Genetic Regulators and Physiological Significance of Glycogen Storage in Candida albicans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Assessing Survival
2.3. Strain Construction
2.4. Determination of Glycogen Content―Iodine Method
2.5. Determination of Glycogen Content―Enzymatic Method
2.6. Determination of Lipid Content―Nile Red Stain
3. Results
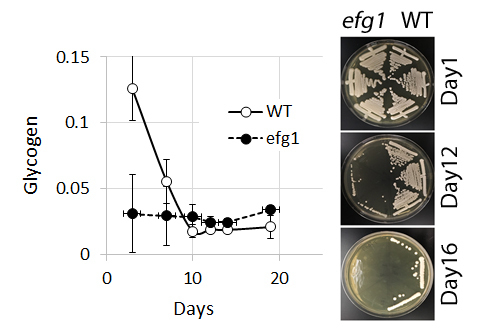
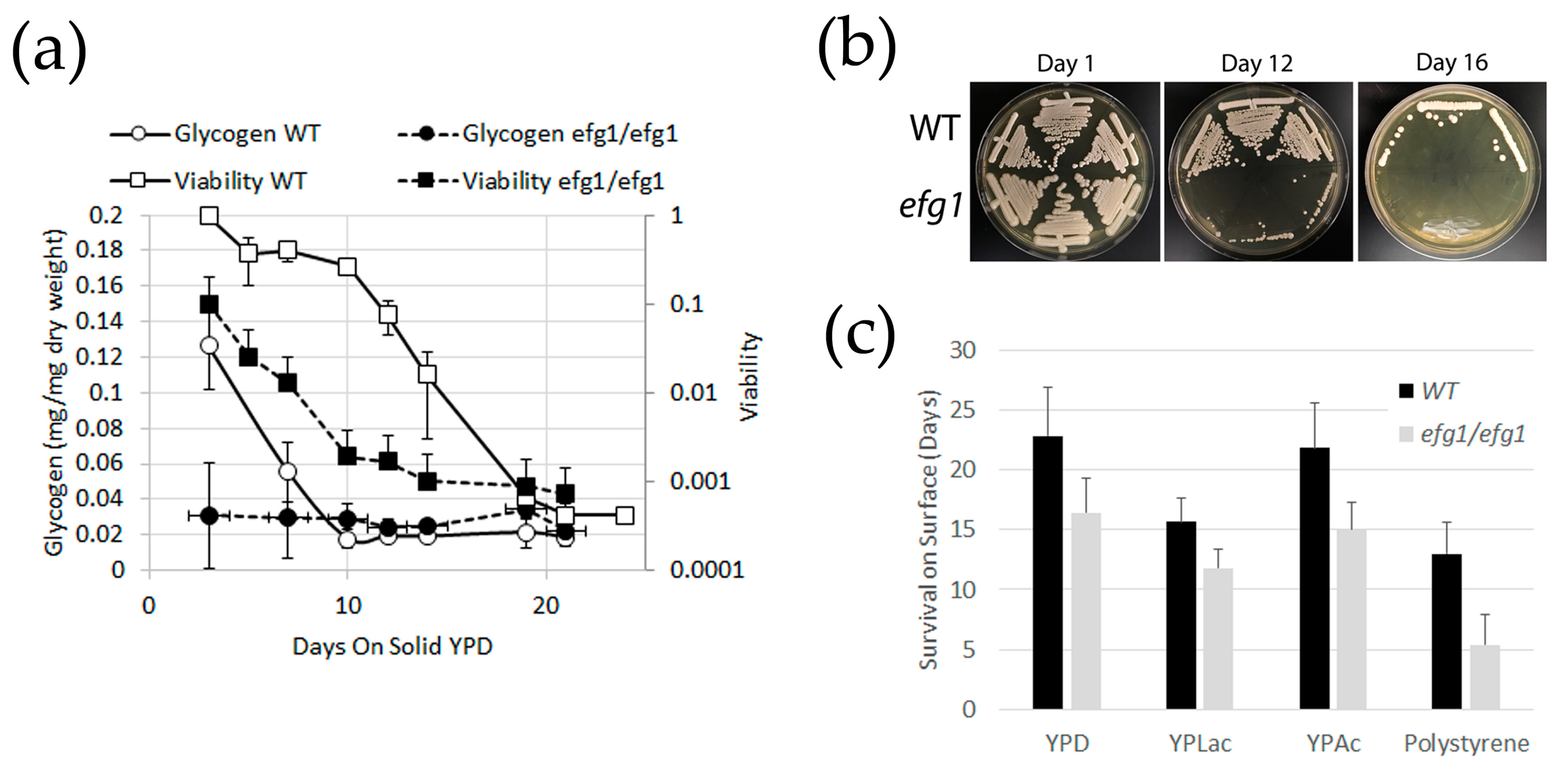
3.1. Glycogen Deposits Decay More Rapidly Than Cell Viability During Long-Term Culture
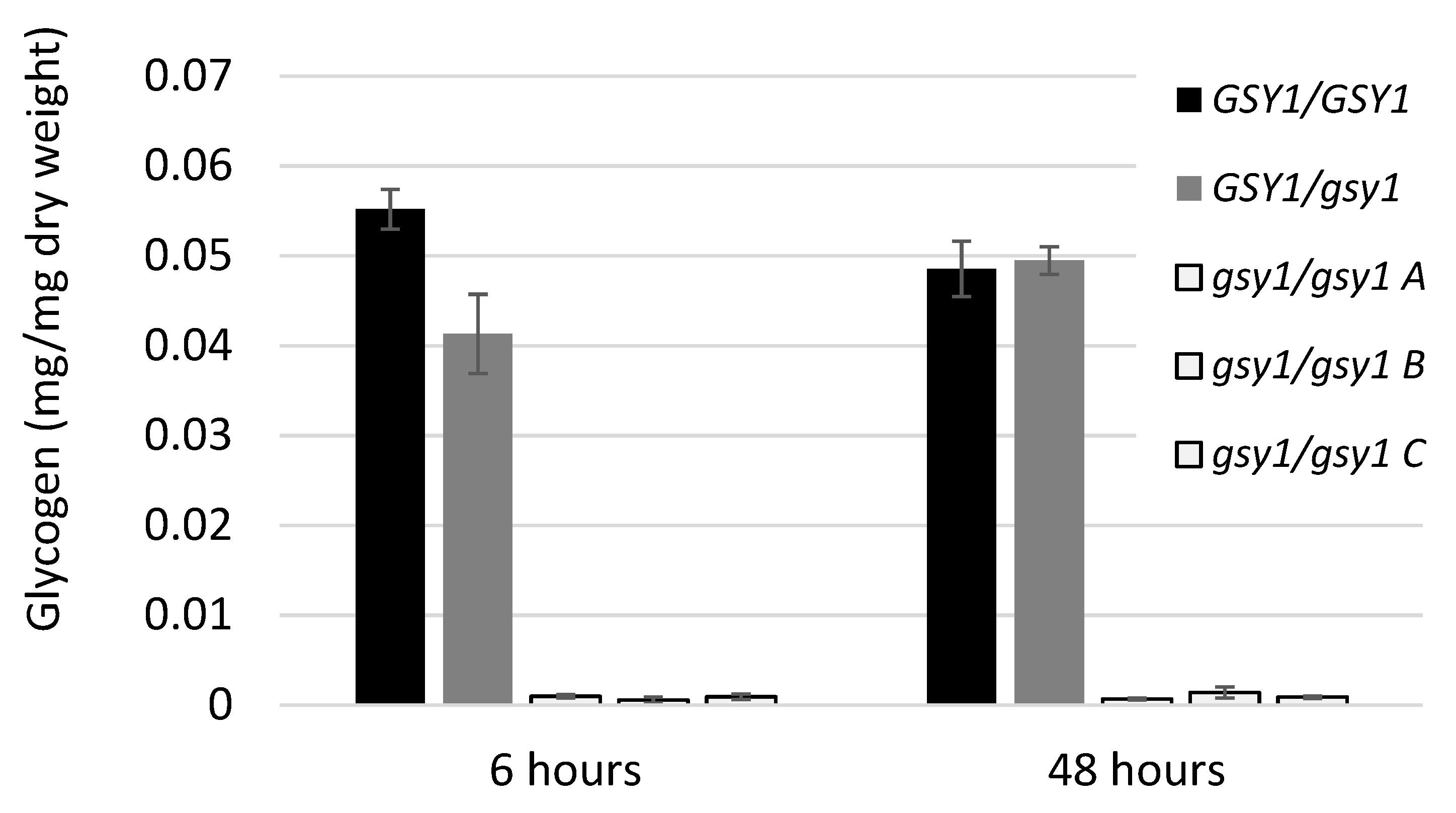
3.2. Construction and Characterization of a Glycogen Synthase 1 Mutant
3.3. Identification of Genetic Factors for Glycogen Synthesis through Mutant Screening
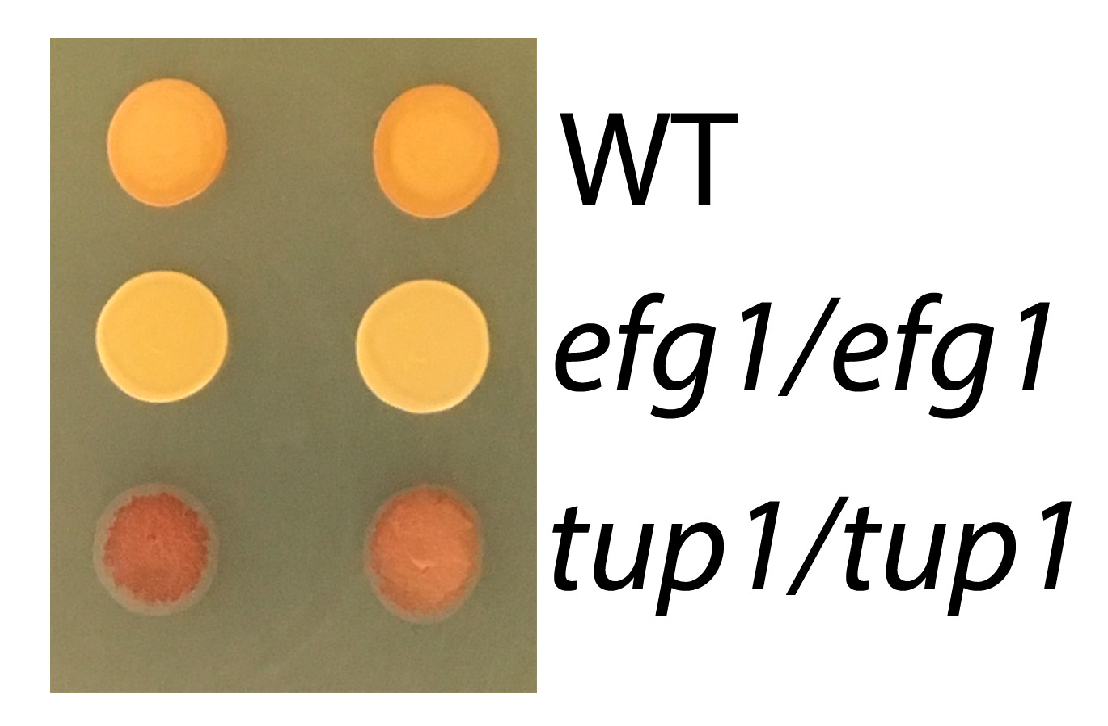
3.4. Role of Efg1 Antagonists Tup1/Nrg1 in the Control of Glycogen Content
3.5. Efg1 Target Genes And Low-Glycogen Strains Have No Long-Term Survival Deficit
4. Discussion
4.1. Glycogen in Yeast
4.2. Glycogen and Viability of C. albicans efg1 Mutants in Long-term Culture
4.3. Glycogen is Consumed During Starvation, But is not Essential for Survival
4.4. Genetic Factors Supporting Glycogen Synthesis in C. albicans
4.5. The Role of Efg1 in Glycogen Storage and Long-Term Survival
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kollar, R.; Reinhold, B.B.; Petrakova, E.; Yeh, H.J.; Ashwell, G.; Drgonova, J.; Kapteyn, J.C.; Klis, F.M.; Cabib, E. Architecture of the yeast cell wall. Beta(1 → 6)-glucan interconnects mannoprotein, beta(1→3)-glucan, and chitin. J. Biol. Chem. 1997, 272, 17762–17775. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.G. Cell envelope of Candida albicans. Crit. Rev. Microbiol. 1987, 15, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.A.; Roach, P.J.; Montero, M.; Baroja-Fernandez, E.; Munoz, F.J.; Eydallin, G.; Viale, A.M.; Pozueta-Romero, J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010, 34, 952–985. [Google Scholar] [CrossRef] [PubMed]
