Hmg1 Gene Mutation Prevalence in Triazole-Resistant Aspergillus fumigatus Clinical Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Aspergillus fumigatus Clinical Isolates and Triazole-Resistance Determination
2.2. hmg1 Gene Sequencing
2.3. Genotyping
2.4. Antifungal Exposure and Clinical Data Management
3. Results
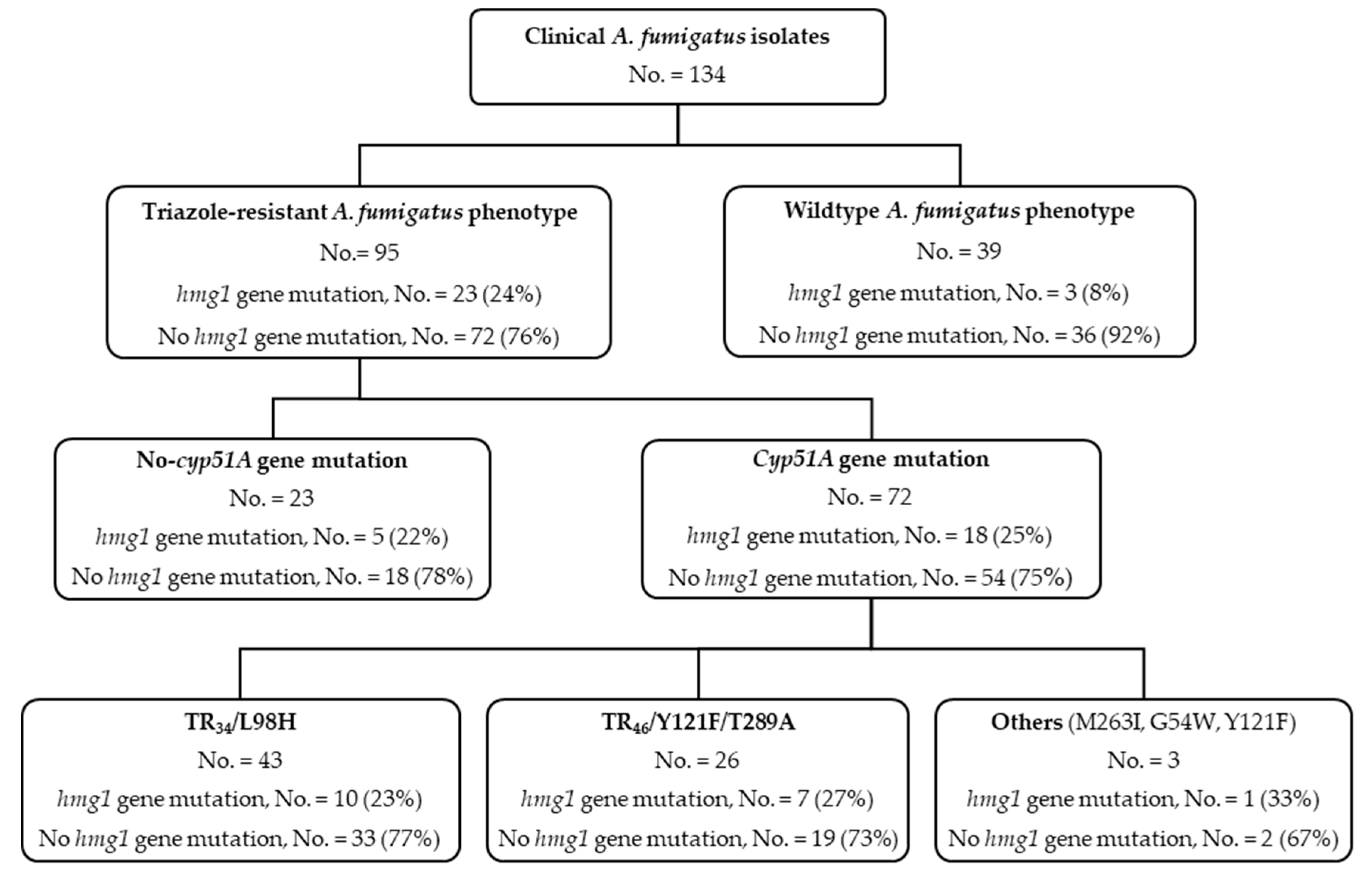
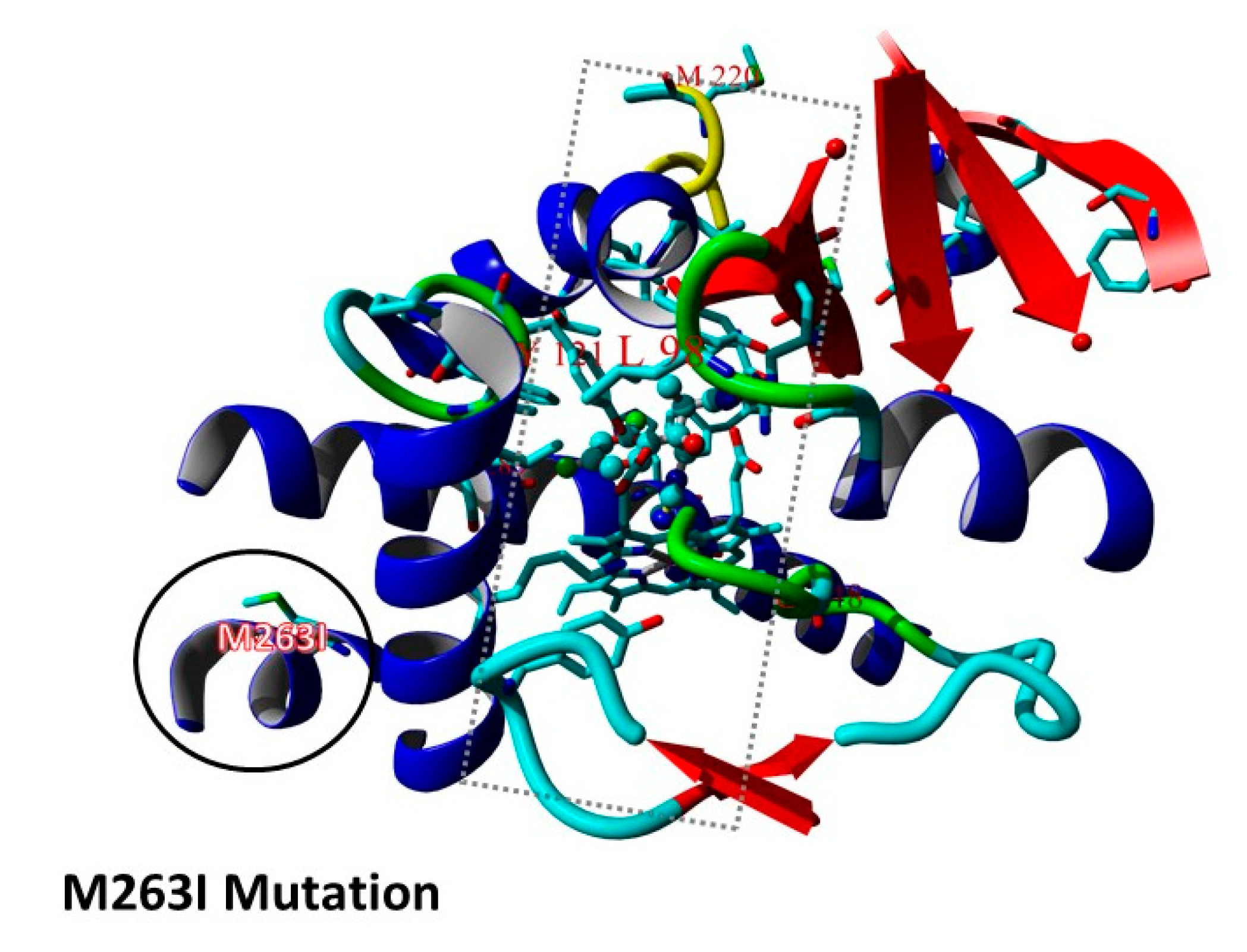
3.1. hmg1 Gene Mutations Are Prevalent among Triazole-Resistant Aspergillus fumigatus Isolates
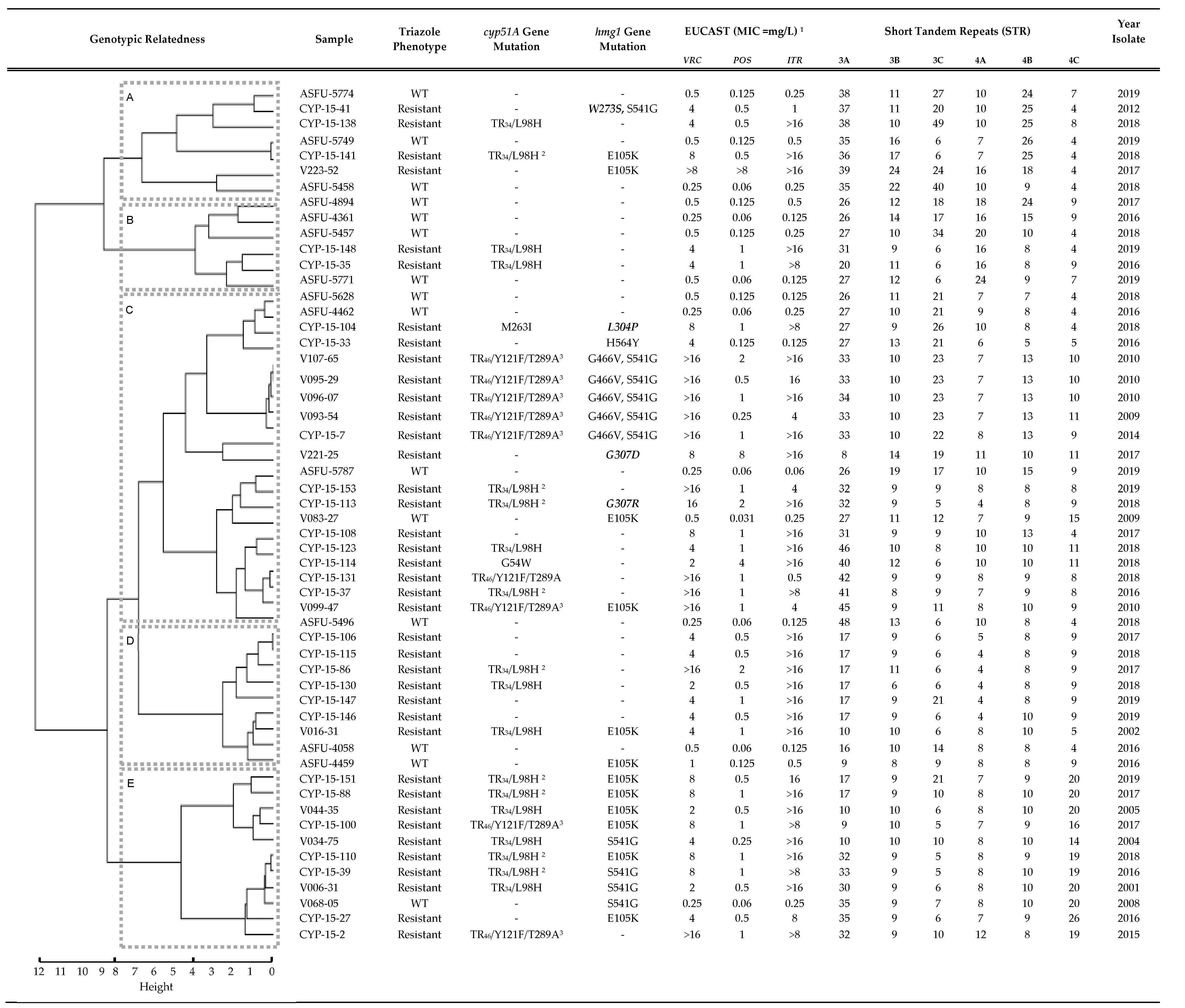
3.2. Triazole Phenotypes in Isolates with Combined cyp51A Gene and hmg1 Gene Mutations
3.3. Previous Exposure to Triazole Antifungals Is Uncommon among Patients Harboring Isolates with hmg1 Gene Mutations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tissot, F.; Agrawal, S.; Pagano, L.; Petrikkos, G.; Groll, A.H.; Skiada, A.; Lass-Flörl, C.; Calandra, T.; Viscoli, C.; Herbrecht, R. ECIL-6 guidelines for the treatment of invasive candidiasis, aspergillosis and mucormycosis in leukemia and hematopoietic stem cell transplant patients. Haematologica 2017, 102, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Patterson, T.F.; Thompson, G.R., III; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, 1–60. [Google Scholar] [CrossRef] [PubMed]
- Resendiz Sharpe, A.; Lagrou, K.; Meis, J.F.; Chowdhary, A.; Lockhart, S.R.; Verweij, P.E. Triazole resistance surveillance in Aspergillus fumigatus. Med. Mycol. 2018, 56, S83–S92. [Google Scholar] [CrossRef] [PubMed]
