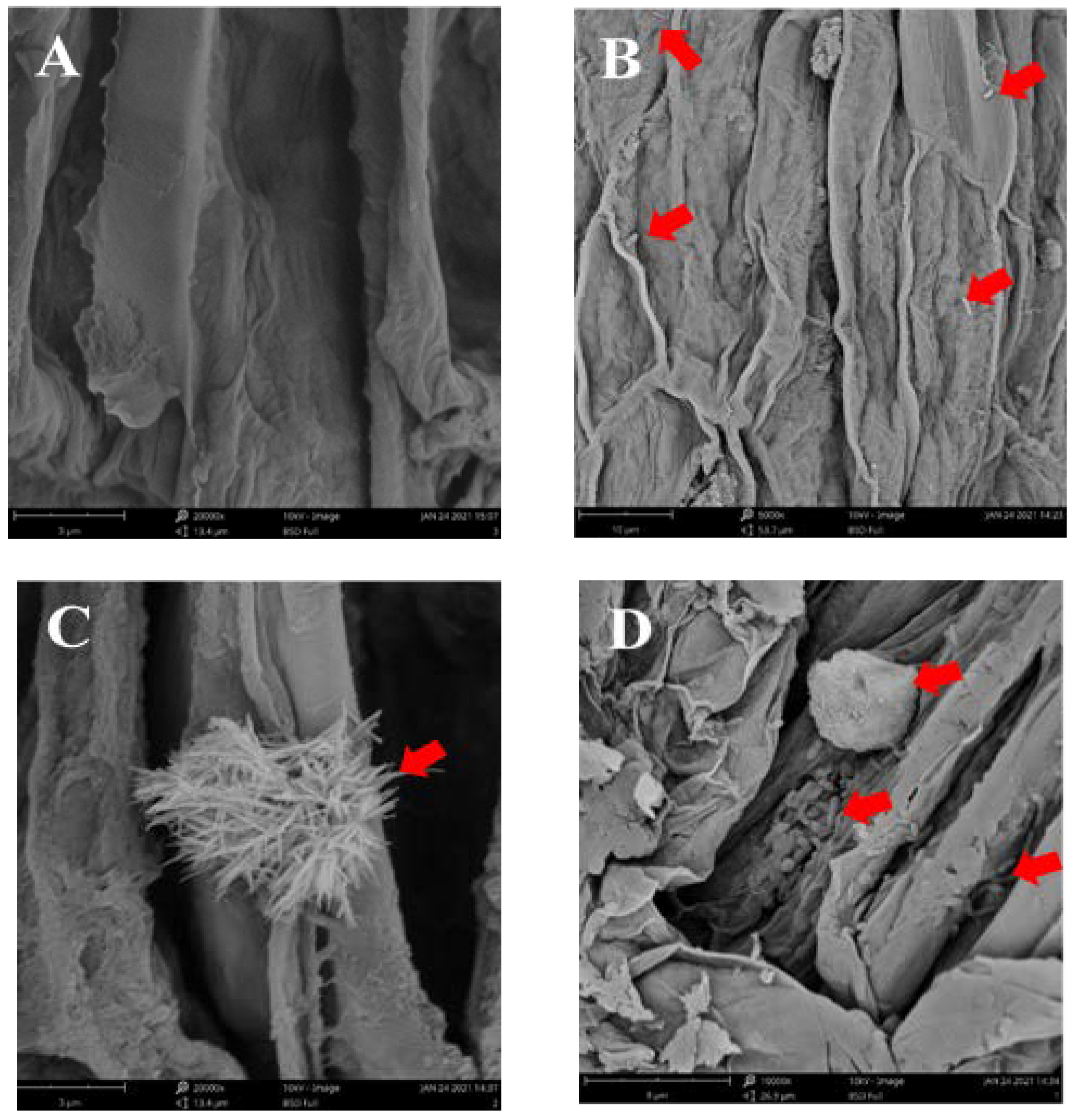
2.8. SEM Studies
2.9. Measurement of Plant Metabolite Content
2.10. Statistical Analysis
3. Results
3.1. Isolation of Endophytic Bacteria and Fungi from S. sphenanthera
3.2. Isolation and Identification of Triterpenoids from S. sphenanthera
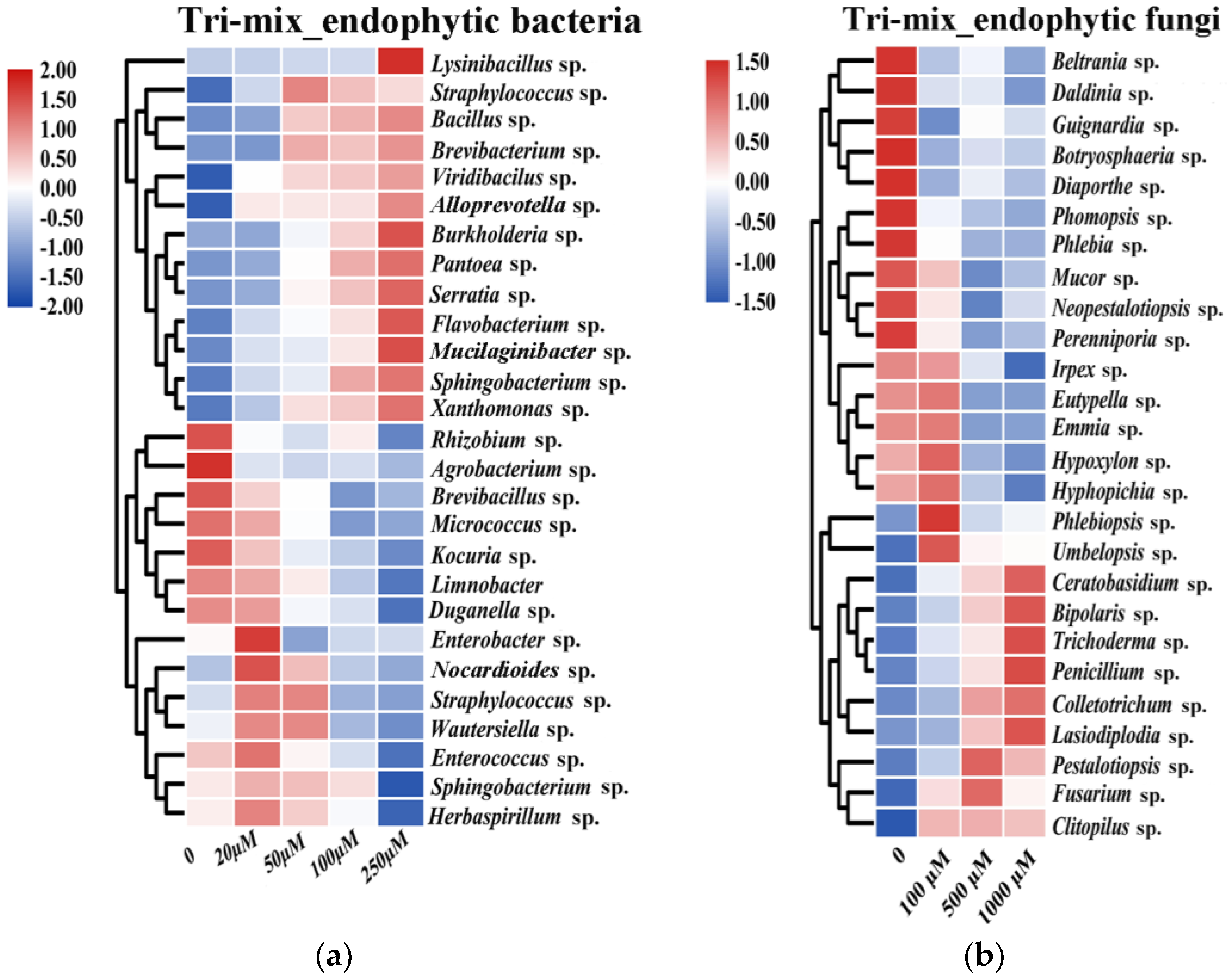
3.3. Effect of Triterpenoid Metabolites on the Growth of Endophytes
3.4. Functional Screening of Endophytes
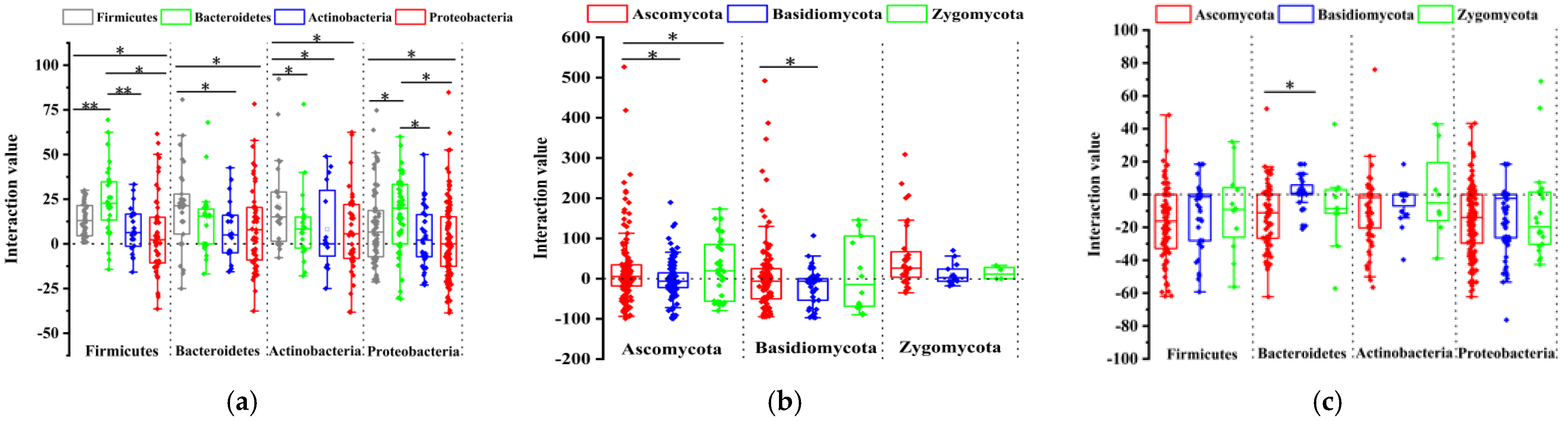
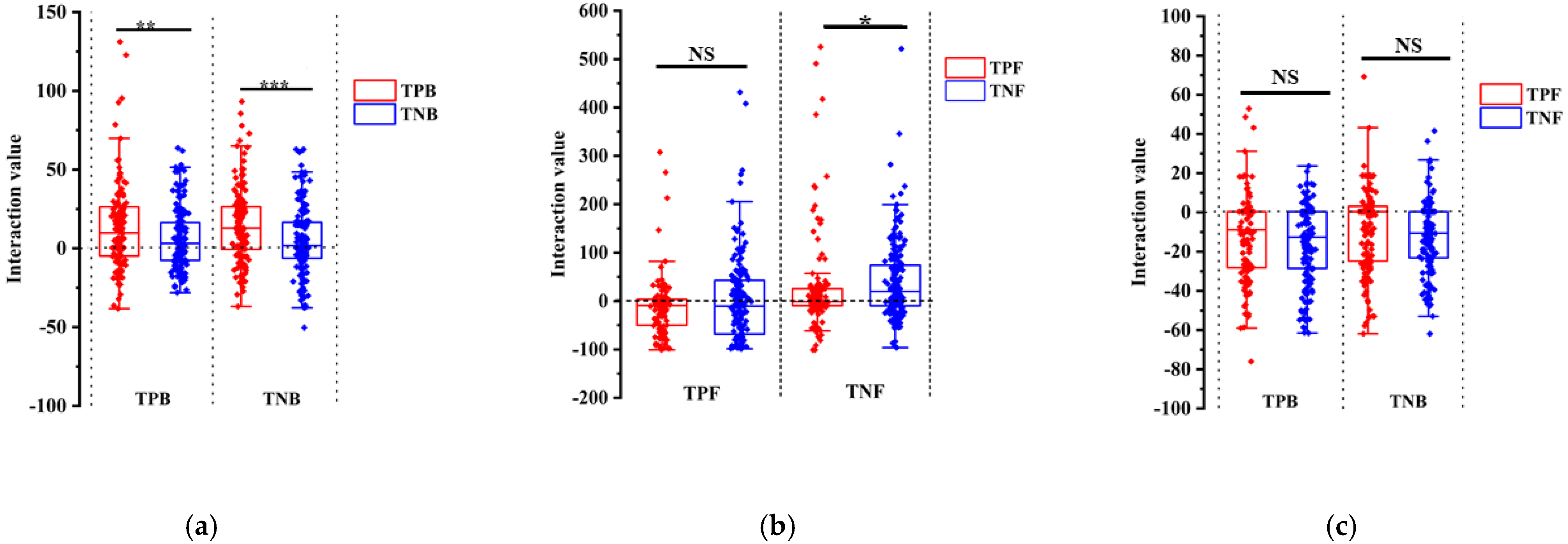
3.5. Plate Interaction Showed TPB and TPF Are More Important in the Interaction Network
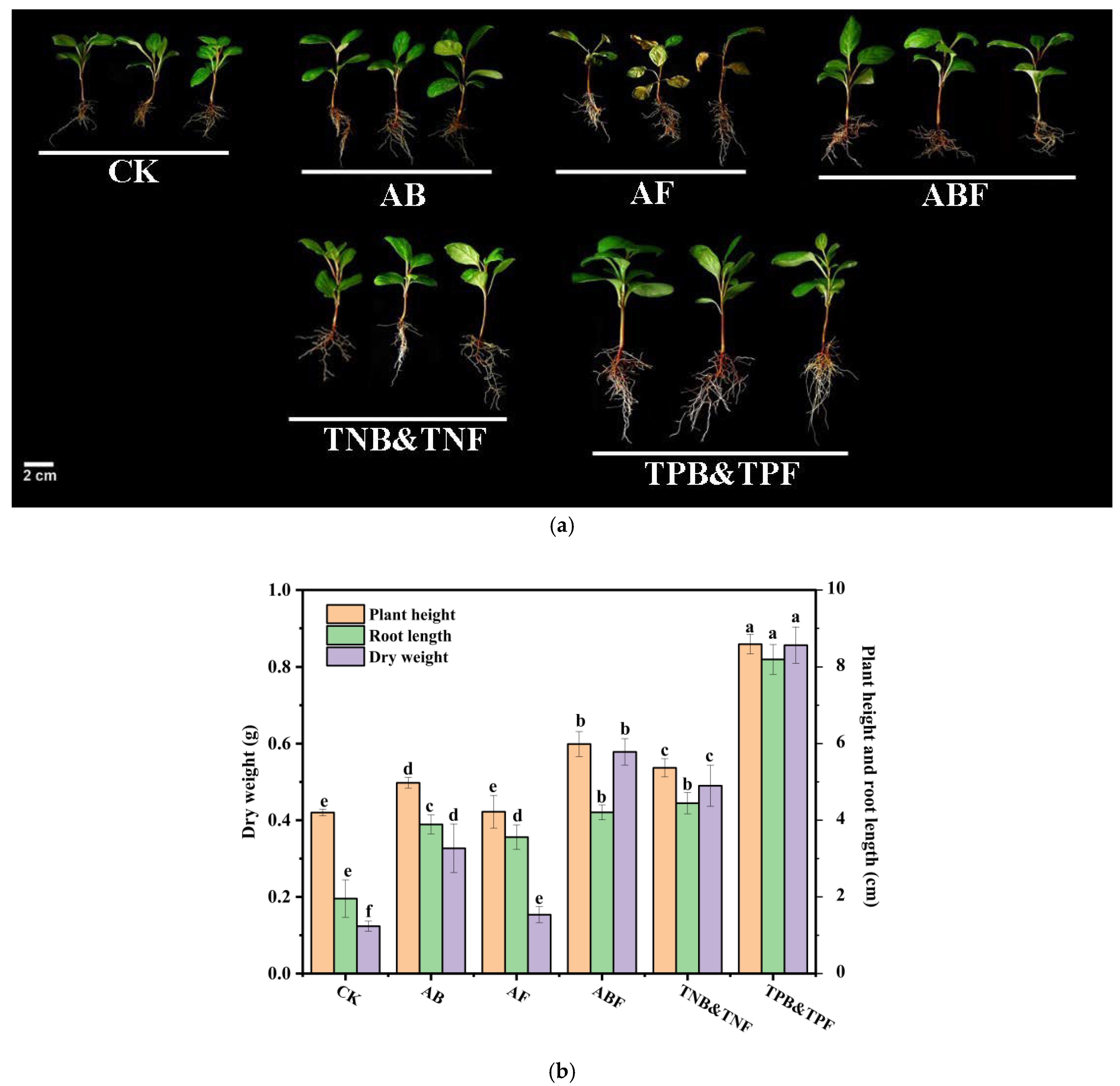
3.6. Evaluation of SynComs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dini-Andreote, F. Endophytes: The second layer of plant defense. Trends Plant Sci. 2020, 25, 4. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Gerlach, N.; Sacristan, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramírez, D.; Bucher, M.; O’Connell, R.J.; et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B.; et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Saha, C.; Nag, M.; Mandal, D.; Naiya, H.; Sen, D.; Mitra, S.; Kumar, M.; Bose, D.; Mukherjee, G.; et al. A Tripartite Interaction among the Basidiomycete Rhodotorula mucilaginosa, N2-Fixing Endobacteria, and Rice Improves Plant Nitrogen Nutrition. Plant Cell 2019, 32, 486–507. [Google Scholar] [CrossRef]
- Zheng, Y.; Gong, X. Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome 2019, 7, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Lopez, B.R.; Tinoco-Ojanguren, C.; Bacilio, M.; Mendoza, A.; Bashan, Y. Endophytic bacteria of the rock-dwelling cactus Mammillaria fraileana affect plant growth and mobilization of elements from rocks. Environ. Exp. Bot. 2012, 81, 26–36. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Koprivova, A.; Kopriva, S. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J. Exp. Bot. 2020, 72, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [Green Version]
