What Do We Know About Cryptococcus spp. in Portugal? One Health Systematic Review in a Comprehensive 13-Year Retrospective Study (2013–2025)
Abstract
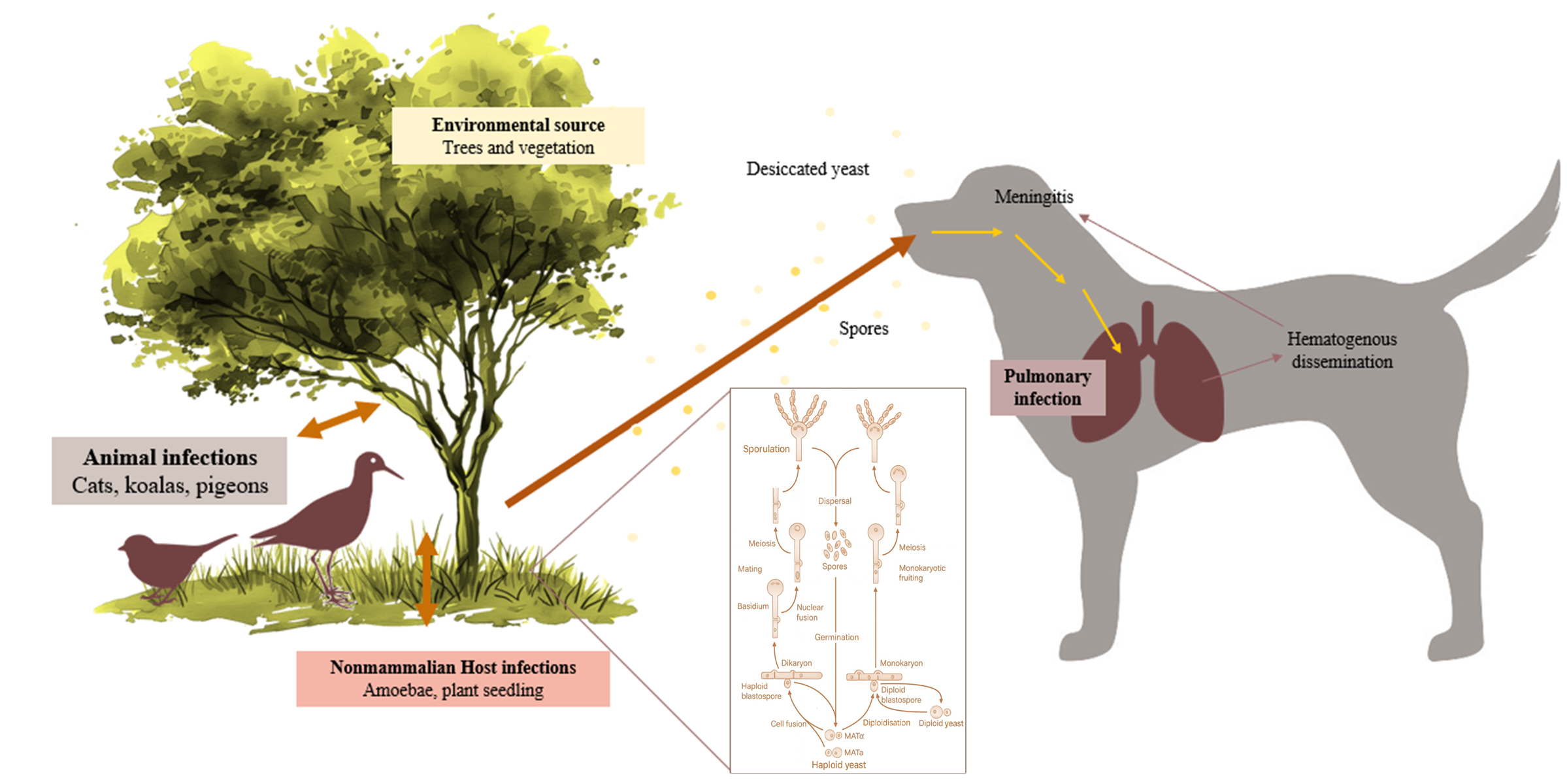
1. Introduction
2. Materials and Methods
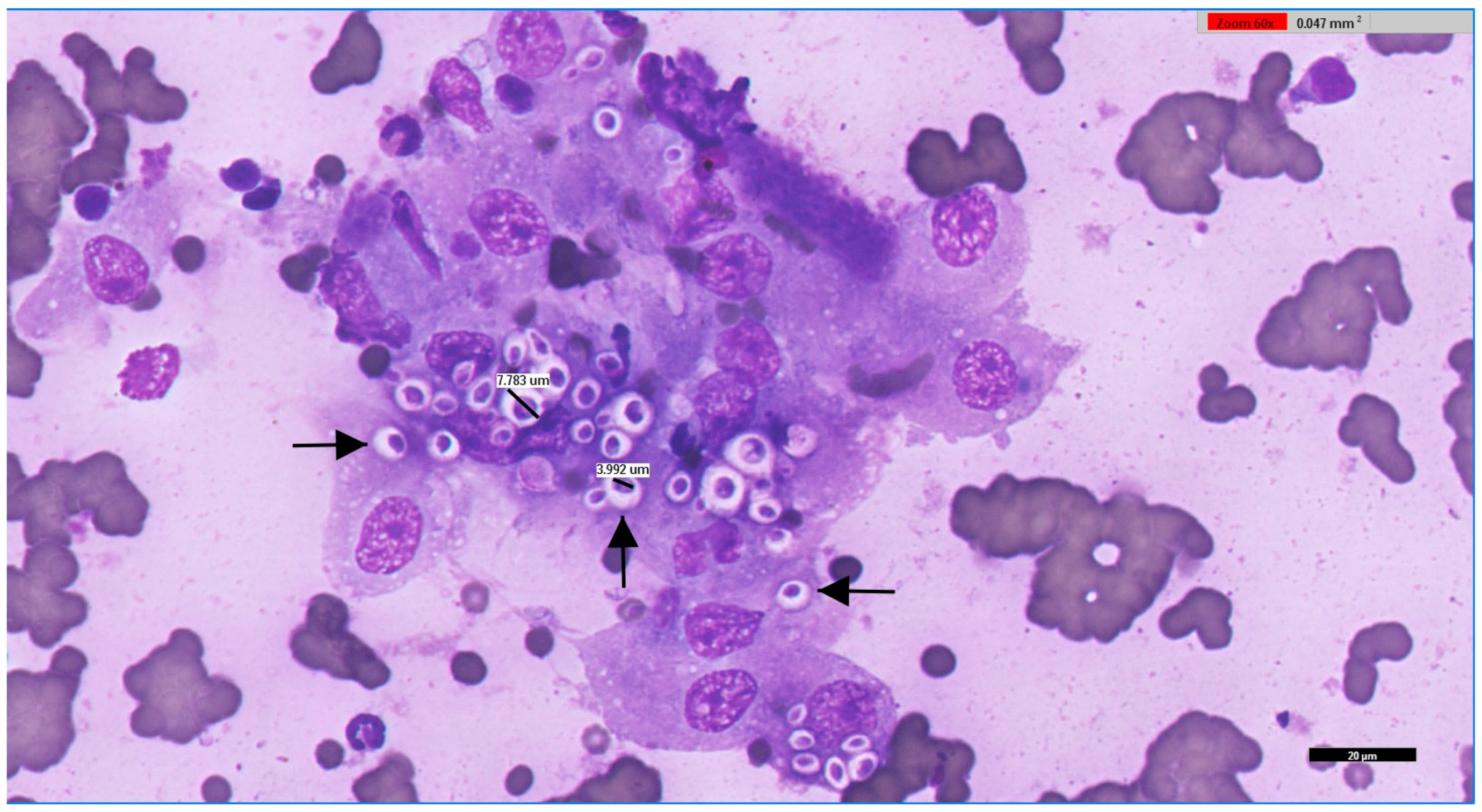
2.1. Data Collection, Sampling, and Microbiological Analysis
2.2. Statistical Analysis
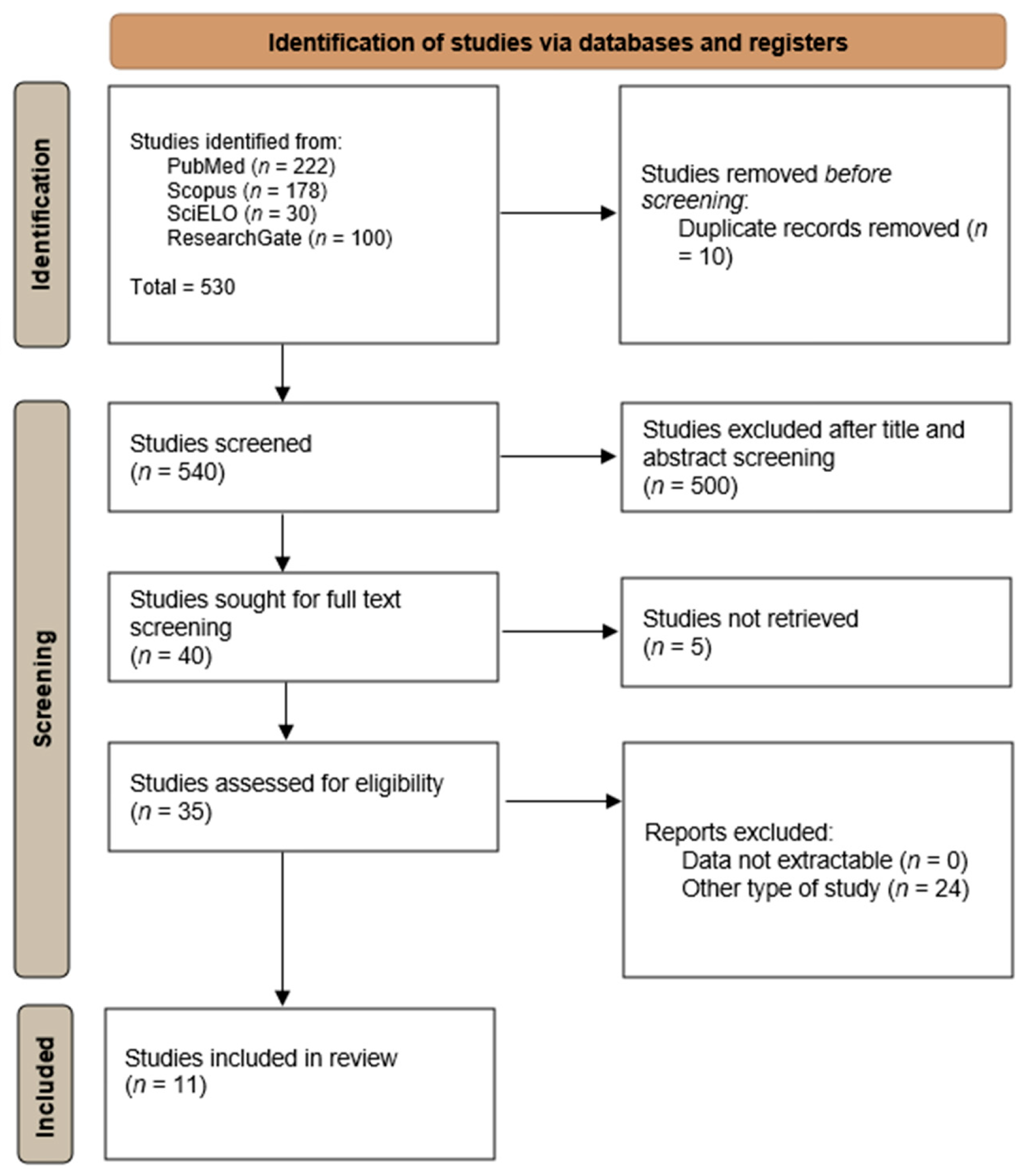
2.3. Systematic Review on Cryptococcus spp. Infections in Portugal