- Lillie, S.H.; Pringle, J.R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: Responses to nutrient limitation. J. Bacteriol. 1980, 143, 1384–1394. [Google Scholar]
- Mundkur, B. Electron microscopical studies of frozen-dried yeast. I. Localization of polysaccharides. Exp. Cell Res. 1960, 20, 28–42. [Google Scholar] [CrossRef]
- Northcote, D.H. The molecular structure and shape of yeast glycogen. Biochem. J. 1953, 53, 348–352. [Google Scholar] [CrossRef]
- Sillje, H.H.; Paalman, J.W.; ter Schure, E.G.; Olsthoorn, S.Q.; Verkleij, A.J.; Boonstra, J.; Verrips, C.T. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J. Bacteriol. 1999, 181, 396–400. [Google Scholar]
- Anderson, C.; Tatchell, K. Hyperactive glycogen synthase mutants of Saccharomyces cerevisiae suppress the glc7-1 protein phosphatase mutant. J. Bacteriol. 2001, 183, 821–829. [Google Scholar] [CrossRef]
- Farkas, I.; Hardy, T.A.; Goebl, M.G.; Roach, P.J. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J. Biol. Chem. 1991, 266, 15602–15607. [Google Scholar]
- Francois, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 125–145. [Google Scholar] [CrossRef]
- Estruch, F.; Carlson, M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993, 13, 3872–3881. [Google Scholar] [CrossRef] [PubMed]
- Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 2000, 24, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pastor, M.T.; Marchler, G.; Schuller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Doedt, T.; Krishnamurthy, S.; Bockmuhl, D.P.; Tebarth, B.; Stempel, C.; Russell, C.L.; Brown, A.J.; Ernst, J.F. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 2004, 15, 3167–3180. [Google Scholar] [CrossRef] [PubMed]
- Bockmuhl, D.P.; Ernst, J.F. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 2001, 157, 1523–1530. [Google Scholar]
