Abstract
Candidiasis caused by species of the Candida haemulonii complex (Candida haemulonii and Candida duobushaemulonii) and closely related species, Candida auris and Candida pseudohaemulonii are increasing. These species often show reduced susceptibility to antifungal drugs, such as azoles and amphotericin B or, less frequently, echinocandins. However, conventional phenotypic identification methods are unable to accurately differentiate these species and, therefore, their prevalence may have been underestimated. In this study, 150 isolates that were probably misidentified were reanalyzed using two novel PCR approaches. We found that one isolate previously identified in 1996 as Candida intermedia was C. duobushaemulonii, being one of the oldest isolates of this species described to date. We also found that this isolate had reduced susceptibility to fluconazole, itraconazole, and amphotericin B.
1. Introduction
The genus Candida is demonstrated to be genetically heterogeneous. For that reason, many Candida species belong to a complex formed by various cryptic species that are phenotypically identical, and only molecular-based methods guarantee their accurate identification [1]. This is the case for Candida haemulonii, an opportunistic pathogen whose first clinical isolation, from a blood sample, was described in 1984 [2]. Prior to 2012, C. haemulonii was separated into two genetically distinct groups based on isoenzyme and protein profiles and DNA relatedness studies [3]. However, in the same year, as further studies revealed the heterogeneity in the DNA sequence of various genes, the complex was reclassified as two species (C. haemulonii sensu stricto and Candida duobushaemulonii) and one variety (C. haemulonii var. vulnera) [4]. Concurrently, two new species phylogenetically close to C. haemulonii complex, namely, Candida pseudohaemulonii and Candida auris, were described in 2006 and 2009, respectively [5,6].
Although infections caused by this complex of species remain rare, since they are not usually involved in outbreaks but in sporadic infections [7,8,9], they are increasingly being isolated worldwide and are reported as important fungi in clinical settings [10,11,12,13]. However, the case of C. auris stands out. Since its first description in 2009 in Japan, this fungus has been identified worldwide in a short time span, suggesting rapid dissemination. As of today, Spain, the United Kingdom, India, Kuwait and USA are among the world countries with the highest number of reported cases (>100) [14,15]. Infections by C. auris cause a wide range of clinical manifestations, but it has mostly been isolated from blood and other deep-seated locations. Moreover, this yeast is associated with nosocomial outbreaks and an estimated high in-hospital mortality rate (30–72%) [14,15].
Both species of the C. haemulonii complex and its relatives C. auris and C. pseudohaemulonii frequently show in vitro resistance to amphotericin B and fluconazole, which often leads to clinical failure [4,5,10,16,17,18,19]. Although less common, low susceptibility to echinocandins has also been reported [4,18,20,21,22]. Besides, accurate identification of these species is troublesome, as conventional panels used in routine microbiology laboratories frequently misidentify them. These species are mistaken not only for other Candida species, but also with close genera, such as Saccharomyces and Rhodotorula [23]. As a result, these pathogens are often unreported as causal agents of fungal infections and, consequently, their actual incidence and global prevalence might be underestimated [24,25].
Currently, only matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and molecular techniques, such as sequencing of genetic loci of the ribosomal DNA¸ including the D1/D2 domain, internal transcribed spacer (ITS) regions, and RPB1 and RPB2, have been demonstrated to provide reliable identification of these species [16,18]. However, these techniques are not available in most routine microbiology laboratories, and there may be issues when testing mixed clinical samples, as they could be not able to distinguish between the species involved [26]. Aiming to achieve simple and easily accessible techniques for accurate identification of C. auris, various species-specific conventional or real-time PCR-based assays have been developed. These assays involve amplification of different regions of the ribosomal gene complex [27,28,29,30] or, alternatively, unique glycosylphosphatidylinositol (GPI) protein-encoding genes [31]. Likewise, given the growing importance of the complex, other groups have come up with assays that allow discrimination between C. auris and its closest relatives [27,32,33,34].