- Koprivova, A.; Schuck, S.; Jacoby, R.P.; Klinkhammer, I.; Welter, B.; Leson, L.; Martyn, A.; Nauen, J.; Grabenhorst, N.; Mandelkow, J.F.; et al. Root-specific camalexin biosynthesis controls the plant growth-promoting effects of multiple bacterial strains. Proc. Natl. Acad. Sci. USA 2019, 116, 15735–15744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudjordjie, E.N.; Sapkota, R.; Steffensen, S.K.; Fomsgaard, I.S.; Nicolaisen, M. Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 2019, 7, 1–17. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Chen, L.; Schwier, M.; Koprivova, A.; Kopriva, S. Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Research 2020, 9, 151. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, Y.; Li, M.; Zhang, K.; Ma, W.; Zheng, L.; Xu, H.; Cui, B.; Liu, R.; Yang, Y.; et al. Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean. J. Integr. Plant Biol. 2021, 63, 1021–1035. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [Green Version]
- Coleman-Derr, D.; Desgarennes, D.; García, C.F.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2015, 209, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, R.S.C.; Okura, V.K.; Armanhi, J.; Jorrín, B.; Lozano, N.; Da Silva, M.J.; Gonzalez-Guerrero, M.; De Araújo, L.M.; Verza, N.C.; Bagheri, H.C.; et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2017, 20, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Millard, P.; Whiteley, A.; Murrell, J. Unravelling rhizosphere–microbial interactions: Opportunities and limitations. Trends Microbiol. 2004, 12, 386–393. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Wei, Z.; Friman, V.-P.; Gu, S.-H.; Wang, X.-F.; Eisenhauer, N.; Yang, T.-J.; Ma, J.; Shen, Q.-R.; Xu, Y.-C.; et al. Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression. mBio 2016, 7, e01790-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, L.; Li, Y.; Wang, Z.; Yu, Y.; Zhang, N.; Yang, C.; Zeng, Q.; Wang, Q. Synthetic community with six Pseudomonas strains screened from garlic rhizosphere microbiome promotes plant growth. Microb. Biotechnol. 2020, 14, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Long, C.; Lam, E. Roles of Plant-Associated Microbiota in Traditional Herbal Medicine. Trends Plant. Sci. 2018, 23, 559–562. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Bai, Y. Reductionist synthetic community approaches in root microbiome research. Curr. Opin. Microbiol. 2019, 49, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, A.; Tan, H.; Cao, L.; Zhang, R. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.-L.; Wu, Y.-L.; Shang, S.-Z.; He, F.; Luo, X.; Yang, G.-Y.; Pu, J.-X.; Chen, G.-Q.; Sun, H.-D. Four New Nortriterpenoids from Schisandra lancifolia. Helvetica Chim. Acta 2010, 93, 1975–1982. [Google Scholar] [CrossRef]
- Shi, Y.-M.; Xiao, W.-L.; Pu, J.-X.; Sun, H.-D. Triterpenoids from the Schisandraceae family: An update. Nat. Prod. Rep. 2014, 32, 367–410. [Google Scholar] [CrossRef]
- Qin, D.; Shen, W.Y.; Gao, T.C.; Zuo, S.H.; Song, H.C.; Xu, J.R.; Yu, B.H.; Peng, Y.J.; Guo, J.L.; Tang, W.W.; et al. Ka-danguslactones A-E, further oxygenated terpenoids from Kadsura angustifolia fermented by a symbiotic endophytic fungi, Penicillium ochrochloron SWUKD4.1850. Phytochemistry 2020, 174, 112335. [Google Scholar] [CrossRef]
- Han, M.; Qin, D.; Ye, T.; Yan, X.; Wang, J.; Duan, X.; Dong, J. An endophytic fungus from Trichoderma harzianum SWUKD3.1610 that produces nigranoic acid and its analogues. Nat. Prod. Res. 2018, 33, 2079–2087. [Google Scholar] [CrossRef]
- Huang, Q.; An, H.; Song, H.; Mao, H.; Shen, W.; Dong, J. Diversity and biotransformative potential of endophytic fungi associated with the medicinal plant Kadsura angustifolia. Res. Microbiol. 2014, 166, 45–55. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Hussain, A.; Hasnain, S. Interactions of bacterial cytokinins and IAA in the rhizosphere may alter phytostimulatory efficiency of rhizobacteria. World J. Microbiol. Biotechnol. 2011, 27, 2645–2654. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Yokozawa, T.; Dong, E.; Nakagawa, T.; Kashiwagi, H.; Nakagawa, H.; Takeuchi, S.; Chung, H.Y. In Vitro and in Vivo Studies on the Radical-Scavenging Activity of Tea. J. Agric. Food Chem. 1998, 46, 2143–2150. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; Kumar, G.N.; Parekh, L.J.; Poole, P.S. Role of soil microorganisms in improving P nutrition of plants. Plant. Soil 2002, 245, 83–93. [Google Scholar] [CrossRef]