- Stoldt, V.R.; Sonneborn, A.; Leuker, C.E.; Ernst, J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997, 16, 1982–1991. [Google Scholar] [CrossRef]
- Lo, H.J.; Kohler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Tandia, F.; Linteau, A.; Sillaots, S.; Marta, C.; et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef]
- Homann, O.R.; Dea, J.; Noble, S.M.; Johnson, A.D. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009, 5, e1000783. [Google Scholar] [CrossRef]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Noble, S.M.; Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 2005, 4, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A.; Irwin, M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993, 134, 717–728. [Google Scholar] [PubMed]
- Wilson, R.B.; Davis, D.; Mitchell, A.P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999, 181, 1868–1874. [Google Scholar] [PubMed]
- Wang, Z.; Wilson, W.A.; Fujino, M.A.; Roach, P.J. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 2001, 21, 5742–5752. [Google Scholar] [CrossRef]
- Rostron, K.A.; Rolph, C.E.; Lawrence, C.L. Nile red fluorescence screening facilitating neutral lipid phenotype determination in budding yeast, Saccharomyces cerevisiae, and the fission yeast Schizosaccharomyces pombe. Antonie van Leeuwenhoek 2015, 108, 97–106. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef]
- Ansari, M.A.; Fatima, Z.; Ahmad, K.; Hameed, S. Monoterpenoid perillyl alcohol impairs metabolic flexibility of Candida albicans by inhibiting glyoxylate cycle. Biochem. Biophys. Res. Commun. 2018, 495, 560–566. [Google Scholar] [CrossRef]