In light of the above, the aim of this study was to deepen our knowledge about the prevalence of this emergent, multidrug-resistant complex of cryptic Candida species by surveying our culture collection for previous potentially misidentified clinical isolates using novel molecular techniques.
2. Materials and Methods
2.1. Fungal Isolates
This study includes 150 clinical isolates from the yeast stock collection of the Medical Mycology Laboratory at the University of the Basque Country (UPV/EHU), collected between 1993 and 2014, which are likely to have been inaccurately identified. These isolates were previously identified by API®ID 32C (bioMérieux, Marcy l’Etoile, France) biochemical panel as: Candida famata (8), C. famata/Candida guilliermondii (8), C. guilliermondii/C. famata (8), C. haemulonii (8), Candida intermedia (8), Candida lipolytica (8), Candida lusitaniae (8), C. lusitaniae/Candida pulcherrima (8), Candida rugosa (8), Candida tropicalis (8), Candida kefyr (7), Candida valida (7), Candida pelliculosa (6), Candida lambica (4), Candida nivariensis (4), Candida sake (4), Candida spp. (4), Candida zeylanoides (4), Candida colliculosa (3), Candida curvata (2), Candida holmii (2), Candida magnoliae (2), Candida norvegensis/Candida inconspicua (2), Candida rubra (2), Candida stellatoidea (2), Debaryomyces polymorphus (2), Saccharomyces cerevisiae (2), Candida catenulata (1), Candida glabrata (1), C. guilliermondii (1), C. lusitaniae/C. tropicalis (1), Candida norvegensis (1), C. pulcherrima/C. lusitaniae (1), C. pulcherrima (1), Candida quercitrusa (1), Candida silvicola (1), Candida sake/C. tropicalis (1), and Candida utilis (1). Additionally, as positive controls, we included two C. auris and one C. duobushaemulonii clinical isolates from Hospital La Fe (Valencia), already confirmed by ITS sequencing, and a C. pseudohaemulonii reference strain (CBS 10004).
2.2. Sample Preparation and DNA Extraction
Both clinical isolates and the reference strain were cultured on Sabouraud dextrose agar (SDA) or yeast extract peptone dextrose (YEPD) medium at 37 °C during 24–48 h. Then, DNA extraction was performed using the DNeasy® UltraClean® Microbial Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. DNA purity and quantity were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. PCR-Based Molecular Identification
The 150 isolates and the four positive controls were analyzed by two different PCR techniques, using primers and amplification conditions described by Ruiz-Gaitan et al. [35] and Arastehfar et al. [32] (Table 1). One was a duplex C. auris-specific PCR that also amplifies the 5.8S rDNA gene as quality control band [35] and the other was a C. haemulonii complex-tetraplex PCR [32] (Figure 1).

Table 1.
Primers used in this study for each PCR-based method and expected amplicon size.
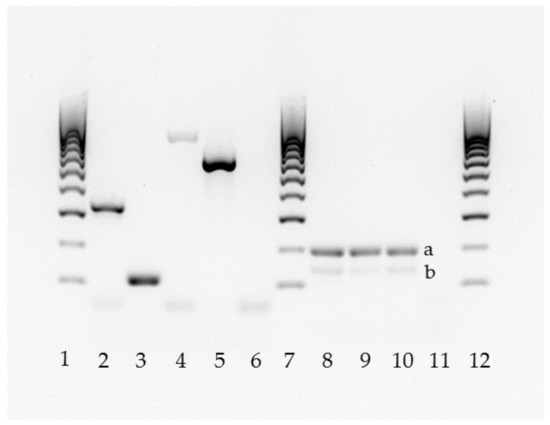
Figure 1.
Molecular identification of the C. haemulonii complex by two PCR methods (lanes 2–6 [32]; lanes 8–11 [35]). Lanes 1, 7, and 12: 100 bp DNA ladder. Lane 2: C. auris; 3: C. duobushaemulonii; 4: C. haemulonii; 5: C. pseudohaemulonii; 6: negative control. Lanes 8–10: C. auris (a), 5.8S rDNA band (b); 11: negative control without DNA.
