A Screen for Small Molecules to Target Candida albicans Biofilms
Abstract
1. Introduction
2. Materials and Methods
2.1. Media and Strains
2.2. Reagents
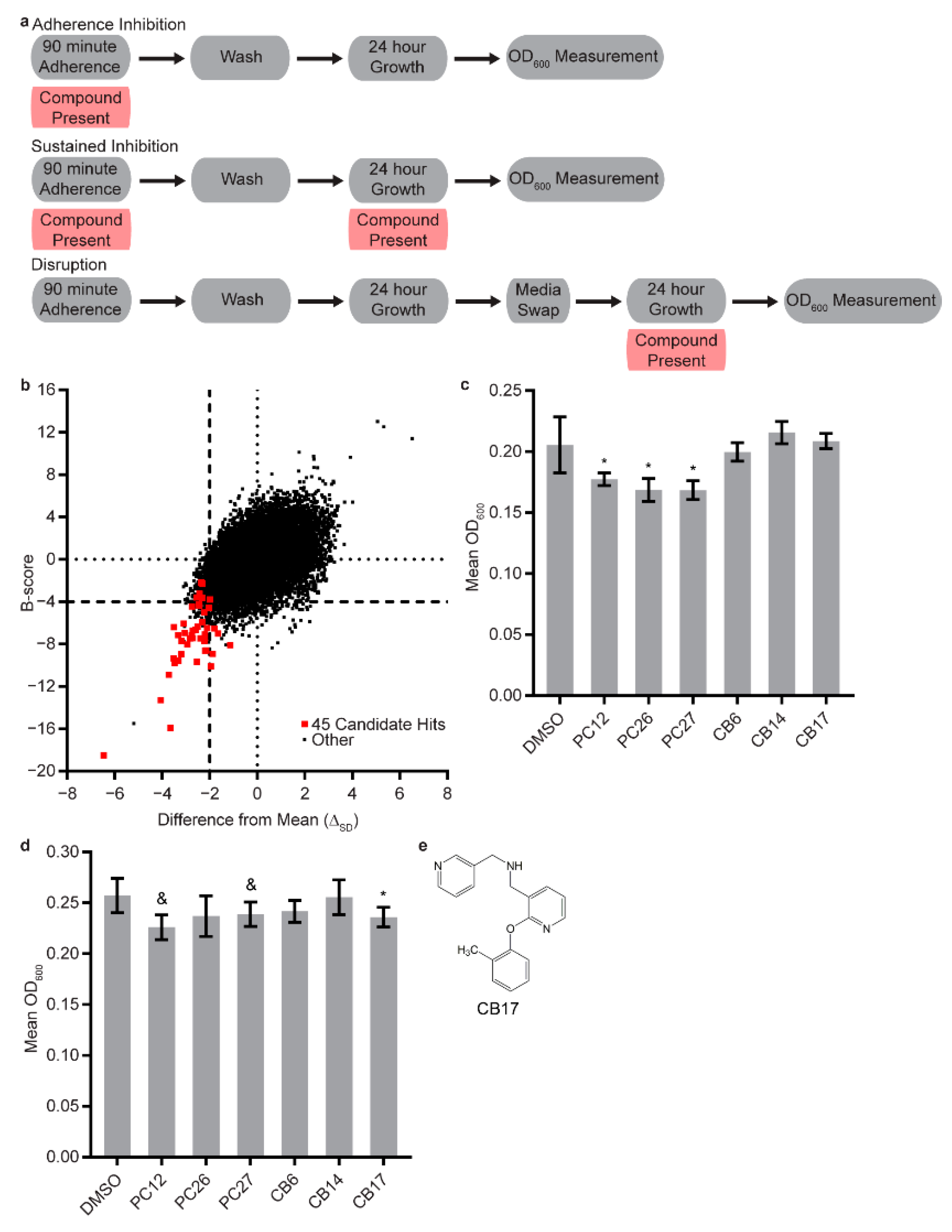
2.3. Biofilm Assays
2.4. Candidate Compound Selection
2.5. Statistical Analysis and “Hit” Calling for the Biofilm Assays
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Douglas, L.J. Candida biofilms and their role in infection. Trends Microbiol. 2003, 11, 30–36. [Google Scholar] [CrossRef]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2018, 16, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Kullberg, B.J.; Oude Lashof, A.M.L. Epidemiology of opportunistic invasive mycoses. Eur. J. Med. Res. 2002, 7, 183–191. [Google Scholar]
- Kim, J.; Sudbery, P. Candida albicans, a major human fungal pathogen. J. Microbiol. 2011, 49, 171–177. [Google Scholar] [CrossRef]
- Achkar, J.M.; Fries, B.C. Candida infections of the genitourinary tract. Clin. Microbiol. Rev. 2010, 23, 253–273. [Google Scholar] [CrossRef]
- Ganguly, S.; Mitchell, A.P. Mucosal biofilms of Candida albicans. Curr. Opin. Microbiol. 2011, 14, 380–385. [Google Scholar] [CrossRef]
- Kennedy, M.J.; Volz, P.A. Ecology of Candida albicans gut colonization: Inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 1985, 49, 654–663. [Google Scholar] [CrossRef]
- Kumamoto, C.A. Candida biofilms. Curr. Opin. Microbiol. 2002, 5, 608–611. [Google Scholar] [CrossRef]
- Kumamoto, C.A. Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 2011, 14, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Pappas, P.G.; Rex, J.H.; Sobel, J.D.; Filler, S.G.; Dismukes, W.E.; Walsh, T.J.; Edwards, J.E. Guidelines for treatment of candidiasis. Clin. Infect. Dis. 2004, 38, 161–189. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, R.P. Nosocomial candidemia: Risk factors and attributable mortality. Clin. Infect. Dis. 1995, 20, 1531–1534. [Google Scholar] [CrossRef]
- López-Ribot, J.L. Candida albicans biofilms: More than filamentation. Curr. Biol. 2005, 15, R453–R455. [Google Scholar] [CrossRef]
- Ramage, G.; Mowat, E.; Jones, B.; Williams, C.; Lopez-Ribot, J. Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 2009, 35, 340–355. [Google Scholar] [CrossRef]
- Douglas, L.J. Medical importance of biofilms in Candida infections. Rev. Iberoam. Micol. 2002, 19, 139–143. [Google Scholar]
- Fox, E.P.; Nobile, C.J. A sticky situation: Untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription 2012, 3, 315–322. [Google Scholar] [CrossRef]
- Chandra, J.; Kuhn, D.M.; Mukherjee, P.K.; Hoyer, L.L.; McCormick, T.; Ghannoum, M.A. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J. Bacteriol. 2001, 183, 5385–5394. [Google Scholar] [CrossRef]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef]