- Cheah, H.L.; Lim, V.; Sandai, D. Inhibitors of the glyoxylate cycle enzyme ICL1 in Candida albicans for potential use as antifungal agents. PLoS ONE 2014, 9, e95951. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Rothman-Denes, L.B.; Cabib, E. Two forms of yeast glycogen synthetase and their role in glycogen accumulation. Proc. Natl. Acad. Sci. USA 1970, 66, 967–974. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.V.; Dignard, D.; Whiteway, M.; Kumamoto, C.A. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot. Cell 2013, 12, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Eschrich, D.; Kotter, P.; Entian, K.D. Gluconeogenesis in Candida albicans. FEMS Yeast Res. 2002, 2, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Adya, A.K.; Wehmeier, S.; Brand, A.C.; MacCallum, D.M.; Gow, N.A.; Brown, A.J. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell. Microbiol. 2012, 14, 1319–1335. [Google Scholar] [CrossRef]
- Askew, C.; Sellam, A.; Epp, E.; Hogues, H.; Mullick, A.; Nantel, A.; Whiteway, M. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009, 5, e1000612. [Google Scholar] [CrossRef]
- Sullivan, P.A.; Yin, C.Y.; Molloy, C.; Templeton, M.D.; Shepherd, M.G. An analysis of the metabolism and cell wall composition of Candida albicans during germ-tube formation. Can. J. Microbiol. 1983, 29, 1514–1525. [Google Scholar] [CrossRef]
- Brown, A.J.; Brown, G.D.; Netea, M.G.; Gow, N.A. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014, 22, 614–622. [Google Scholar] [CrossRef]
- Rocha, C.R.; Schroppel, K.; Harcus, D.; Marcil, A.; Dignard, D.; Taylor, B.N.; Thomas, D.Y.; Whiteway, M.; Leberer, E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 2001, 12, 3631–3643. [Google Scholar] [CrossRef]
- Setiadi, E.R.; Doedt, T.; Cottier, F.; Noffz, C.; Ernst, J.F. Transcriptional response of Candida albicans to hypoxia: Linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 2006, 361, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.E.; Kelly, M.N.; Sturtevant, J.E. Autophagy in the pathogen Candida albicans. Microbiology 2007, 153, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhao, H.; Yin, Y.; Liang, C.; Mao, X.; Liu, Y.; Yu, Q.; Li, M. Function of Atg11 in non-selective autophagy and selective autophagy of Candida albicans. Biochem. Biophys. Res. Commun. 2019, 516, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