2.4. ITS Region Sequencing and Species Identification
When robust amplification in any of the PCR or low-intensity bands were detected, sequencing of the ITS1-5.8S-ITS2 gene was performed for confirmation. This region was amplified with ITS1 and ITS4 primers as described previously [36]. Unpurified PCR products were sent to the Advanced Research Facilities of the University of the Basque Country (SGIker), where samples were purified and sequenced. Received sequences from both DNA strands were analyzed by Chromas v. 2.6.4 (Technelysium Pty Ltd., South Brisbane, Australia) and BioEdit v. 7.2.6.1 (Tom Hall, Raleigh, NC, USA), and sequence identity for species identification was determined using the Basic Local Alignment Search Tool (BLAST) at the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.5. In Vitro Antifungal Susceptibility
In vitro susceptibility testing of the identified isolates related to the C. haemulonii complex (n = 9) as well as the positive controls (n = 4) against nine different antifungal agents (anidulafungin, micafungin, caspofungin, posaconazole, voriconazole, itraconazole, fluconazole, amphotericin B, and 5-fluorocytosine) was tested using SensititreTM YeastOne YO10 (Thermo Scientific, Waltham, MA, USA), following the manufacturer’s instructions. Candida parapsilosis ATCC 22019 and C. krusei ATCC 6258 were included as quality controls.
2.6. Phenotypical Characterization
All the isolates belonging to the C. haemulonii complex (n = 13) were plated on CHROMagar Candida medium supplemented with Pal’s medium at both 37 °C and 42 °C during 24–48 h, in order to assess growth ability and colony characteristics [37].
3. Results
3.1. Molecular Identification
Except for the two confirmed C. auris clinical isolates, no other C. auris was found using the C. auris-specific PCR. Nevertheless, as the quality control band of 5/8 C. rugosa, 7/8 C. lipolytica, 1/2 C. magnoliae, and 1/8 C. intermedia was absent, ITS sequencing was performed for these isolates. The identification of the seven C. lipolytica and the only C. magnoliae isolate was confirmed. On the contrary, the five C. rugosa and the only C. intermedia were all re-identified as Candida pararugosa (Table 2). The C. haemulonii complex-tetraplex PCR method accurately identified the four isolates used as positive controls. Neither C. auris nor C. pseudohaemulonii were detected in the collection and the eight isolates previously identified as C. haemulonii were confirmed by the tetraplex PCR, as they all produced a 696 bp sized band. Interestingly, one isolate identified in 1996 as C. intermedia by API®ID 32C produced a 115 bp sized band, corresponding to C. duobushaemulonii. Additionally, we re-analyzed this isolate by the current API®ID 32C panel, which identified it as C. sake (87.4%, acceptable profile). The eight C. haemulonii isolates, the new C. duobushaemulonii, as well as the four positive controls, were all confirmed by ITS sequencing showing a percentage of identity with the reference strains higher than 99%.

Table 2.
Profiles of identification and susceptibility of clinical and control isolates of C. haemulonii complex against nine antifungal agents.
3.2. Fungal Susceptibility
Although the C. duobushaemulonii isolate that was used as positive control showed susceptibility to all azole agents, the re-identified C. duobushaemulonii isolate was less susceptible to itraconazole (MIC = 0.5 μg/mL) and fluconazole (MIC = 32 μg/mL). Six C. haemulonii isolates also showed a lower susceptibility to itraconazole (MIC = 0.5 μg/mL) and fluconazole (MIC = 16–32 μg/mL), other one was inhibited by 1 μg/mL of itraconazole, and another one by 64 μg/mL of fluconazole, and another showed reduced susceptibility to both agents (MIC = 1 and 64 μg/mL, respectively, of itraconazole and fluconazole). Both C. auris isolates were less susceptible to fluconazole (MIC > 256 μg/mL) and itraconazole (MIC = 0.25 μg/mL). One of them also showed decreased susceptibility to voriconazole (MIC = 2 μg/mL). With regard to amphotericin B, all but one C. haemulonii isolate showed reduced susceptibility (MIC > 2 μg/mL). Finally, no reduced susceptibility was found to either echinocandins or 5-flucytosine (Table 2).