- Snelders, E.; Van Der Lee, H.A.L.; Kuijpers, J.; Rijs, A.J.M.M.; Varga, J.; Samson, R.A.; Mellado, E.; Donders, A.R.T.; Melchers, W.J.G.; Verweij, P.E. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008, 5, 1629–1637. [Google Scholar] [CrossRef]
- Snelders, E.; Karawajczyk, A.; Schaftenaar, G.; Verweij, P.E.; Melchers, W.J.G. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob. Agents Chemother. 2010, 54, 2425–2430. [Google Scholar] [CrossRef]
- Fraczek, M.G.; Bromley, M.; Buied, A.; Moore, C.B.; Rajendran, R.; Rautemaa, R.; Ramage, G.; Denning, D.W.; Bowyer, P. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013, 68, 1486–1496. [Google Scholar] [CrossRef]
- Pérez-Cantero, A.; López-Fernández, L.; Guarro, J.; Capilla, J. Azole resistance mechanisms in Aspergillus: Update and recent advances. Int. J. Antimicrob. Agents 2020, 55, 105807. [Google Scholar] [CrossRef]
- Hagiwara, D.; Arai, T.; Takahashi, H.; Kusuya, Y.; Watanabe, A.; Kamei, K. Non-cyp51A azole-resistant aspergillus fumigatus isolates with mutation in HMG-CoA reductase. Emerg. Infect. Dis. 2018, 24, 1889–1897. [Google Scholar] [CrossRef]
- Rybak, J.M.; Ge, W.; Wiederhold, N.P.; Parker, J.E.; Kelly, S.L.; Rogers, P.D.; Fortwendel, J.R. Mutations in hmg1, Challenging the Paradigm of Clinical Triazole Resistance in Aspergillus fumigatus. MBio 2019, 10, e00437-19. [Google Scholar] [CrossRef]
- Sharma, C.; Nelson-Sathi, S.; Singh, A.; Pillai, M.R.; Chowdhary, A. Genomic perspective of triazole resistance in clinical and environmental Aspergillus fumigatus isolates without cyp51A mutations. Fungal Genet. Biol. 2019, 132, 103265. [Google Scholar] [CrossRef]