3. Results
3.1. Microbiological Study of Cryptococcus spp. in Portugal Between 2013 and 2025
3.1.1. General Positivity and Distribution by Host Species and Breed
3.1.2. Animal Species and Breeds
3.1.3. Sex
3.1.4. Age
3.1.5. Geographical Localisation
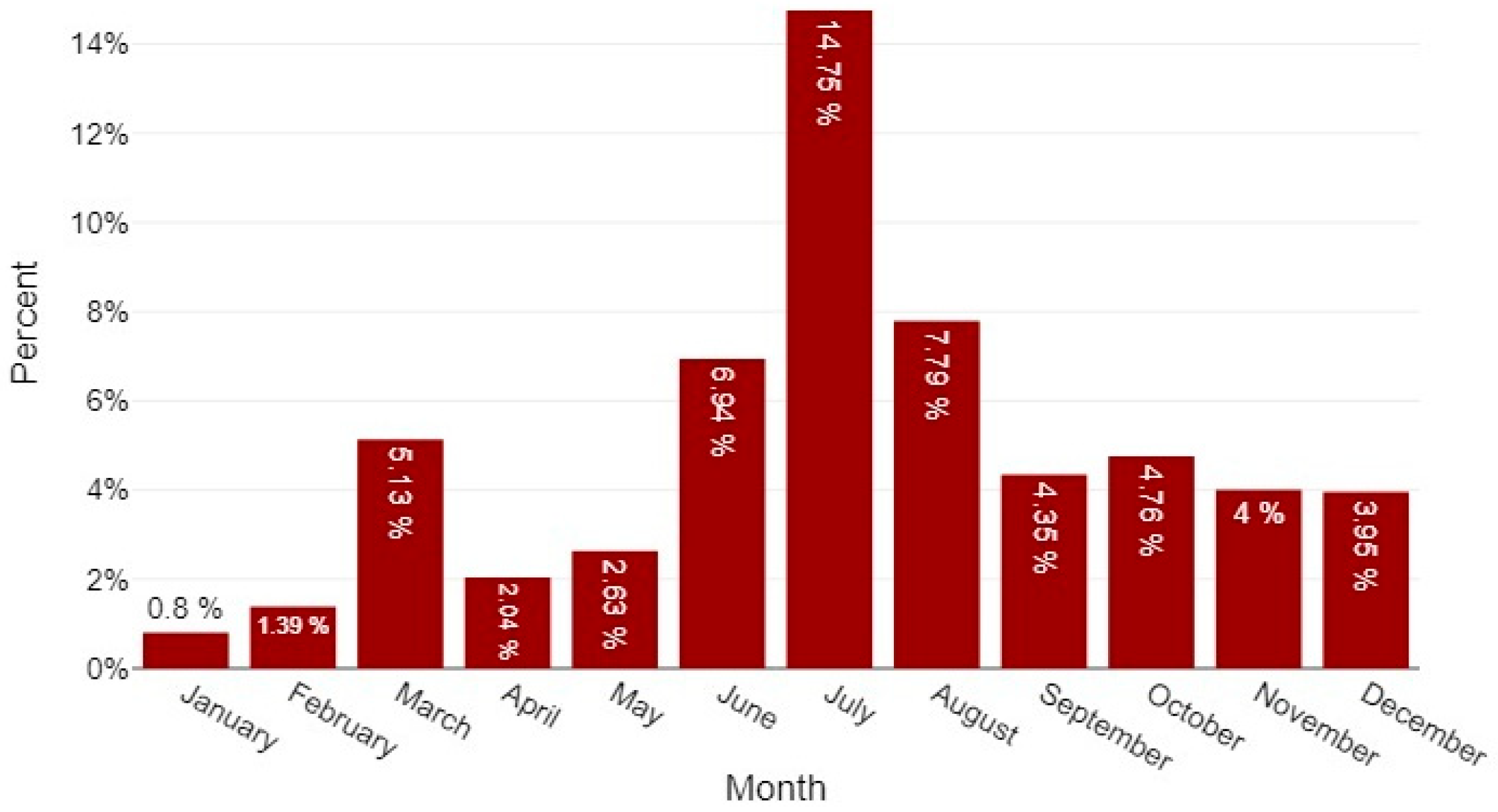
3.1.6. Month and Seasons
3.2. Systematic Review on Cryptococcus spp. in Portugal
4. Discussion
4.1. Cryptococcus Species Distribution and Host Compartments
4.2. Ecological and Geographic Patterns
4.3. Seasonality and Transmission Implications
4.4. Diagnostic Approach and Laboratory Considerations
4.5. One Health, Planetary Health, and Public Health Relevance
4.6. Limitations of This Study
4.7. Recommendations and Future Directions
- Routine molecular typing (MLST/AFLP or whole-genome sequencing where feasible) of clinical and environmental Cryptococcus isolates to map molecular types and detect emergent strains.
- Systematic, seasonally stratified environmental sampling (pigeon roosts, eucalyptus and other tree hollows, soil, and compost) in regions with veterinary or human cases to identify reservoirs and temporal dynamics.
- Integration of veterinary diagnostic data with human public-health laboratories (shared databases and joint investigations) under a One Health umbrella.
- Incorporation of antifungal susceptibility testing for clinical isolates to inform veterinary therapy and to monitor resistance trends.
