Development of a Field-Deployable Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Erysiphe corylacearum in Hazelnut
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Primer Design for LAMP Assay
2.3. Optimization of the LAMP Assay
2.4. Sensitivity and Specificity Evaluation
2.5. Comparison of LAMP with PCR Specific Assay
2.6. Development of a Rapid Crude Extract Method for LAMP
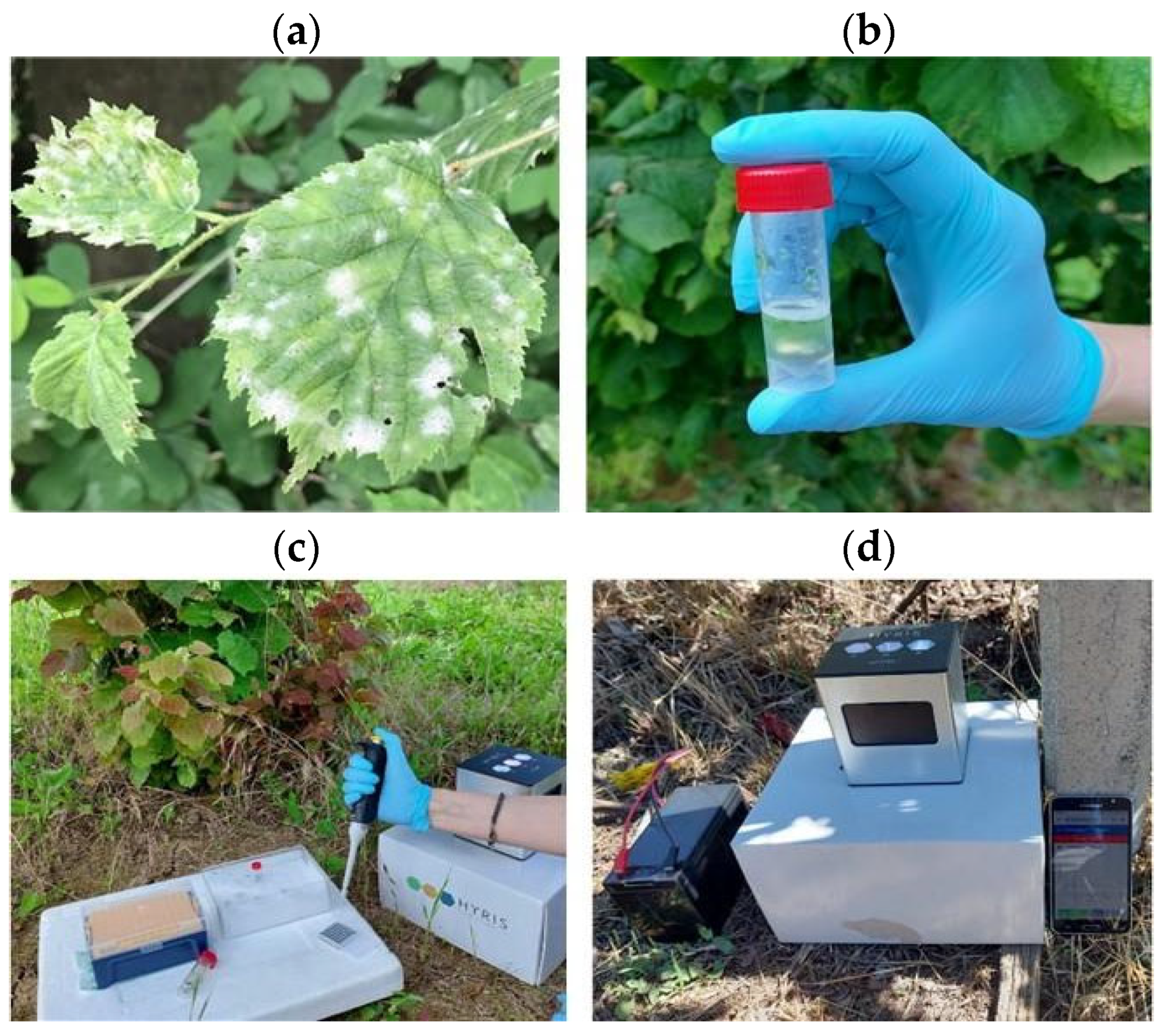
2.7. On-Site LAMP Assay
3. Results
3.1. Design of LAMP Primers
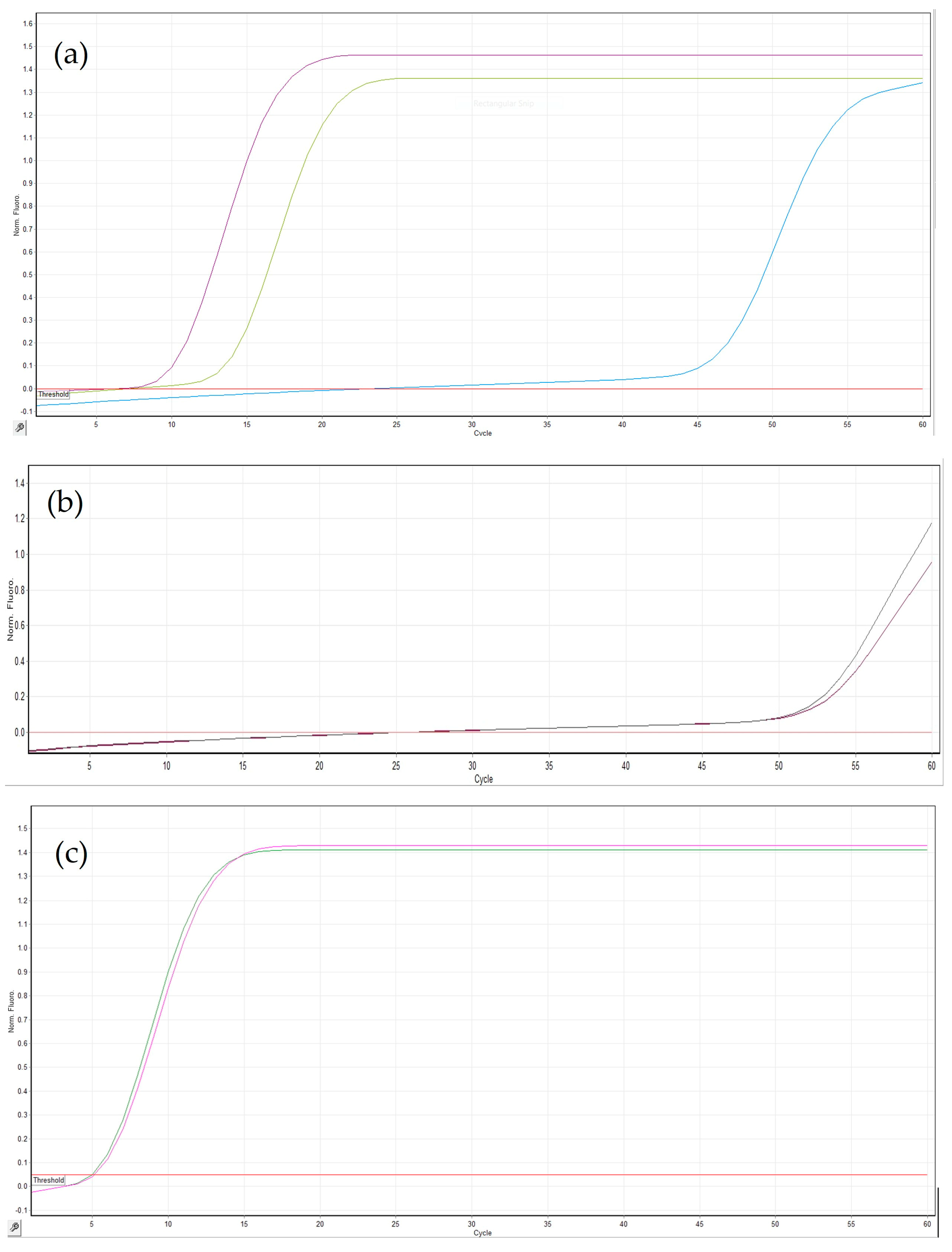
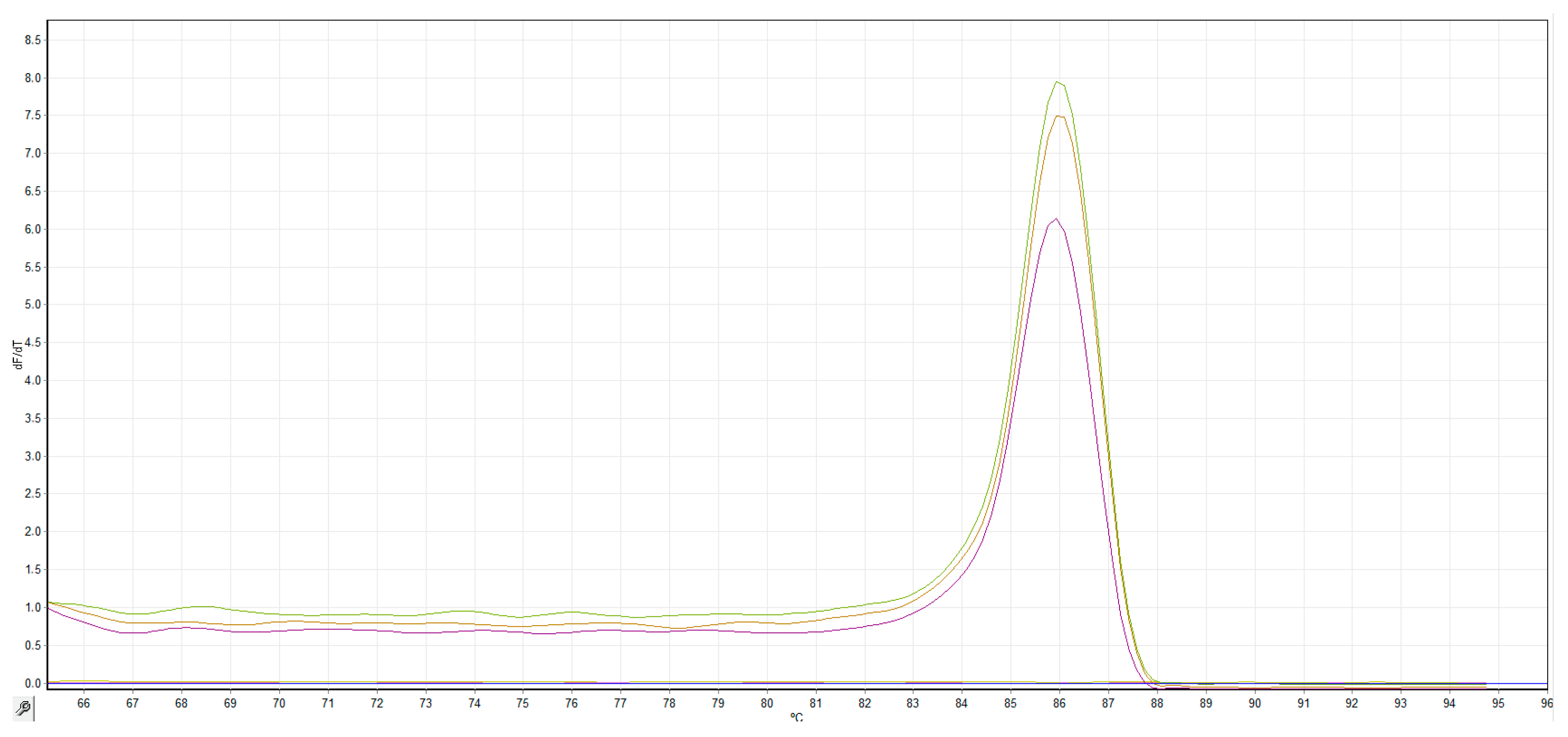