- Wu, C.-J.; Liu, W.-L.; Lai, C.-C.; Chao, C.-M.; Ko, W.-C.; Wang, H.-C.; Dai, C.-T.; Hsieh, M.-I.; Choi, P.-C.; Yang, J.-L.; et al. Multicenter Study of Azole-Resistant Aspergillus fumigatus Clinical Isolates, Taiwan. Emerg. Infect. Dis. 2020, 26, 804–806. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Suzuki, J.; Watanabe, A.; Arai, T.; Koiwa, T.; Shinfuku, K.; Narumoto, O.; Kawashima, M.; Fukami, T.; Tamura, A.; et al. OUP accepted manuscript. Med. Mycol. 2020. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 2008, 73, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Theesfeld, C.L.; Pourmand, D.; Davis, T.; Garza, R.M.; Hampton, R.Y. The Sterol-sensing Domain (SSD) Directly Mediates Signal-regulated Endoplasmic Reticulum-associated Degradation (ERAD) of 3-Hydroxy-3-methylglutaryl (HMG)-CoA Reductase Isozyme Hmg2. J. Biol. Chem. 2011, 286, 26298–26307. [Google Scholar] [CrossRef] [PubMed]
- Gil, G.; Faust, J.R.; Chin, D.J.; Goldstein, J.L.; Brown, M.S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell 1985, 41, 249–258. [Google Scholar] [CrossRef]
- Resendiz-Sharpe, A.; Mercier, T.; Lestrade, P.P.A.; Van Der Beek, M.T.; Von Dem Borne, P.A.; Cornelissen, J.J.; De Kort, E.; Rijnders, B.J.A.; Schauwvlieghe, A.F.A.D.; Verweij, P.E.; et al. Prevalence of voriconazole-resistant invasive aspergillosis and its impact on mortality in haematology patients. J. Antimicrob. Chemother. 2019, 74, 2759–2766. [Google Scholar] [CrossRef] [PubMed]