- Targeted education for veterinarians and pet owners, particularly during summer months, on cryptococcosis recognition and strategies to reduce environmental exposure.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFLP | amplified fragment length polymorphism |
| CI | confidence interval |
| CNS | central nervous system |
| CrAg | cryptococcal antigen |
| CSF | cerebrospinal fluid |
| FeLV | feline leukaemia virus |
| FIV | feline immunodeficiency virus |
| IQR | interquartile range |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption/Ionisation Time-of-Flight Mass Spectrometry |
| MLST | multilocus sequence typing |
| NUTS | Nomenclature of Territorial Units for Statistics |
| p | p-value |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| SD | standard deviation |
| U | Mann–Whitney U statistic |
| VG | Cryptococcus gattii molecular type |
| VN | Cryptococcus neoformans molecular type |
References
- da Godinho, R.M.C.; Oliveira, D.L.; Albuquerque, P.C.; Dutra, F.F.; de Almeida-Paes, R.; Rodrigues, M.L.; Fonseca, F.L. Cryptococcus and cryptococcosis. In Current Progress in Medical Mycology; Springer International Publishing: Cham, Switzerland, 2017; pp. 169–214. [Google Scholar]
- Sabino, R.; Antunes, F.; Araujo, R.; Bezerra, A.R.; Brandão, J.; Carneiro, C.; Carvalho, A.; Carvalho, D.; Conceição, I.C.; Cota Medeiros, F.; et al. Addressing critical fungal pathogens under a One Health perspective: Key insights from the Portuguese Association of Medical Mycology. Mycopathologia 2025, 190, 73. [Google Scholar] [CrossRef]
- Mada, P.K.; Jamil, R.T.; Alam, M.U. Cryptococcus; StatPearls: Petersburg, FL, USA, 2025. [Google Scholar]
- Seffah, K.; Agyeman, W.; Madeo, J.L.; Ahmad, A. Cryptococcus infection in an immunocompetent patient. Cureus 2022, 14, e27635. [Google Scholar] [CrossRef]
- Bermann, C.D.S.; Braga, C.Q.; Ianiski, L.B.; de Botton, S.A.; Pereira, D.I.B. Cryptococcosis in domestic and wild animals: A review. Med. Mycol. 2023, 61, myad016. [Google Scholar] [CrossRef]
- Danesi, P.; Falcaro, C.; Schmertmann, L.J.; de Miranda, L.H.M.; Krockenberger, M.; Malik, R. Cryptococcus in wildlife and free-living mammals. J. Fungi 2021, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- do Carmo, F.N.; de Camargo Fenley, J.; Garcia, M.T.; Rossoni, R.D.; Junqueira, J.C.; de Barros, P.P.; Scorzoni, L. Cryptococcus spp. and Cryptococcosis: Focusing on the infection in Brazil. Braz. J. Microbiol. 2022, 53, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Z.; Wang, Q.-M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an integrated phylogenetic classification of the tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef]
- Beardsley, J.; Dao, A.; Keighley, C.; Garnham, K.; Halliday, C.; Chen, S.C.-A.; Sorrell, T.C. What’s New in Cryptococcus gattii: From bench to bedside and beyond. J. Fungi 2022, 9, 41. [Google Scholar] [CrossRef]
- Takashima, M.; Sugita, T. Taxonomy of pathogenic yeasts Candida, Cryptococcus, Malassezia, and Trichosporon. Med. Mycol. J. 2022, 63, 119–132. [Google Scholar] [CrossRef]
- Montoya, M.C.; Magwene, P.M.; Perfect, J.R. Associations between Cryptococcus genotypes, phenotypes, and clinical parameters of human disease: A review. J. Fungi 2021, 7, 260. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.-A.; Meyer, W.; Sorrell, T.C. Cryptococcus gattii infections. Clin. Microbiol. Rev. 2014, 27, 980–1024. [Google Scholar] [CrossRef]
- Sugita, T.; Cho, O.; Takashima, M. Current status of taxonomy of pathogenic yeasts. Med. Mycol. J. 2017, 58, J77–J81. [Google Scholar] [CrossRef] [PubMed]