- Moron-Rios, A.; Gómez-Cornelio, S.; Morales, B.O.O.; De la Rosa-García, S.; Partida-Martinez, L.P.; Quintana, P.; Alayón-Gamboa, J.A.; Cappello-García, S.; González-Gómez, S. Interactions between abundant fungal species influence the fungal community assemblage on limestone. PLoS ONE 2017, 12, e0188443. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.H. Plant Tissue Culture, 3rd ed.; Academic Press: Cambridge, MA, USA, 2013; Chapter 12 In Vitro Propagation for Commercial Production of Ornamentals; pp. 127–145. [Google Scholar] [CrossRef]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983. [Google Scholar] [CrossRef]
- Tilahun, S.; An, H.S.; Solomon, T.; Baek, M.W.; Choi, H.R.; Lee, H.C.; Jeong, C.S. Indices for the Assessment of Glycoalkaloids in Potato Tubers Based on Surface Color and Chlorophyll Content. Horticulturae 2020, 6, 107. [Google Scholar] [CrossRef]
- Kamonwannasit, S.; Nantapong, N.; Kumkrai, P.; Luecha, P.; Kupittayanant, S.; Chudapongse, N. Antibacterial activity of Aqularia crassna leaf extract against Staphylococcus epidermis by disruption of cell wall. Ann. Clin. Microbiol. 2013, 12, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulet, J.-C.; Trarieux, C.; Souquet, J.-M.; Ducasse, M.-A.; Caillé, S.; Samson, A.; Williams, P.; Doco, T.; Cheynier, V. Models based on ultraviolet spectroscopy, polyphenols, oligosaccharides and polysaccharides for prediction of wine astringency. Food Chem. 2016, 190, 357–363. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, Z. Spectrophotometric methods to determine the grand total of the triterpenoids in Betula. J. Nanjing For. Univ. 2005, 29, 110–112. [Google Scholar] [CrossRef]
- Lin, Y.; Lu, M.-F.; Liao, H.-B.; Li, Y.-X.; Han, W.; Yuan, K. Content determination of the flavonoids in the different parts and different species of Abelmoschus esculentus L. by reversed phase-high performance liquid chromatograph and colorimetric method. Pharmacogn. Mag. 2014, 10, 278–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Viticult. 1965, 16, 144–158. [Google Scholar]
- Sun, H.-D.; Qiu, S.-X.; Lin, L.-Z.; Wang, Z.-Y.; Lin, Z.-W.; Pengsuparp, T.; Pezzuto, J.M.; Fong, H.H.S.; Cordell, G.A.; Farnsworth, N.R. Nigranoic Acid, a Triterpenoid from Schisandra sphaerandra That Inhibits HIV-1 Reverse Transcriptase. J. Nat. Prod. 1996, 59, 525–527. [Google Scholar] [CrossRef]