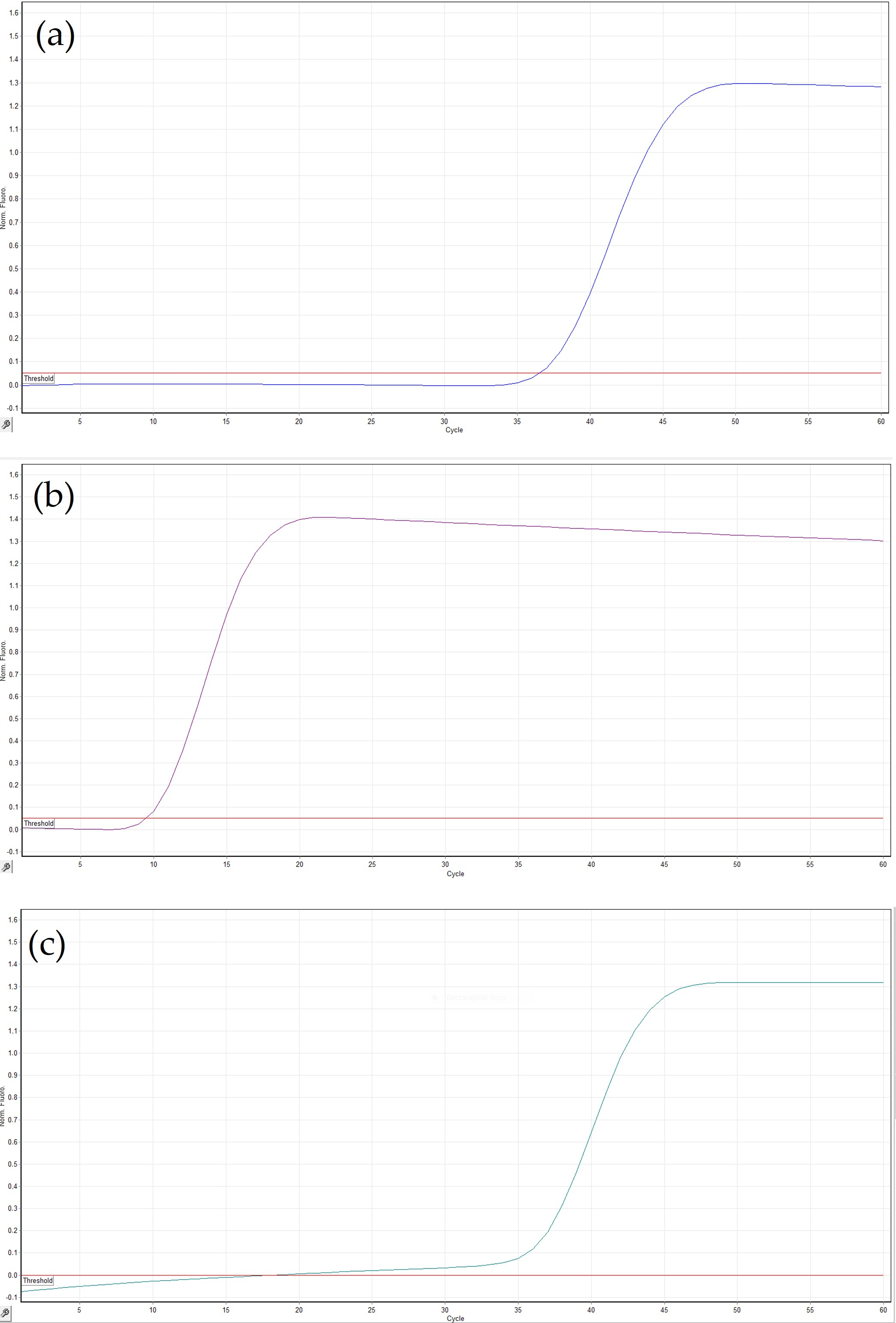
3.2. Optimization of the LAMP Assay
3.3. Sensitivity, Specificity and Repeatability of the LAMP Assay
3.4. Comparison of LAMP with a Specific PCR Assay
3.5. Comparison of Different Crude Extract Methods
3.6. LAMP Assay with DNA Extracted from Field Samples
3.7. On-Site LAMP Assay with Crude Plant Extracts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBIF Secretariat. GBIF Backbone Taxonomy. Checklist Dataset. 2023. Available online: https://doi.org/10.15468/39omei (accessed on 28 November 2025).