- Ramage, G.; Martínez, J.P.; López-Ribot, J.L. Candida biofilms on implanted biomaterials: A clinically significant problem. FEMS Yeast Res. 2006, 6, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Posteraro, B.; Trecarichi, E.M.; Fiori, B.; Rossi, M.; Porta, R.; de Gaetano Donati, K.; La Sorda, M.; Spanu, T.; Fadda, G.; et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J. Clin. Microbiol. 2007, 45, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef]
- Tumbarello, M.; Fiori, B.; Trecarichi, E.M.; Posteraro, P.; Losito, A.R.; De Luca, A.; Sanguinetti, M.; Fadda, G.; Cauda, R.; Posteraro, B. Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital. PLoS ONE 2012, 7, e33705. [Google Scholar] [CrossRef]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Playford, G.; Reboli, A.C.; Rex, J.H.; Sobel, J.D.; Pappas, P.G.; Kullberg, B.J. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: A patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012, 54, 1110–1122. [Google Scholar] [CrossRef]
- Fox, E.P.; Singh-Babak, S.D.; Hartooni, N.; Nobile, C.J. Biofilms and Antifungal Resistance. In Antifungals: From Genomics to Resistance and the Development of Novel Agents; Caister Academic Press: Poole, UK, 2015; pp. 71–90. [Google Scholar]
- Shinde, R.B.; Chauhan, N.M.; Raut, J.S.; Karuppayil, S.M. Sensitization of Candida albicans biofilms to various antifungal drugs by cyclosporine A. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 27. [Google Scholar] [CrossRef]
- Bink, A.; Kucharíková, S.; Neirinck, B.; Vleugels, J.; Van Dijck, P.; Cammue, B.P.A.; Thevissen, K. The Nonsteroidal Antiinflammatory Drug Diclofenac Potentiates the In Vivo Activity of Caspofungin Against Candida albicans Biofilms. J. Infect. Dis. 2012, 206, 1790–1797. [Google Scholar] [CrossRef]
- Barchiesi, F.; Spreghini, E.; Maracci, M.; Fothergill, A.W.; Baldassarri, I.; Rinaldi, M.G.; Scalise, G. In Vitro Activities of Voriconazole in Combination with Three Other Antifungal Agents against Candida glabrata. Antimicrob. Agents Chemother. 2004, 48, 3317–3322. [Google Scholar] [CrossRef]
- Chatzimoschou, A.; Katragkou, A.; Simitsopoulou, M.; Antachopoulos, C.; Georgiadou, E.; Walsh, T.J.; Roilides, E. Activities of Triazole-Echinocandin Combinations against Candida Species in Biofilms and as Planktonic Cells. Antimicrob. Agents Chemother. 2011, 55, 1968–1974. [Google Scholar] [CrossRef]
- Uppuluri, P.; Nett, J.; Heitman, J.; Andes, D. Synergistic Effect of Calcineurin Inhibitors and Fluconazole against Candida albicans Biofilms. Antimicrob. Agents Chemother. 2008, 52, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, S.P.; Ramage, G.; VandeWalle, K.; Patterson, T.F.; Wickes, B.L.; López-Ribot, J.L. Antifungal Combinations against Candida albicans Biofilms In Vitro. Antimicrob. Agents Chemother. 2003, 47, 3657–3659. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; McCarthy, M.; Alexander, E.L.; Antachopoulos, C.; Meletiadis, J.; Jabra-Rizk, M.A.; Petraitis, V.; Roilides, E.; Walsh, T.J. In vitro interactions between farnesol and fluconazole, amphotericin B or micafungin against Candida albicans biofilms. J. Antimicrob. Chemother. 2015, 70, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Troskie, A.M.; Rautenbach, M.; Delattin, N.; Vosloo, J.A.; Dathe, M.; Cammue, B.P.A.; Thevissen, K. Synergistic Activity of the Tyrocidines, Antimicrobial Cyclodecapeptides from Bacillus aneurinolyticus, with Amphotericin B and Caspofungin against Candida albicans Biofilms. Antimicrob. Agents Chemother. 2014, 58, 3697–3707. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Xu, X.; Wu, C.D. In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol. 2011, 56, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.A.; Ahmad, I. Antibiofilm activity of certain phytocompounds and their synergy with fluconazole against Candida albicans biofilms. J. Antimicrob. Chemother. 2012, 67, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.; Fukazawa, H.; Ohno, H.; Miyazaki, Y. Combinatory effect of fluconazole and FDA-approved drugs against Candida albicans. J. Infect. Chemother. 2013, 19, 1141–1145. [Google Scholar] [CrossRef]
- De Cremer, K.; Lanckacker, E.; Cools, T.L.; Bax, M.; De Brucker, K.; Cos, P.; Cammue, B.P.A.; Thevissen, K. Artemisinins, New Miconazole Potentiators Resulting in Increased Activity against Candida albicans Biofilms. Antimicrob. Agents Chemother. 2015, 59, 421–426. [Google Scholar] [CrossRef]
- Delattin, N.; De Brucker, K.; Vandamme, K.; Meert, E.; Marchand, A.; Chaltin, P.; Cammue, B.P.A.; Thevissen, K. Repurposing as a means to increase the activity of amphotericin B and caspofungin against Candida albicans biofilms. J. Antimicrob. Chemother. 2014, 69, 1035–1044. [Google Scholar] [CrossRef]
- LaFleur, M.D.; Lucumi, E.; Napper, A.D.; Diamond, S.L.; Lewis, K. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J. Antimicrob. Chemother. 2011, 66, 820–826. [Google Scholar] [CrossRef]
- Lohse, M.B.; Gulati, M.; Craik, C.S.; Johnson, A.D.; Nobile, C.J. Combination of Antifungal Drugs and Protease Inhibitors Prevent Candida albicans Biofilm Formation and Disrupt Mature Biofilms. Front. Microbiol. 2020, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Ennis, C.L.; Hartooni, N.; Johnson, A.D.; Lohse, M.B. A Selective Serotonin Reuptake Inhibitor, a Proton Pump Inhibitor, and Two Calcium Channel Blockers Inhibit Candida albicans Biofilms. Microorganisms 2020, 8, 756. [Google Scholar] [CrossRef] [PubMed]