- Dai, P.; Han, G.Q.; Arison, B.H. A new triterpenoid form Kadsura heteroclita (Roxb) Craib. Chem. Res. Chin. Univ. 1990, 11, 423–424. [Google Scholar]
- Li, R.-T.; Han, Q.-B.; Zheng, Y.-T.; Wang, R.-R.; Yang, L.-M.; Lu, Y.; Sang, S.-Q.; Zheng, Q.-T.; Zhao, Q.-S.; Sun, H.-D. Structure and anti-HIV activity of micrandilactones B and C, new nortriterpenoids possessing a unique skeleton from Schisandra micrantha. Chem. Commun. 2005, 2936–2938. [Google Scholar] [CrossRef]
- Li, R.T.; Xiang, W.; Li, S.H.; Lin, Z.W.; Sun, H.D. Lancifodilactones B-E, New Nortriterpenes from Schisandra lancifolia. J. Nat. Prod. 2004, 67, 94–97. [Google Scholar] [CrossRef]
- Szucs, Z.; Plaszko, T.; Cziaky, Z.; Kiss-Szikszai, A.; Emri, T.; Bertoti, R.; Sinka, L.T.; Vasas, G.; Gonda, S. Endophytic fungi from the roots of horseradish (Armoracia rusticana) and their interactions with the defensive metabolites of the glucosinolate-my-rosinase-isothiocyanate system. BMC Plant. Biol. 2018, 18, 85. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Amand, S.; Buisson, D.; Kunz, C.; Hachette, F.; Dupont, J.; Nay, B.; Prado, S. The fungal leaf endophyte Paraconiothyrium variabile specifically metabolizes the host-plant metabolome for its own benefit. Phytochemistry 2014, 108, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Haack, F.S.; Poehlein, A.; Kröger, C.; Voigt, C.; Piepenbring, M.; Bode, H.B.; Daniel, R.; Schäfer, W.; Streit, W.R. Molecular Keys to the Janthinobacterium and Duganella spp. Interaction with the Plant Pathogen Fusarium graminearum. Front. Microbiol. 2016, 7, 1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, R.; Sharma, K.C.; Gulati, A.; Sud, R.K.; Gulati, A. Stress-Tolerant Viridibacillus arenosi Strain IHB B 7171 from Tea Rhizosphere as a Potential Broad-Spectrum Microbial Inoculant. Indian J. Microbiol. 2017, 57, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, R.; Niu, Y.; Lin, H.; Ye, W.; Guo, L.; Hu, X. Whole genome sequence of Pantoea ananatis R100, an antagonistic bacterium isolated from rice seed. J. Biotechnol. 2016, 225, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Sajitha, K.L.; Dev, S.A.; Florence, E.J.M. Biocontrol potential of Bacillus subtilis B1 against sapstain fungus in rubber wood. Eur. J. Plant Pathol. 2017, 150, 237–244. [Google Scholar] [CrossRef]