- Agents, A. European Committee on Antimicrobial Susceptibility Testing Antifungal Agents Breakpoint Tables for Interpretation of MICs. 2017, pp. 1–5. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/Antifungal_breakpoints_v_8.1_March_2017.pdf (accessed on 1 December 2019).
- Vermeulen, E.; Maertens, J.; De Bel, A.; Nulens, E.; Boelens, J.; Surmont, I.; Mertens, A.; Boel, A.; Lagrou, K. Nationwide surveillance of azole resistance in Aspergillus diseases. Antimicrob. Agents Chemother. 2015, 59, 4569–4576. [Google Scholar] [CrossRef]
- De Valk, H.; Meis, J.F.G.M.; Curfs, I.M.; Muehlethaler, K.; Mouton, J.W.; Corné, H.W. Use of a Novel Panel of Nine Short Tandem Repeats for Exact and High-Resolution Fingerprinting of Aspergillus fumigatus Isolates Use of a Novel Panel of Nine Short Tandem Repeats for Exact and High-Resolution Fingerprinting of Aspergillus fumigatus Isolat. J. Clin. Microbiol. 2005, 43, 4112–4120. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing Version 4.0.2; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Snelders, E.; Karawajczyk, A.; Verhoeven, R.J.A.; Venselaar, H.; Schaftenaar, G.; Verweij, P.E.; Melchers, W.J.G. The structure-function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: The mechanism of L98H azole resistance. Fungal Genet. Biol. 2011, 48, 1062–1070. [Google Scholar] [CrossRef]
- Snelders, E.; Camps, S.M.T.; Karawajczyk, A.; Rijs, A.J.M.M.; Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet. Biol. 2015, 82, 129–135. [Google Scholar] [CrossRef]