3.3. Phenotypic Characteristics
When grown on chromogenic agar, all but the C. pseudohaemulonii reference strain, whose colonies were purple, showed smooth, matte, and light-pink colored colonies (Figure 2). When cultured at 42 °C, only the two C. auris isolates were able to grow.
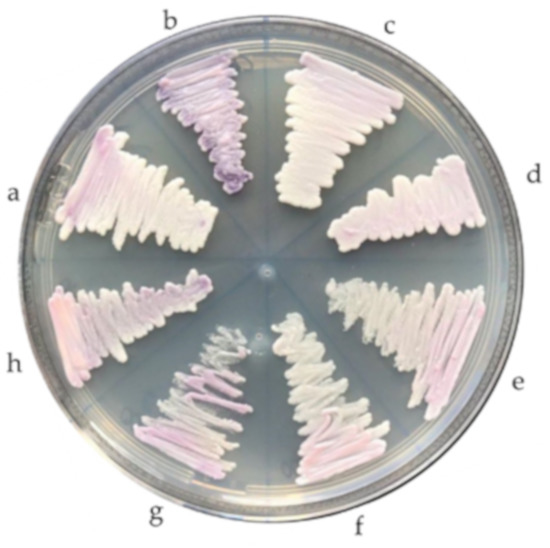
Figure 2.
Phenotypic characteristics of Candida haemulonii complex species grown on CHROMagar Candida medium: (a) Candida duobushaemulonii, (b) Candida pseudohaemulonii, (c,d) Candida auris, and (e–h) Candida haemulonii.
4. Discussion
Although Candida albicans remains the most often isolated species, a remarkable shift in the etiology of candidiasis is ongoing, as previously uncommon non-C. albicans species are arising or increasing in prevalence [38]. Part of this increase in prevalence of uncommon species of Candida might be due to the broadened use of molecular methods, such as PCR and MALDI-TOF for identification of clinical isolates. Examples of such pathogens are species related to the C. haemulonii complex, whose frequency has increased over the years. Between 1997 and 2007, the prevalence of C. haemulonii was very low (<0.01%), as observed by the ARTEMIS DISK Global Surveillance Study [39]. In a different retrospective study that was carried out between 2001 and 2005, this species was the sixth most common Candida species isolated (1.5%). Ramos et al. reported that C. haemulonii was the fourth most common species (12.1%) causing cutaneous candidiasis in Brazil from 2008 to 2009 [10]. A study in India reported that the prevalence of this species increased from 5.5% in 2006 to 18.2% in 2008 [40]. However, the identifications in those studies were performed using commercially available biochemical methods, such as API, MicroScan, and Vitek, which have been proved to misidentify these species. In addition, C. duobushaemulonii was not described until 2012, when Cendejas-Bueno et al. established C. haemulonii as a complex of species [4]. Therefore, whether candidiasis caused by these species is relatively new, as suggested by Frías-de-León et al. [13], or longstanding but unidentified remains unknown. Our results support this latter idea. We carried out a retrospective study in which we surveyed for species related to this complex in a culture collection that contains clinical isolates from 1993 to 2014 using novel molecular methods. From that year onwards, all the isolates with questionable identifications were sequenced. Interestingly, we discovered that one isolate that was identified in 1996 as C. intermedia by API®ID 32C in fact was C. duobushaemulonii, and this type of misidentification has not been described before. In accordance with literature, this result shows the inaccuracy of biochemical methods towards correct identification of some infrequent Candida species and, consequently, that the real clinical incidence of the C. haemulonii complex might have been underestimated. For instance, Ramos et al. found that four C. haemulonii isolates that were collected between 2009 and 2013 in fact were C. duobushaemulonii [10,23].