- Naureen, Z.; Rehman, N.U.; Hussain, H.; Hussain, J.; Gilani, S.A.; Al Housni, S.K.; Mabood, F.; Khan, A.L.; Farooq, S.; Abbas, G.; et al. Exploring the Potentials of Lysinibacillus sphaericus ZA9 for Plant Growth Promotion and Biocontrol Activities against Phytopathogenic Fungi. Front. Microbiol. 2017, 8, 1477. [Google Scholar] [CrossRef]
- Chopra, A.; Vandana, U.K.; Rahi, P.; Satpute, S.; Mazumder, P.B. Plant growth promoting potential of Brevibacterium sediminis A6 isolated from the tea rhizosphere of Assam, India. Biocatal. Agric. Biotechnol. 2020, 27, 101610. [Google Scholar] [CrossRef]
- Mosquera-Espinosa, A.T.; Bayman, P.; Prado, G.A.; Gómez-Carabalí, A.; Otero, J.T. The double life of Ceratobasidium: Orchid mycorrhizal fungi and their potential for biocontrol of Rhizoctonia solani sheath blight of rice. Mycologia 2013, 105, 141–150. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Chen, X.; Meng, Z.; Guo, S. Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages. Int. J. Mol. Sci. 2020, 21, 564. [Google Scholar] [CrossRef] [Green Version]
- Oztopuz, O.; Sarigul, N.; Liaqat, F.; Park, R.-D.; Eltem, R. Chitinolytic Bacillus subtilis Ege-B-1.19 as a biocontrol agent against mycotoxigenic and phytopathogenic fungi. Turk. J. Biochem. 2018, 44, 323–331. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Correction: Author Correction: Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1117. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Khan, R.A.A. Biological control of bacterial wilt in tomato through the metabolites produced by the biocontrol fungus, Trichoderma harzianum. Egypt. J. Biol. Pest. Control. 2021, 31, 1–9. [Google Scholar] [CrossRef]