- Ballard, E.; Weber, J.; Melchers, W.J.G.; Tammireddy, S.; Whitfield, P.D.; Brakhage, A.A.; Brown, A.J.P.; Verweij, P.E.; Warris, A. Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism. Fungal Genet. Biol. 2019, 130, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Sewell, T.R.; Zhu, J.; Rhodes, J.; Hagen, F.; Meis, J.F.; Fisher, M.C.; Jombart, T. Nonrandom distribution of azole resistance across the global population of aspergillus fumigatus. MBio 2019, 10, e00392-19. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Tashiro, M.; Urano, R.; Kikuchi, M.; Ito, N.; Moriya, E.; Shirahige, T.; Mishima, M.; Takazono, T.; Miyazaki, T.; et al. Characteristics of azole-resistant Aspergillus fumigatus attached to agricultural products imported to Japan. J. Infect. Chemother. 2020, 26, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
| Triazole-Resistant A. fumigatus Cases | Triazole Antifungal Exposure 1 | No Triazole Antifungal Exposure 2 | Total Cases |
|---|---|---|---|
| Hmg1 gene mutation, no. (%) | 3 (25) | 9 (75) | 12 (100) |
| No hmg1 gene mutation, no. (%) | 7 (18) | 32 (82) | 39 (100) |
| hmg1 Gene Mutation (Location) | Number of Reported Clinical A. fumigatus Isolates * | ||||||
|---|---|---|---|---|---|---|---|
| Hagiwara et al.* | Rybak et al.* | Sharma et al.* | Chi-Jung et al.* | Takeda et al. * | This study | Total | |
| E105K | 1 | 12 | 13 | ||||
| S269F, F390Y | 1 | 1 | |||||
| S269P, H564Y | 4 2 | 4 | |||||
| Y250H | 1 | 1 | |||||
| F261del | 1 | 1 | |||||
| F262del | 1 | 1 | |||||
| F262del, H564Y | 2 2 | 2 | |||||
| S269F | 18 2 | 18 | |||||
| S269Y | 1 | 1 | |||||
| L273F | 1 | 1 | |||||
| W273S, S541G | 1 | 1 | |||||
| L304P | 1 | 1 | |||||
| S305P | 1 | 1 | 2 | ||||
| S305P, V995I | 1 | 1 | |||||
| G307D | 1 | 1 | 2 | ||||