- Voglmayr, H.; Zankl, T.; Krisai-Greilhuber, I.; Kirisits, T. First report of Erysiphe corylacearum on Corylus avellana and C. colurna in Austria. New Dis. Rep. 2020, 42, 14. [Google Scholar] [CrossRef]
- Beenken, L.; Kruse, J.; Schmidt, A.; Braun, U. Epidemic spread of Erysiphe corylacearum in Europe—First records from Germany. Schlechtendalia 2022, 39, 112–118. [Google Scholar]
- Sezer, A.; Dolar, F.S.; Lucas, S.J.; Köse, Ç.; Gümüş, E. First report of Erysiphe corylacearum on hazelnut in Turkey. Phytoparasitica 2017, 45, 577–581. [Google Scholar] [CrossRef]
- Arzanlou, M.; Torbati, M.; Golmohammadi, H. Powdery mildew on hazelnut (Corylus avellana) caused by Erysiphe corylacearum in Iran. For. Pathol. 2018, 48, e12450. [Google Scholar] [CrossRef]
- Gur, L. Occurrence of Powdery Mildew Caused by Erysiphe corylacearum on Hazelnuts in Israel. Plant Dis. 2023, 108, 1096. [Google Scholar] [CrossRef]
- Mezzalama, M.; Guarnaccia, V.; Martano, G.; Spadaro, D. Presence of Powdery Mildew Caused by Erysiphe corylacearum on Hazelnut (Corylus avellana) in Italy. Plant Dis. 2020, 105, 1565. [Google Scholar] [CrossRef]
- Matić, S.; Caruso, A.G.; D’Errico, C.; Botto, C.S.; Noris, E.; Trkulja, V.; Panno, S.; Davino, S.; Moizio, M. Powdery mildew caused by E. corylacearum: An emerging problem in Italy. PLoS ONE 2024, 19, e0301941. [Google Scholar] [CrossRef]
- Cunnington, J.H.; Takamatsu, S.; Lawrie, A.C.; Pascoe, I.G. Molecular identification of anamorphic powdery mildews (Erysiphales). Australas. Plant Pathol. 2003, 32, 421–428. [Google Scholar] [CrossRef]
- Takamatsu, S.; Kano, Y. PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fung. Mycoscience 2001, 42, 135–139. [Google Scholar] [CrossRef]
- Bradshaw, M.; Braun, U.; Meeboon, J.; Tobin, P. Phylogeny and taxonomy of powdery mildew caused by Erysiphe species on Corylus hosts. Mycologia 2021, 113, 459–475. [Google Scholar] [CrossRef]
- Panno, S.; Matić, S.; Tiberini, A.; Caruso, A.G.; Bella, P.; Torta, L.; Stassi, R.; Davino, S. Loop Mediated Isothermal Amplification: Principles and Applications in Plant Virology. Plants 2020, 9, 461. [Google Scholar] [CrossRef]
- Yang, L.; Sun, Y.; Sun, L.; Wang, Z.; Feng, J.; Liang, Y. Application of Loop-Mediated Isothermal Amplification in Plant Pathogen Detection. Phytopathology 2025, 115, 6–13. [Google Scholar] [CrossRef]
- Aglietti, C.; Benigno, A.; Cacciola, S.O.; Moricca, S. LAMP Reaction in Plant Disease Surveillance: Applications, Challenges, and Future Perspectives. Life 2024, 14, 1549. [Google Scholar] [CrossRef]
- Caruso, A.; Ragona, A.; Agrò, G.; Bertacca, S.; Yahyaoui, E.; Galipienso, L.; Rubio, L.; Panno, S.; Davino, S. Rapid detection of tomato spotted wilt virus by real-time RT-LAMP and in-field application. J. Plant Pathol. 2024, 106, 697–712. [Google Scholar] [CrossRef]