- Srikanta, D.; Santiago-Tirado, F.H.; Doering, T.L. Cryptococcus neoformans: Historical curiosity to modern pathogen. Yeast 2014, 31, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Camargo, M.; Ramírez, J.D. Estimating the intra-taxa diversity, population genetic structure, and evolutionary pathways of Cryptococcus neoformans and Cryptococcus gatti. Front. Genet. 2018, 9, 148. [Google Scholar] [CrossRef]
- Reagan, K.L.; Krockenberger, M.; Sykes, J.E. 82—Cryptococcosis. In Greene’s Infectious Diseases of the Dog and Cat, 5th ed.; Saunders, W.B., Sykes, J.E., Eds.; Saunders: Philadelphia, PA, USA, 2021; pp. 1014–1029. ISBN 978-0-323-50934-3. [Google Scholar]
- Siqueira, N.P.; Favalessa, O.C.; Maruyama, F.H.; Dutra, V.; Nakazato, L.; Hagen, F.; Hahn, R.C. Domestic birds as source of Cryptococcus deuterogattii (AFLP6/VGII): Potential risk for cryptococcosis. Mycopathologia 2022, 187, 103–111. [Google Scholar] [CrossRef]
- McGill, S.; Malik, R.; Saul, N.; Beetson, S.; Secombe, C.; Robertson, I.; Irwin, P. Cryptococcosis in domestic animals in western Australia: A retrospective study from 1995–2006. Med. Mycol. 2009, 47, 625–639. [Google Scholar] [CrossRef]
- de Abreu, D.P.B.; Machado, C.H.; Makita, M.T.; Botelho, C.F.M.; Oliveira, F.G.; da Veiga, C.C.P.; Martins, M.D.A.; Baroni, F. de A. Intestinal lesion in a dog due to Cryptococcus gattii type VGII and review of published cases of canine gastrointestinal cryptococcosis. Mycopathologia 2017, 182, 597–602. [Google Scholar] [CrossRef]
- Touhali, I.S. Effect of Cryptococcus neoformans isolated from pigeon on some hematological, biochemical and histopathological parameters by experimental infection in BALB/c mice. Adv. Anim. Vet. Sci. 2018, 6, 201–206. [Google Scholar] [CrossRef]
- Refai, M.; El-Hariri, M.; Alarousy, R. Cryptococcosis in animals and birds: A review. EJAE 2017, 4, 202–223. [Google Scholar]
- Nosanchuk, J.D.; Shoham, S.; Fries, B.C.; Shapiro, D.S.; Levitz, S.M.; Casadevall, A. Evidence of zoonotic transmission of Cryptococcus neoformans from a pet cockatoo to an immunocompromised patient. Ann. Intern. Med. 2000, 132, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R.; Bicanic, T. Cryptococcosis diagnosis and treatment: What do we know now. Fungal. Genet. Biol. 2015, 78, 49–54. [Google Scholar] [CrossRef]
- Reagan, K.L.; McHardy, I.; Thompson, G.R.; Sykes, J.E. Evaluation of the clinical performance of 2 point-of-care cryptococcal antigen tests in dogs and cats. J Vet. Intern. Med. 2019, 33, 2082–2089. [Google Scholar] [CrossRef]
- Langner, K.F.A.; Yang, W.-J. Clinical performance of the IMMY cryptococcal antigen lateral flow assay in dogs and cats. J. Vet. Intern. Med. 2022, 36, 1966–1973. [Google Scholar] [CrossRef]
- Krockenberger, M.B.; Marschner, C.; Martin, P.; Reppas, G.; Halliday, C.; Schmertmann, L.J.; Harvey, A.M.; Malik, R. Comparing immunochromatography with latex antigen agglutination testing for the diagnosis of cryptococcosis in cats, dogs and koalas. Med. Mycol. 2020, 58, 39–46. [Google Scholar] [CrossRef]
- Huang, H.-R.; Fan, L.-C.; Rajbanshi, B.; Xu, J.-F. Evaluation of a new cryptococcal antigen lateral flow immunoassay in serum, cerebrospinal fluid and urine for the diagnosis of cryptococcosis: A meta-analysis and systematic review. PLoS ONE 2015, 10, e0127117. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, M.G.; Hartmann, K.; Lloret, A.; Ferrer, L.; Addie, D.; Belák, S.; Boucraut-Baralon, C.; Egberink, H.; Frymus, T.; Gruffydd-Jones, T.; et al. Cryptococcosis in cats: ABCD guidelines on prevention and management. J. Feline. Med. Surg. 2013, 15, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Maziarz, E.K.; Perfect, J.R. Cryptococcosis. Infect. Dis. Clin. N. Am. 2016, 30, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Petrie, A.; Watson, P. Statistics for Veterinary and Animal Science, 3rd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; ISBN 978-0-470-67075-0. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Viviani, M.A.; Nikolova, R.; Esposto, M.C.; Prinz, G.; Cogliati, M. First european case of serotype A MATa Cryptococcus neoformans infection. Emerg. Infect. Dis. 2003, 9, 1179–1180. [Google Scholar] [CrossRef]