Moreover, we show that the current API®ID 32C panel is still unable to identify this species, which reinforces the importance of using molecular methods or MALDI-TOF to detect pathogens related to the complex. The PCR proposed by Ruiz-Gaitán et al. [31] that was tested in our study accurately identified both C. auris isolates and did not yield any C. auris-specific amplicon with other tested species. The 5.8S rDNA control amplicon was not produced with C. pararugosa, C. magnoliae, and C. lipolytica isolates. Accordingly, Ruiz-Gaitán et al. already reported that these control primers do not yield amplicons and therefore cannot be used in more distinct species such as C. lipolytica [31], which we confirmed in our study. However, as there are no studies of reported misidentifications of C. auris with those three species and their incidence of causing candidiasis is utterly low, we believe this PCR design is a powerful approach for clinicians to effectively detect C. auris infections. Likewise, the tetraplex-PCR proposed by Arastehfar et al. [32] is also an accurate and useful molecular technique. This was also demonstrated by Frias-de-León et al. [13], who showed correct identification of four C. auris control isolates, eight C. haemulonii isolates, and two C. duobushaemulonii, whereas no other isolate yielded any amplicon.
The re-identified C. duobushaemulonii isolate in the present study was obtained in 1996 from a toenail of a patient from Bizkaia (Spain), while the eight C. haemulonii isolates had been obtained in 2000 from various sources, including both invasive and superficial infections. Our results support the idea that species of this complex were primarily associated to superficial infections, prior to its emergence related to invasive candidiasis. Even though C. haemulonii was first clinically isolated in 1984 from the blood of a patient [2], most of the invasive infections caused by species belonging to this complex were reported in more recent years. In 1991, Gargeya et al. reported onychomycoses cases in the USA caused by C. haemulonii [41], and the five isolates used by Lehmann et al. in 1993 were also obtained from skin and nails [3]. In the study carried out by Ramos et al., all the isolates obtained from non-invasive sources (nails and skin) were collected prior to 2009, whereas all isolates collected between 2010 and 2013 originated from invasive sites [10]. Likewise, Hou et al. collected 31 clinical isolates (26 C. haemulonii and five C. duobushaemulonii) from 2010 to 2014, which were all recovered from invasive sources, mainly from blood and cerebrospinal fluid [22]. Although Lehmann et al. had already described in 1993 that C. haemulonii was a complex of two genetically different groups (I and II) [3], our re-identified isolate, together with the first isolate from a foot ulcer in 1990 [42] are, to our knowledge, the oldest C. duobushaemulonii isolates found. This underlines the importance of our study to understand the origin and spread of cryptic emergent species.
The C. duobushaemulonii isolate re-identified in our study, the eight C. haemulonii isolates and the two control C. auris isolates had reduced susceptibility to fluconazole and itraconazole, whereas seven C. haemulonii isolates had also reduced susceptibility to amphotericin B. This concurs with other studies, as clinical isolates of this complex usually show in vitro reduced susceptibility or resistance to these antifungal drugs [4,5,10,16,17,18,19], often being associated with clinical failure [4,10,17]. Recently, a potential mechanism of resistance among the species of C. haemulonii complex to amphotericin B has been described. The membrane of the species of the complex is composed mainly of ergosterol pathway intermediates, thus there is no target for amphotericin B [43]. As all clinical isolates were susceptible to echinocandins, these antifungal drugs might be the best treatment option for infections caused by species related to the complex, as was also suggested elsewhere [12,17]. Nevertheless, the tendency of the complex to show resistance to azoles, echinocandins, and amphotericin B reinforces the necessity to accurately identify these uncommon species.
Finally, morphological characteristics such as colony color are not very useful for the identification of these species. Although in our study C. haemulonii, C. auris, and C. duobushaemulonii isolates showed smooth, light-pink colored colonies and the C. pseudohaemulonii isolate purple colonies on CHROMagar, other authors have reported that C. haemulonii complex species and C. auris can also show a dark pink color [21]. On the other hand, the ability of only C. auris isolates to grow well at 42 °C, which has also been described before [10,18,37], could comprise a helpful characteristic to differentiate C. auris from the other species.