- Kalmár, K.; Borostyán, K.; Molnár, O.; Ágoston, J.; Preininger, É.; Németh, M.Z. A Species-Specific PCR Differentiates Two Causal Agents of Hazel Powdery Mildew and Reveals the Occurrence of Erysiphe corylacearum. Horticulturae 2024, 10, 763. [Google Scholar] [CrossRef]
- Matić, S.; Myrta, A. Development of Loop-Mediated Isothermal Amplification (LAMP) Assay for In-Field Detection of American Plum Line Pattern Virus. Viruses 2024, 16, 1572. [Google Scholar] [CrossRef]
- Basha, J.S.; Kamalakannan, A.; Saraswathy, S.; Johnson, I.; Ganapati, P.S.; Swarna Lakshmi, K.R. Rapid detection of airborne inocula of grapevine mildews using PCR and LAMP assay. Int. J. Plant Soil Sci. 2021, 33, 12–21. [Google Scholar] [CrossRef]
- Thiessen, L.D.; Keune, J.A.; Neill, T.M.; Turechek, W.W.; Grove, G.G.; Mahaffee, W.F. Development of a grower-conducted inoculum detection assay for management of grape powdery mildew. Plant Pathol. 2016, 65, 238–249. [Google Scholar] [CrossRef]
- Thiessen, L.D.; Neill, T.M.; Mahaffee, W.F. Development of a quantitative loop-mediated isothermal amplification assay for the field detection of Erysiphe necator. PeerJ 2018, 6, e4639. [Google Scholar] [CrossRef]
- Vettraino, A.M.; Luchi, N.; Rizzo, D.; Pepori, A.L.; Pecori, F.; Santini, A. Rapid diagnostics for Gnomoniopsis smithogilvyi (syn. Gnomoniopsis castaneae) in chestnut nuts: New challenges by using LAMP and real-time PCR methods. AMB Express 2021, 11, 105. [Google Scholar] [CrossRef]
- Németh, M.Z.; Kovács, G.M. A comprehensive guide to loop-mediated isothermal amplification, an emerging diagnostic tool for plant pathogenic fungi. Front. Plant Sci. 2025, 16, 1568657. [Google Scholar] [CrossRef]
- Rizzo, D.; Aglietti, C.; Benigno, A.; Bracalini, M.; Da Lio, D.; Bartolini, L.; Cappellini, G.; Aronadio, A.; Francia, C.; Luchi, N. Loop-mediated isothermal amplification (LAMP) and SYBR Green qPCR for fast and reliable detection of Geosmithia morbida (Kolařík) in infected walnut. Plants 2022, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Candian, V.; D’Errico, C.; Pierro, R.; Panno, S.; Davino, S.; Noris, E.; Tedeschi, R. In-Field LAMP Detection of Flavescence Dorée Phytoplasma in Crude Extracts of the Scaphoideus titanus Vector. Agronomy 2022, 12, 1645. [Google Scholar] [CrossRef]
- Yaseen, T.; Drago, S.; Valentini, F.; Elbeaino, T.; Stampone, G.; Digiaro, M.; D’Onghia, A.M. On-site detection of Xylella fastidiosa in host plants and in “spy insects” using the real-time loop-mediated isothermal amplification method. Phytopathol. Mediterr. 2015, 54, 488–496. [Google Scholar]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Duan, Y.; Ge, C.; Zhang, X.; Wang, J.; Zhou, M. A rapid detection method for the plant pathogen Sclerotinia sclerotiorum based on loop-mediated isothermal amplification (LAMP). Australas. Plant Pathol. 2014, 43, 61–66. [Google Scholar] [CrossRef]