- Wei, F.; Zhang, Y.; Shi, Y.; Feng, H.; Zhao, L.; Feng, Z.; Zhu, H. Evaluation of the Biocontrol Potential of Endophytic Fungus Fusarium solani CEF559 against Verticillium dahliae in Cotton Plant. BioMed Res. Int. 2019, 2019, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Q.; Ma, Q.; Chen, Y.; Tian, B.; Xu, L.; Bai, Y.; Chen, W.; Li, X. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020, 14, 1915–1928. [Google Scholar] [CrossRef] [Green Version]
- Gohil, N.; Bhattacharjee, G.; Singh, V. Synergistic bactericidal profiling of prodigiosin extracted from Serratia marcescens in combination with antibiotics against pathogenic bacteria. Microb. Pathog. 2020, 149, 104508. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Pérez, M.M.; Torres-Mendoza, D.; Vásquez, R.; Rios, N.; Cubilla-Rios, L. Exploring the Antibacterial Activity of Pestalotiopsis spp. under Different Culture Conditions and Their Chemical Diversity Using LC–ESI–Q–TOF–MS. J. Fungi 2020, 6, 140. [Google Scholar] [CrossRef]



| Bacteria | Fungi | ||||
|---|---|---|---|---|---|
| Phylum | Genus | Numbers | Phylum | Genus | Numbers |
| Firmicutes | Bacillus sp. | SLB12 | Ascomycota | Pestalotiopsis sp. | SRF21 |
| Firmicutes | Lysinibacillus sp. | SRB6 | Ascomycota | Trichoderma sp. | SSF13 |
| Firmicutes | Viridibacilus sp. | SFB11 | Ascomycota | Colletotrichum sp. | SSF14 |
| Firmicutes | Romboutsia sp. | SRB23 | Ascomycota | Fusarium sp. | SFF11 |
| Firmicutes | Straphylococcus sp. | SSB39 | Ascomycota | Lasiodiplodia sp. | SLF1 |
| Firmicutes | Enterococcus sp. | SLB1 | Ascomycota | Bipolaris sp. | SSF12 |
| Firmicutes | Brevibacillus sp. | SFB12 | Ascomycota | Penicillium sp. | SRF9 |
| Bacteroidetes | Flavobacterium sp. | SSB1 | Ascomycota | Diaporthe sp. | SLF18 |
| Bacteroidetes | Sphingobacterium sp. | SRB12 | Ascomycota | Daldinia sp. | SLF4 |
| Bacteroidetes | Mucilaginibacter sp. | SSB4 | Ascomycota | Neopestalotiopsis sp. | SLF5 |
| Bacteroidetes | Wautersiella sp. | SSB19 | Ascomycota | Beltrania sp. | SLF21 |
| Bacteroidetes | Alloprevotella sp. | SLB9 | Ascomycota | Guignardia sp. | SLF6 |
| Actinobacteria | Nocardioides sp. | SSB16 | Ascomycota | Phomopsis sp. | SFF9 |
| Actinobacteria | Micrococcus sp. | SLB31 | Ascomycota | Botryosphaeria sp. | SRF16 |
| Actinobacteria | Brevibacterium sp. | SLB15 | Ascomycota | Hypoxylon sp. | SLF24 |