5. Conclusions
Species of the C. haemulonii complex are emerging invasive pathogens that often show resistance to commonly used antifungal drugs. Therefore, their accurate and quick identification using novel molecular methods is essential as they can cause challenging invasive infections that pose difficulties in their therapeutic management. Although initially described as etiological agents of superficial candidiasis, surveillance should be conducted to clarify the mechanisms involved in developing invasive candidiasis.
Author Contributions
Conceptualization, C.M.-A., G.Q., and E.E.; methodology, I.J.-M. and C.M.-A.; validation, P.W.J.d.G. and E.E.; formal analysis, E.T. and A.G.; investigation, I.J.-M. and C.M.-A.; data curation, I.J.-M. and C.M.-A.; writing—original draft preparation, I.J.-M. and C.M.-A.; writing—review and editing, E.T., A.G., P.W.J.d.G., G.Q., and E.E.; supervision, G.Q. and E.E.; project administration, P.W.J.d.G., G.Q., and E.E.; funding acquisition, C.M.-A., P.W.J.d.G., G.Q., and E.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) [SAF2017-86188-P] and from the Consejería de Educación, Universidades e Investigación of Gobierno Vasco-Eusko Jaurlaritza [GIC15/78 IT-990-16]. C.M.-A. is recipient of a research grant from the program Reina Letizia para la inclusión of the Real Patronato de Discapacidad.
Acknowledgments
Alba Ruiz-Gaitán and Javier Pemán (Hospital Universitario y Politécnico La Fe of Valencia, Spain) are thanked for kindly providing the C. auris clinical isolates.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Brandt, M.E.; Lockhart, S.R. Recent taxonomic developments with Candida and other opportunistic yeasts. Curr. Fungal Infect. Rep. 2012, 6, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Lavarde, V.; Daniel, F.; Saez, H.; Arnold, M.; Faguer, B. Peritonite mycosique a Torulopsis haemulonii. Bull. Soc. Fr. Mycol. Med. 1984, 13, 173–176. [Google Scholar]
- Lehmann, P.F.; Wu, L.C.; Pruitt, W.R.; Meyer, S.A.; Ahearn, D.G. Unrelatedness of groups of yeasts within the Candida haemulonii complex. J. Clin. Microbiol. 1993, 31, 1683–1687. [Google Scholar] [CrossRef] [PubMed]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gómez-López, A.; Boekhout, T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii varvulnera var. nov.: Three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar] [CrossRef] [PubMed]
- Sugita, T.; Takashima, M.; Poonwan, N.; Mekha, N. Candida pseudohaemulonii Sp. Nov., an amphotericin B-and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol. Immunol. 2006, 50, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- González, G.M.; Treviño-Rangel, R.J.; Palma-Nicolás, J.P.; Martínez, C.; González, J.G.; Ayala, J.; Caballero, A.; Morfín-Otero, R.; Rodríguez-Noriega, E.; Velarde, F.; et al. Species distribution and antifungal susceptibility of bloodstream fungal isolates in paediatric patients in Mexico: A nationwide surveillance study. J. Antimicrob. Chemother. 2013, 68, 2847–2851. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Dzajic, E.; Jensen, R.H.; Johansen, H.K.; Kjaeldgaard, P.; Knudsen, J.D.; Kristensen, L.; Leitz, C.; Lemming, L.E.; Nielsen, L.; et al. Epidemiological changes with potential implication for antifungal prescription recommendations for fungaemia: Data from a nationwide fungaemia surveillance programme. Clin. Microbiol. Infect. 2013, 19, E343–E353. [Google Scholar] [CrossRef]
- Guinea, J.; Zaragoza, Ó.; Escribano, P.; Martín-Mazuelos, E.; Pemán, J.; Sánchez-Reu, S.F.; Cuenca-Estrella, M. CANDIPOP Project, GEIH-GEMICOMED (SEIMC), and REIPI. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob. Agents Chemother. 2014, 58, 1529–1537. [Google Scholar] [CrossRef]