- Marra, M.; D’Errico, C.; Montemurro, C.; Ratti, C.; Baldoni, E.; Matic, S.; Accotto, G.P. Fast and Sensitive Detection of Soil-Borne Cereal Mosaic Virus in Leaf Crude Extract of Durum Wheat. Viruses 2023, 15, 140. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Nagavardhini, A.; Sengupta, A.; Sharma, M. Development of Loop-Mediated Isothermal Amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris-wilt pathogen of chickpea. BMC Res. Notes 2015, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Marra, M.; Mussano, P.; Pinton, E.; Montemurro, C.; Baldoni, E.; Ratti, C.; Matić, S.; D’Errico, C.; Accotto, G.P. Rapid and specific detection of wheat spindle streak mosaic virus using RT-LAMP in durum wheat crude leaf extract. PLoS ONE 2024, 19, e0299078. [Google Scholar] [CrossRef]
- Zhang, J.; Borth, W.; Lin, B.; Melzer, M.; Shen, H.; Pu, X.; Sun, D.; Nelson, S.; Hu, J. Multiplex detection of three banana viruses by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Trop. Plant Pathol. 2018, 43, 543–551. [Google Scholar] [CrossRef]
- Hieno, A.; Li, M.; Otsubo, K.; Suga, H.; Kageyama, K. Multiplex LAMP detection of the genus Phytophthora and four Phytophthora species (P. ramorum, P. lateralis, P. kernoviae, and P. nicotianae), with a plant internal control. Microbes Environ. 2021, 36, ME21019. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gutierrez, S.V.; Goodwin, S.B. Loop-Mediated Isothermal Amplification for Detection of Plant Pathogens in Wheat (Triticum aestivum). Front. Plant Sci. 2022, 13, 857673. [Google Scholar] [CrossRef] [PubMed]
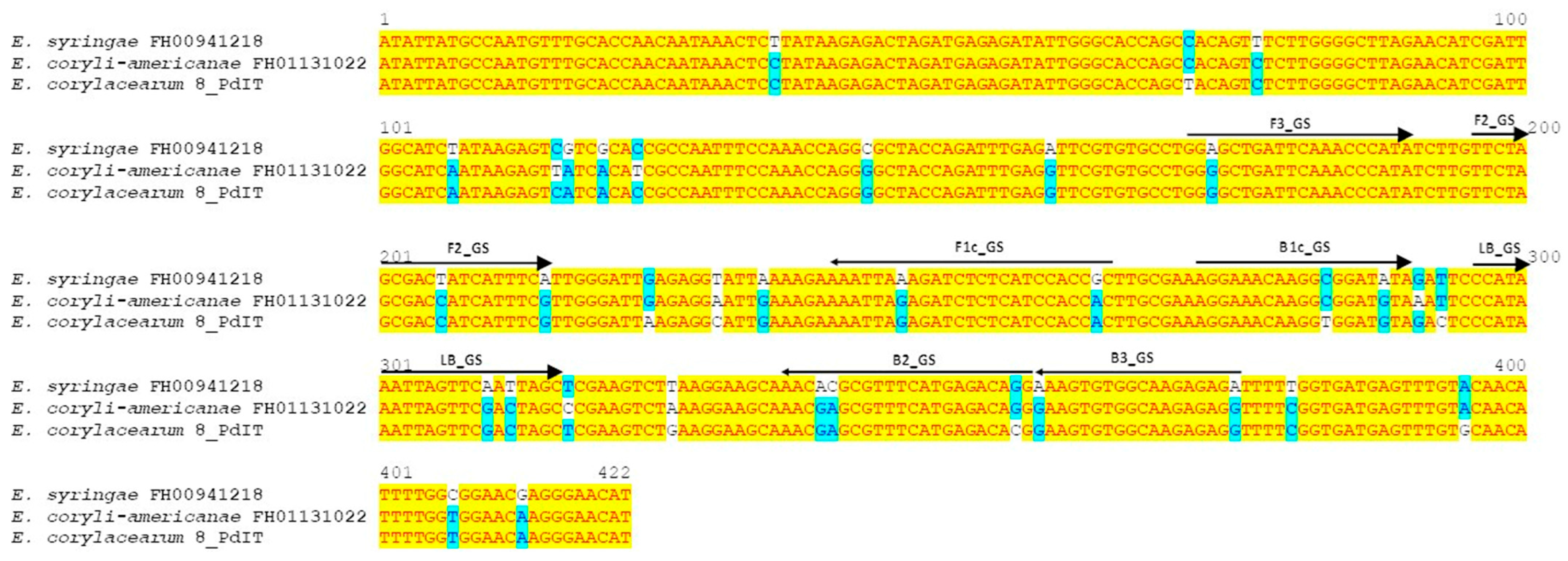
| Primer Sets | Name | Sequence (5′–3′) | Gene | Primer Position | Final Concentration in LAMP Reaction |
|---|---|---|---|---|---|
| 1 | F3_Ecr1 | AGACAGTTCAAAATTTACGCC | rpb2 | 350–370 | 0.2 µM |
| B3_Ecr1 | TGATCGTCCTCAAGCCTA | 537–554 | 0.2 µM | ||
| FIP_Ecr1 | ACACGACCTGCATCTGTAAATATTT-GGTCTCATTTGATCTCTCACG | 46 | 2.0 µM | ||
| BIP_Ecr1 | TGTTATTGATAATGACCCTGAGAGC-CGAATATGCTCTTTATTAAGAACC | 49 | 2.0 µM | ||
| 2 | F3_Ecg | GGGCTGATTCAAACCCATA | GS | 172–190 | 0.2 µM |
| B3_Ecg | CCTCTCTTGCCACACTTC | 358–375 | 0.2 µM | ||
| FIP_Ecg | AGTGGTGGATGAGAGATCTCTAATT-TTCTAGCGACCATCATTTCG | 45 | 2.0 µM | ||
| BIP_Ecg | AGGAAACAAGGTGGATGTAGACT-GTCTCATGAAACGCTCGT | 41 | 2.0 µM | ||
| LB_Ecg | CCCATAAATTAGTTCGACTAGCTCG | 295–319 | 1.0 µM | ||
| 3 | F3_Eci | TGTTCGAGCGTCATAACACC | ITS | 440–459 | 0.2 µM |
| B3_Eci | GGTCAACCTGTGATCCATGT | 618–637 | 0.2 µM | ||
| FIP_Eci | CTGTCTTTAAGGGCCGCCGC-CCTCCAGCTGCCTTTGTG | 38 | 2.0 µM | ||
| BIP_Eci | GCGTGGGCTCTACGCGTAGTA-TTTTGGCAAGCCACCGTC | 39 | 2.0 µM | ||
| LF_Eci | CCCCAACACCGCAACCA | 479–495 | 1.0 µM | ||