- Kessel, J.; Rossaert, A.-C.; Lingscheid, T.; Grothe, J.; Harrer, T.; Wyen, C.; Tominski, D.; Bollinger, T.; Kehr, A.K.; Kalbitz, S.; et al. Survival after cryptococcosis in Germany: A retrospective multicenter cohort study of patients diagnosed between 2004 and 2021. Int. J. Med. Microbiol. 2024, 314, 151614. [Google Scholar] [CrossRef]
- Maduro, A.P.; Mansinho, K.; Teles, F.; Silva, I.; Meyer, W.; Martins, M.L.; Inácio, J. Insights on the genotype distribution among Cryptococcus neoformans and C. gattii Portuguese clinical isolates. Curr. Microbiol. 2014, 68, 199–203. [Google Scholar] [CrossRef]
- Cogliati, M.; D’Amicis, R.; Zani, A.; Montagna, M.T.; Caggiano, G.; De Giglio, O.; Balbino, S.; De Donno, A.; Serio, F.; Susever, S.; et al. Environmental distribution of Cryptococcus neoformans and C. gattii around the mediterranean basin. FEMS Yeast Res. 2016, 16, fow045. [Google Scholar] [CrossRef]
- Pimenta, P.; Alves-Pimenta, S.; Barros, J.; Pereira, M.J.; Maltez, L.; Maduro, A.P.; Cardoso, L.; Coelho, A.C. Blepharitis due to Cryptococcus neoformans in a cat from northern Portugal. JFMS Open Rep. 2015, 1, 2055116915593963. [Google Scholar] [CrossRef]
- Maduro, A.P.; Gonçalves, L.; Inácio, J.; Faria, N.C.G.; Martins, M.L.; Teles, F.R.R. HIV/AIDS-Associated cryptococcosis in Portugal spanning the pre- to post-HAART era: A retrospective assessment at the genotypic level based on URA5-RFLP. Curr. Microbiol. 2015, 71, 449–457. [Google Scholar] [CrossRef]
- Morera, N.; Hagen, F.; Juan-Sallés, C.; Artigas, C.; Patricio, R.; Serra, J.I.; Colom, M.F. Ferrets as sentinels of the presence of pathogenic Cryptococcus species in the Mediterranean Environment. Mycopathologia 2014, 178, 145–151. [Google Scholar] [CrossRef]
- Stilwell, G.; Pissarra, H. Cryptococcal meningitis in a goat—A case report. BMC Vet. Res. 2014, 10, 84. [Google Scholar] [CrossRef]
- Cuetara, M.S.; Jusdado Ruiz-Capillas, J.J.; Nuñez-Valentin, M.P.; Rodríguez Garcia, E.; Garcia-Benayas, E.; Rojo-Amigo, R.; Rodriguez-Gallego, J.C.; Hagen, F.; Colom, M.F. Successful isavuconazole salvage therapy for a Cryptococcus deuterogattii (AFLP6/VGII) disseminated infection in a european immunocompetent patient. Mycopathologia 2021, 186, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Juan-Sallés, C.; Mendes, J.; Mendes, A.; Ruivo, M.; Abad, J.L.; Hagen, F.; Colom, M.F. Cryptococcus bacillisporus causing cryptococcoma of the beak of an African Grey Parrot (Psittacus erithacus), Portugal. Med. Mycol. Case. Rep. 2021, 34, 8–12. [Google Scholar] [CrossRef]
- Freire, S.; Grilo, T.; Poirel, L.; Aires-de-Sousa, M. Urban pigeons (Columba livia) as a source of broad-spectrum β-lactamase-producing Escherichia coli in Lisbon, Portugal. Antibiotics 2022, 11, 1368. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.S.; Sampaio, A.; Maduro, A.P.; Silva, I.; Teles, F.; da Martins, M.L.; Inácio, J. Genotypic diversity of environmental Cryptococcus neoformans isolates from northern Portugal. Mycoses 2014, 57, 98–104. [Google Scholar] [CrossRef]