| Strain/Collection | Genotype | Author |
|---|---|---|
| Gene Replacement and Conditional Expression (GRACE) CaSS1-based (2356 conditional mutants); Enriched for putatively essential genes | One allele deleted with HIS3, second allele under control of tetracycline-repressible promoter | [18] |
| Homan et al. Transcriptional Regulator Knockout collection (165 knockout mutants) | LEU2 and HIS1 deletion of target genes in SN152 background arg4∆/arg4∆ leu2∆/leu2∆ his1∆/his1∆ URA3/ura3∆::imm434 IRO1/iro1∆::imm434 Collection includes wildtype control with LEU2 and HIS1 integration | [19] |
| Noble et al. Morphogenetic switching/pathogenicity collection (674 knockout mutants) | LEU2 and HIS1 deletion of target genes in SN152 background arg4∆/arg4∆ leu2∆/leu2∆ his1∆/his1∆ URA3/ura3∆::imm434 IRO1/iro1∆::imm434 Collection includes wildtype control with LEU2 and HIS1 integration | [20] |
| SN152 | arg4∆/arg4∆ leu2∆/leu2∆ his1∆/his1∆ URA3/ura3∆::imm434 IRO1/iro1∆::imm434 | [21] |
| CaMS626 | SN152 with gsy1::ARG4 | This study |
| CaMS627 | SN152 with gsy1::ARG4/gsy1::HIS1 | This study |
| CAI4 | ura3::imm434/ura3::imm434 | [22] |
| HLC52 (efg1) | ura3::imm434/ura3::imm434 efg1::hisG/efg1::hisG::URA3::hisG | [17] |
| NAME/SYST ORF | Collection | Glycogen mg/mg (% of WT) |
|---|---|---|
| CaSS1/WT | GRACE | 0.046 +/− 0.007 |
| SN250/WT | H, N | 0.048 +/− 0.01 |
| PGK1/C6_00750C_A | GRACE | 0.019 +/− 0.001 (42%) |
| C3_06700C_A | GRACE | 0.016 +/− 0.0001 (35%) |
| COQ1/CR_00570W_A | GRACE | 0.018 +/− 0.004 (39%) |
| C4_03410W_A | GRACE | 0.022 +/− 0.009 (48%) |
| C4_00660W_A | GRACE | 0.019 +/− 0 (41%) |
| MSU1/C2_08550C_A | GRACE | 0.019 +/− 0.004 (41%) |