| LB_Eci | ACTTGCTTCTCGCGACAGA | 559–577 | 1.0 µM |
| Isolate | Time to positive (Tp) in minutes | Isothermal Temperature | ||
| 60 °C | 63 °C | 65 °C | ||
| Positive control | 5 | 32 | 34 | |
| 8.2_PdIT | 6 | 37 | 35 | |
| 8.3_PdIT | 6.5 | 40 | 37 | |
| Isolate | Sample Type | Tp * (Min) ± SD ** at Different Dilutions | ||||||
|---|---|---|---|---|---|---|---|---|
| 20 ng/μL | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | ||
| 1.1_PdIT | DNA from epiphytic fungal structures | 4.05 ± 0.34 | 5.04 ± 0.8 | 36.24 ± 0.80 | 37.37 ± 1.20 | 45.43 ± 2.82 | 47.43 ± 1.26 | ND *** |
| 7.1_PdIT | 4.13 ± 0.23 | 4.9 ± 1.09 | 36.57 ± 2.32 | 41.06 ± 0.82 | 46.99 ± 1.47 | 49.87 ± 0.18 | ND | |
| Positive control | 4.7 ± 0.15 | 5.98 ± 0.05 | 32.32 ± 1.14 | 36.11 ± 4.22 | 39.74 ± 0.91 | 49.41 ± 3.42 | ND | |
| Isolate | Tp * (Min) ± SD ** | ||
|---|---|---|---|
| DNA | Crude Plant Extract | p-Value | |
| 1_PdIT.1 (S) *** | 4.05 ± 0.34 a | 5.92 ± 1.37 a | 0.0878 n.s. **** |
| 7_PdIT.1 (S) | 4.13 ± 0.23 a | 7.43 ± 0.24 b | 0.0002 |
| 8_PdIT.1 (S) | 4.7 ± 0.15 a | 8.05 ± 2.10 a | 0.1060 n.s. |
| 5_PdIT.1 (AS) ***** | 9.74 ± 0.06 a | 12.60 ± 1.50 a | 0.0777 n.s. |
| 49_PdIT (S) | 8.63 ± 0.01 a | 11.33 ± 0.50 b | 0.0002 |
| 50_PdIT (S) | 9.52 ± 0.17 a | 13.97 ± 0.29 b | 0.0002 |
| 51_PdIT (AS) | 10.03 ± 0.39 a | 12.87 ± 0.37 b | <0.0001 |
| 52_PdIT (AS) | 10.61 ± 0.92 a | 5.84 ± 0.68 b | 0.0008 |
| 53_PdIT (S) | 7.94 ± 0.20 a | 8.02 ± 0.09 a | 0.6893 n.s. |
| 54_PdIT (AS) | 12.6 ± 1.07 b | 10.80 ± 0.80 a | 0.0074 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Barone, M.M.; Moizio, M.; Choudhary, R.; D’Errico, C.; Trkulja, V.; Torta, L.; Davino, S.; Matić, S. Development of a Field-Deployable Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Erysiphe corylacearum in Hazelnut. J. Fungi 2026, 12, 79. https://doi.org/10.3390/jof12010079
Barone MM, Moizio M, Choudhary R, D’Errico C, Trkulja V, Torta L, Davino S, Matić S. Development of a Field-Deployable Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Erysiphe corylacearum in Hazelnut. Journal of Fungi. 2026; 12(1):79. https://doi.org/10.3390/jof12010079
Chicago/Turabian StyleBarone, Marta Maria, Marco Moizio, Ravish Choudhary, Chiara D’Errico, Vojislav Trkulja, Livio Torta, Salvatore Davino, and Slavica Matić. 2026. "Development of a Field-Deployable Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Erysiphe corylacearum in Hazelnut" Journal of Fungi 12, no. 1: 79. https://doi.org/10.3390/jof12010079
APA StyleBarone, M. M., Moizio, M., Choudhary, R., D’Errico, C., Trkulja, V., Torta, L., Davino, S., & Matić, S. (2026). Development of a Field-Deployable Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Erysiphe corylacearum in Hazelnut. Journal of Fungi, 12(1), 79. https://doi.org/10.3390/jof12010079