- Ramos, L.S.; Figueiredo-Carvalho, M.H.; Barbedo, L.S.; Ziccardi, M.; Chaves, A.L.; Zancopé-Oliveira, R.M.; Pinto, M.R.; Sgarbi, D.B.; Dornelas-Ribeiro, M.; Branquinha, M.H.; et al. Candida haemulonii complex: Species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J. Antimicrob. Chemother. 2015, 70, 111–115. [Google Scholar] [CrossRef]
- Boatto, H.F.; Cavalcanti, S.D.; Del Negro, G.M.; Girão, M.J.; Francisco, E.C.; Ishida, K.; Gompertz, O.F. Candida duobushaemulonii: An emerging rare pathogenic yeast isolated from recurrent vulvovaginal candidiasis in Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 407–410. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, J.N., Jr.; Assy, J.G.; Levin, A.S.; Del Negro, G.M.; Giudice, M.C.; Tringoni, M.P.; Thomaz, D.Y.; Motta, A.L.; Abdala, E.; Pierroti, L.C.; et al. Candida haemulonii complex species, Brazil, January 2010-March 2015. Emerg. Infect. Dis. 2016, 22, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Frías-De-León, M.G.; Martínez-Herrera, E.; Acosta-Altamirano, G.; Arenas, R.; Rodríguez-Cerdeira, C. Superficial candidosis by Candida duobushaemulonii: An emerging microorganism. Infect. Genet. Evol. 2019, 75, 103960. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2019, e00901. [Google Scholar] [CrossRef]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef]
- Kim, M.N.; Shin, J.H.; Sung, H.; Lee, K.; Kim, E.C.; Ryoo, N.; Lee, J.S.; Jung, S.I.; Park, K.H.; Kee, S.J.; et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: Identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 2009, 48, e57–e61. [Google Scholar] [CrossRef]
- Ruan, S.Y.; Kuo, Y.W.; Huang, C.T.; Hsiue, H.C.; Hsueh, P.R. Infections due to Candida haemulonii: Species identification, antifungal susceptibility and outcomes. Int. J. Antimicrob. Agents 2010, 35, 85–88. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Muro, M.D.; Motta, F.d.A.; Burger, M.; Melo, A.S.; Dalla-Costa, L.M. Echinocandin resistance in two Candida haemulonii isolates from pediatric patients. J. Clin. Microbiol. 2012, 50, 3783–3785. [Google Scholar] [CrossRef]
- Kumar, A.; Prakash, A.; Singh, A.; Kumar, H.; Hagen, F.; Meis, J.F.; Chowdhary, A. Candida haemulonii species complex: An emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg. Microves. Infect. 2012, 5, e49. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xiao, M.; Chen, S.C.; Wang, H.; Cheng, J.W.; Chen, X.X.; Xu, Z.P.; Fan, X.; Kong, F.; Xu, Y.C. Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J. Clin. Microbiol. 2016, 54, 2676–2680. [Google Scholar] [CrossRef] [PubMed]
- Desnos-Ollivier, M.; Ragon, M.; Robert, V.; Raoux, D.; Gantier, J.C.; Dromer, F. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 2008, 46, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Navalkele, B.D.; Revankar, S.; Chandrasekar, P. Candida auris: A worrisome, globally emerging pathogen. Expert Rev. AntiInfect. Ther. 2017, 15, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Sarma, S.; Upadhyay, S. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect. Drug Resist. 2017, 10, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Arias, C.; Mateo, E.; Jurado-Martín, I.; Pena-Fernández, N.; Cantón, E.; Pemán, J.; Quindós, G.; Eraso, E. Utility of two PCR-RFLP-based techniques for identification of Candida parapsilosis complex blood isolates. Mycoses 2020, 63, 461–470. [Google Scholar] [CrossRef]
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef]
- Martínez-Murcia, A.; Navarro, A.; Bru, G.; Chowdhary, A.; Hagen, F.; Meis, J.F. Internal validation of GPS MONODOSE CanAur dtec-qPCR kit following the UNE/EN ISO/IEC 17025:2005 for detection of the emerging yeast Candida auris. Mycoses 2018, 61, 877–884. [Google Scholar] [CrossRef]