| Actinobacteria | Kocuria sp. | SLB14 | Ascomycota | Eutypella sp. | SSF11 |
| Proteobacteria | Pantoea sp. | SSB8 | Ascomycota | Hyphopichia sp. | SFF7 |
| Proteobacteria | Burkholderia sp. | SRB18 | Basidiomycota | Clitopilus sp. | SSF19 |
| Proteobacteria | Serratia sp. | SFB13 | Basidiomycota | Ceratobasidium sp. | SFF6 |
| Proteobacteria | Xanthomonas sp. | SFB7 | Basidiomycota | Phlebiopsis sp. | SSF18 |
| Proteobacteria | Limnobacter sp. | SLB7 | Basidiomycota | Emmia sp. | SFF12 |
| Proteobacteria | Duganella sp. | SRB22 | Basidiomycota | Irpex sp. | SRF17 |
| Proteobacteria | Rhizobium sp. | SLB15 | Basidiomycota | Phlebia sp. | SLF27 |
| Proteobacteria | Agrobacterium sp. | SSB31 | Basidiomycota | Perenniporia sp. | SLF26 |
| Proteobacteria | Herbaspirillum sp. | SFB6 | Zygomycota | Umbelopsis sp. | SLF28 |
| Proteobacteria | Stenotrophomonas sp. | SSB25 | Zygomycota | Mucor sp. | SRF1 |
| Proteobacteria | Enterobacter sp. | SLB11 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, C.; Qin, D.; Wang, Y.; Lan, W.; Li, Y.; Yu, B.; Peng, Y.; Xu, J.; Dong, J. Plant Triterpenoids Regulate Endophyte Community to Promote Medicinal Plant Schisandra sphenanthera Growth and Metabolites Accumulation. J. Fungi 2021, 7, 788. https://doi.org/10.3390/jof7100788
You C, Qin D, Wang Y, Lan W, Li Y, Yu B, Peng Y, Xu J, Dong J. Plant Triterpenoids Regulate Endophyte Community to Promote Medicinal Plant Schisandra sphenanthera Growth and Metabolites Accumulation. Journal of Fungi. 2021; 7(10):788. https://doi.org/10.3390/jof7100788
Chicago/Turabian StyleYou, Chuan, Dan Qin, Yumeng Wang, Wenyi Lan, Yehong Li, Baohong Yu, Yajun Peng, Jieru Xu, and Jinyan Dong. 2021. "Plant Triterpenoids Regulate Endophyte Community to Promote Medicinal Plant Schisandra sphenanthera Growth and Metabolites Accumulation" Journal of Fungi 7, no. 10: 788. https://doi.org/10.3390/jof7100788
APA StyleYou, C., Qin, D., Wang, Y., Lan, W., Li, Y., Yu, B., Peng, Y., Xu, J., & Dong, J. (2021). Plant Triterpenoids Regulate Endophyte Community to Promote Medicinal Plant Schisandra sphenanthera Growth and Metabolites Accumulation. Journal of Fungi, 7(10), 788. https://doi.org/10.3390/jof7100788