| C1_13150W_A | GRACE | 0.02 +/− 0.002 (43%) |
| C6_02150C_A | GRACE | 0.017 +/− 0.002 (37%) |
| MSW1/C5_02770W_A | GRACE | 0.016 +/− 0.004 (35%) |
| TPS2/C1_03380W_A | GRACE | 0.025 +/− 0.01 (54%) |
| GPM1/C2_03270W_A | GRACE | 0.02 +/− 0.006 (44%) |
| C3_03100C_A | GRACE | 0.016 +/− 0.0005 (35%) |
| WAL1/CR_09650W_A | GRACE | 0.015 +/− 0.001 (33%) |
| C1_01140C_A | GRACE | 0.006 +/− 0.002 (12%) |
| COQ5/C2_05470W_A | GRACE | 0.023 +/− 0.003 (49%) |
| CR_01120C_A | GRACE | 0.016 +/− 0.002 (35%) |
| C5_05230C_A | GRACE | 0.011 +/− 0.001 (25%) |
| C4_06000W_A | GRACE | 0.02 +/− 0.003 (43%) |
| C1_01010W_A | GRACE | 0.023 +/− 0.005 (49%) |
| TSC11/CR_07580C_A | GRACE | 0.033 +/− 0.001 (71%) |
| SSK1/C1_13930W_A | GRACE | 0.02 +/− 0.016 (44%) |
| EFG1/CR_07890W_A | H | 0.005 +/− 0.001 (11%) |
| COX4/C2_01620W_A | N | 0.024 +/− 0.0004 (49%) |
| SNF4/C6_03920W_A | N | 0.009 +/− 0.0025 (18%) |
| PSR1/C3_00570C_A | N | 0.018 +/− 0.007 (36%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeitz, M.A.; Tanveer, Z.; Openshaw, A.T.; Schmidt, M. Genetic Regulators and Physiological Significance of Glycogen Storage in Candida albicans. J. Fungi 2019, 5, 102. https://doi.org/10.3390/jof5040102
Zeitz MA, Tanveer Z, Openshaw AT, Schmidt M. Genetic Regulators and Physiological Significance of Glycogen Storage in Candida albicans. Journal of Fungi. 2019; 5(4):102. https://doi.org/10.3390/jof5040102
Chicago/Turabian StyleZeitz, Marcus A., Zainab Tanveer, Anatole T. Openshaw, and Martin Schmidt. 2019. "Genetic Regulators and Physiological Significance of Glycogen Storage in Candida albicans" Journal of Fungi 5, no. 4: 102. https://doi.org/10.3390/jof5040102
APA StyleZeitz, M. A., Tanveer, Z., Openshaw, A. T., & Schmidt, M. (2019). Genetic Regulators and Physiological Significance of Glycogen Storage in Candida albicans. Journal of Fungi, 5(4), 102. https://doi.org/10.3390/jof5040102