- Sexton, D.J.; Kordalewska, M.; Bentz, M.L.; Welsh, R.M.; Perlin, D.S.; Litvintseva, A.P. Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR Green-based quantitative PCR assay. J. Clin. Microbiol. 2018, 56, e01337-18. [Google Scholar] [CrossRef]
- Ahmad, A.; Spencer, J.E.; Lockhart, S.R.; Singleton, S.; Petway, D.J.; Bagarozzi, D.A., Jr.; Herzegh, O.T. A high-throughput and rapid method for accurate identification of emerging multidrug-resistant Candida auris. Mycoses 2019, 62, 513–518. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.C.; Fernández-Pereira, J.; Valentin, E.; Tormo-Mas, M.A.; Eraso, E.; Pemán, J.; de Groot, P.W.J. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 2018, 308, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Fang, W.; Badali, H.; Vaezi, A.; Jiang, W.; Liao, W.; Pan, W.; Hagen, F.; Boekhout, T. Low-cost tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front. Microbiol. 2018, 9, 1119. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Fang, W.; Daneshnia, F.; Al-Hatmi, A.M.; Liao, W.; Pan, W.; Khan, Z.; Ahmad, S.; Rosam, K.; Lackner, M.; et al. Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol. 2019, 14, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Theill, L.; Dudiuk, C.; Morales-Lopez, S.; Berrio, I.; Rodríguez, J.Y.; Marin, A.; Gamarra, S.; Garcia-Effron, G. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev. Iberoam. Micol. 2018, 35, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Gaitán, A.C.; Moret, A.; López Hontangas, J.L.; Molina, J.M.; Aleixandre López, A.I.; Cabezas, A.H.; Mollar Maseres, J.; Arcas, R.C.; Gómez Ruiz, M.D.; Chiveli, M.Á.; et al. Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Rev. Iberoam. Micol. 2017, 34, 23–27. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics PCR—Protocols and Applications—A Laboratory Manual; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Kumar, A.; Sachu, A.; Mohan, K.; Vinod, V.; Dinesh, K.; Karim, S. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal’s medium. Rev. Iberoam. Micol. 2017, 34, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Quindós, G.; Marcos-Arias, C.; San-Millán, R.; Mateo, E.; Eraso, E. The continuous changes in the aetiology and epidemiology of invasive candidiasis: From familiar Candida albicans to multiresistant Candida auris. Int. Microbiol. 2018, 21, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Global Antifungal Surveillance Group. Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2007: A 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef]
- Oberoi, J.K.; Wattal, C.; Goel, N.; Raveendran, R.; Datta, S.; Prasad, K. Non-albicans Candida species in blood stream infections in a tertiary care hospital at New Delhi, India. Indian J. Med. Res. 2012, 136, 997–1003. [Google Scholar]
- Gargeya, I.B.; Pruitt, W.R.; Meyer, S.A.; Ahearn, D.G. Candida haemulonii from clinical specimens in the USA. J. Med. Vet. Mycol. 1991, 29, 335–338. [Google Scholar] [CrossRef]
- Gade, L.; Muñoz, J.F.; Sheth, M.; Wagner, D.; Berkow, E.L.; Forsberg, K.; Jackson, B.R.; Ramos-Castro, R.; Escandón, P.; Dolande, M.; et al. Understanding the emergence of multidrug-resistant Candida: Using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front. Genet. 2020, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Oliveira, S.S.C.; Magalhães, L.B.; Andrade Neto, V.V.; Torres-Santos, E.C.; Carvalho, M.D.C.; Pereira, M.D.; Branquinha, M.H.; Santos, A.L.S. Unmasking the Amphotericin B resistance mechanisms in Candida haemulonii species complex. ACS Infect. Dis. 2020, 6, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).