- Watamoto, T.; Egusa, H.; Sawase, T.; Yatani, H. Screening of Pharmacologically Active Small Molecule Compounds Identifies Antifungal Agents Against Candida Biofilms. Front. Microbiol. 2015, 6, 1453. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.G.; Chaturvedi, A.K.; Lazzell, A.L.; Powell, A.T.; Saville, S.P.; McHardy, S.F.; Lopez-Ribot, J.L. A novel small molecule inhibitor of Candida albicans biofilm formation, filamentation and virulence with low potential for the development of resistance. NPJ Biofilms Microbiomes 2015, 1, 15012. [Google Scholar] [CrossRef]
- Gulati, M.; Lohse, M.B.; Ennis, C.L.; Gonzalez, R.E.; Perry, A.M.; Bapat, P.; Arevalo, A.V.; Rodriguez, D.L.; Nobile, C.J. In Vitro Culturing and Screening of Candida albicans Biofilms. Curr. Protoc. Microbiol. 2018, 50, e60. [Google Scholar] [CrossRef]
- Lohse, M.B.; Gulati, M.; Arevalo, A.V.; Fishburn, A.; Johnson, A.D.; Nobile, C.J. Assessment and optimizations of Candida albicans in vitro biofilm assays. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef]
- Nobile, C.J.; Fox, E.P.; Hartooni, N.; Mitchell, K.F.; Hnisz, D.; Andes, D.R.; Kuchler, K.; Johnson, A.D. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio 2014, 5, e01201–e01214. [Google Scholar] [CrossRef]
- Brideau, C.; Gunter, B.; Pikounis, B.; Liaw, A. Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 2003, 8, 634–647. [Google Scholar] [CrossRef]
- Malo, N.; Hanley, J.A.; Cerquozzi, S.; Pelletier, J.; Nadon, R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 2006, 24, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Dandapani, S.; Rosse, G.; Southall, N.; Salvino, J.M.; Thomas, C.J. Selecting, Acquiring, and Using Small Molecule Libraries for High-Throughput Screening. Curr. Protoc. Chem. Biol. 2012, 4, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Huggins, D.J.; Venkitaraman, A.R.; Spring, D.R. Rational methods for the selection of diverse screening compounds. ACS Chem. Biol. 2011, 6, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.J.D.; Isidro-Llobet, A.; Spring, D.R. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat. Commun. 2010, 1, 80. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lohse, M.B.; Ennis, C.L.; Hartooni, N.; Johnson, A.D.; Nobile, C.J. A Screen for Small Molecules to Target Candida albicans Biofilms. J. Fungi 2021, 7, 9. https://doi.org/10.3390/jof7010009
Lohse MB, Ennis CL, Hartooni N, Johnson AD, Nobile CJ. A Screen for Small Molecules to Target Candida albicans Biofilms. Journal of Fungi. 2021; 7(1):9. https://doi.org/10.3390/jof7010009
Chicago/Turabian StyleLohse, Matthew B., Craig L. Ennis, Nairi Hartooni, Alexander D. Johnson, and Clarissa J. Nobile. 2021. "A Screen for Small Molecules to Target Candida albicans Biofilms" Journal of Fungi 7, no. 1: 9. https://doi.org/10.3390/jof7010009
APA StyleLohse, M. B., Ennis, C. L., Hartooni, N., Johnson, A. D., & Nobile, C. J. (2021). A Screen for Small Molecules to Target Candida albicans Biofilms. Journal of Fungi, 7(1), 9. https://doi.org/10.3390/jof7010009