| G307R | 1 | 1 | |||||
| IP309L | 1 | 1 | |||||
| F390Y | 1 | 1 | |||||
| I412S | 2 | 2 | |||||
| I412T | 1 | 1 | |||||
| L413P | 1 | 1 | |||||
| G466V, S541G | 1 | 5 | 6 | ||||
| S541G 1 | 4 | 4 | |||||
| H564Y 1 | 1 | 3 | 1 | 5 | |||
| V995I | 1 | 1 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Resendiz-Sharpe, A.; Hokken, M.W.J.; Mercier, T.; Merckx, R.; Verhagen, K.; Dewitte, L.; Melchers, W.J.G.; Verweij, P.E.; Maertens, J.; Lagrou, K. Hmg1 Gene Mutation Prevalence in Triazole-Resistant Aspergillus fumigatus Clinical Isolates. J. Fungi 2020, 6, 227. https://doi.org/10.3390/jof6040227
Resendiz-Sharpe A, Hokken MWJ, Mercier T, Merckx R, Verhagen K, Dewitte L, Melchers WJG, Verweij PE, Maertens J, Lagrou K. Hmg1 Gene Mutation Prevalence in Triazole-Resistant Aspergillus fumigatus Clinical Isolates. Journal of Fungi. 2020; 6(4):227. https://doi.org/10.3390/jof6040227
Chicago/Turabian StyleResendiz-Sharpe, Agustin, Margriet W.J. Hokken, Toine Mercier, Rita Merckx, Kamiel Verhagen, Lisa Dewitte, Willem J.G. Melchers, Paul E. Verweij, Johan Maertens, and Katrien Lagrou. 2020. "Hmg1 Gene Mutation Prevalence in Triazole-Resistant Aspergillus fumigatus Clinical Isolates" Journal of Fungi 6, no. 4: 227. https://doi.org/10.3390/jof6040227
APA StyleResendiz-Sharpe, A., Hokken, M. W. J., Mercier, T., Merckx, R., Verhagen, K., Dewitte, L., Melchers, W. J. G., Verweij, P. E., Maertens, J., & Lagrou, K. (2020). Hmg1 Gene Mutation Prevalence in Triazole-Resistant Aspergillus fumigatus Clinical Isolates. Journal of Fungi, 6(4), 227. https://doi.org/10.3390/jof6040227