- Lemos, L.S.; Siqueira de O dos Santos, A.; Vieira-da-Motta, O.; Texeira, G.N.; Queiroz de Carvalho, E.C. Pulmonary cryptococcosis in slaughtered sheep: Anatomopathology and culture. Vet. Microbiol. 2007, 125, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Pllana-Hajdari, D.; Cogliati, M.; Čičmak, L.; Pleško, S.; Mlinarić-Missoni, E.; Mareković, I. First Isolation, Antifungal susceptibility, and molecular characterization of Cryptococcus neoformans from the environment in Croatia. J. Fungi 2019, 5, 99. [Google Scholar] [CrossRef]
- Fernandes, F.F.; Dias, A.L.T.; Ramos, C.L.; Ikegaki, M.; de Siqueira, A.M.; Franco, M.C. The “in vitro” antifungal activity evaluation of propolis G12 ethanol extract on Cryptococcus neoformans. Rev. Inst. Med. Trop. Sao Paulo 2007, 49, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Duncan, C.; Stephen, C.; Lester, S.; Bartlett, K.H. Sub-clinical infection and asymptomatic carriage of Cryptococcus gattii in dogs and cats during an outbreak of cryptococcosis. Med. Mycol. 2005, 43, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Mbangiwa, T.; Sturny-Leclère, A.; Lechiile, K.; Kajanga, C.; Boyer-Chammard, T.; Hoving, J.C.; Leeme, T.; Moyo, M.; Youssouf, N.; Lawrence, D.S.; et al. Development and validation of quantitative PCR assays for HIV-associated cryptococcal meningitis in sub-Saharan Africa: A diagnostic accuracy study. Lancet Microbe 2024, 5, e261–e271. [Google Scholar] [CrossRef]
- Skipper, C.P.; Hullsiek, K.H.; Stadelman, A.; Williams, D.A.; Ssebambulidde, K.; Okafor, E.; Tugume, L.; Nuwagira, E.; Akampurira, A.; Musubire, A.K.; et al. Sterile cerebrospinal fluid culture at cryptococcal meningitis diagnosis is associated with high mortality. J. Fungi 2022, 9, 46. [Google Scholar] [CrossRef]
- Kwizera, R.; Akampurira, A.; Kandole, T.K.; Nielsen, K.; Kambugu, A.; Meya, D.B.; Boulware, D.R.; Rhein, J.; ASTRO-CM Study Team. Evaluation of trypan blue stain in a haemocytometer for rapid detection of cerebrospinal fluid sterility in HIV patients with cryptococcal meningitis. BMC Microbiol. 2017, 17, 182. [Google Scholar] [CrossRef] [PubMed]
- Howard-Jones, A.R.; Sparks, R.; Pham, D.; Halliday, C.; Beardsley, J.; Chen, S.C.-A. Pulmonary cryptococcosis. J. Fungi 2022, 8, 1156. [Google Scholar] [CrossRef]
- Setianingrum, F.; Rautemaa-Richardson, R.; Denning, D.W. Pulmonary cryptococcosis: A review of pathobiology and clinical aspects. Med. Mycol. 2019, 57, 133–150. [Google Scholar] [CrossRef]
- O’Brien, C.R.; Krockenberger, M.B.; Wigney, D.I.; Martin, P.; Malik, R. Retrospective study of feline and canine cryptococcosis in Australia from 1981 to 2001: 195 cases. Med. Mycol. 2004, 42, 449–460. [Google Scholar] [CrossRef]
- Malik, R.; Wigney, D.I.; Muir, D.B.; Love, D.N. Asymptomatic carriage of Cryptococcus neoformans in the nasal cavity of dogs and cats. J. Med. Vet. Mycol. 1997, 35, 27–31. [Google Scholar] [CrossRef]
- Reis, R.S.; Bonna, I.C.F.; da Antonio, I.M.S.; Pereira, S.A.; do Nascimento, C.R.S.; Ferraris, F.K.; Brito-Santos, F.; Ferreira Gremião, I.D.; Trilles, L. Cryptococcus neoformans VNII as the main cause of cryptococcosis in domestic cats from Rio de Janeiro, Brazil. J. Fungi 2021, 7, 980. [Google Scholar] [CrossRef] [PubMed]
| Distribution of Cryptococcus spp.-Positive Cases by Animal Species | |||||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | Total | |||||
| Species | n | % Within Species | 95% CI (%) | n | % Within Species | 95% CI (%) | n |
| Avian | 691 | 96.0% | 94.3–97.2 | 29 | 4.0% | 2.8–5.8 | 720 |
| Canine | 177 | 93.2% | 88.7–96.0 | 13 | 6.8% | 4.0–11.4 | 190 |
| Feline | 107 | 94.7% | 88.9–97.5 | 6 | 5.3% | 2.5–11.1 | 113 |
| Equine | 9 | 100% | 0.7–100 | 0 | 0% | - | 9 |
| Lagomorphs | 7 | 100% | 64.6–100 | 0 | 0% | - | 7 |
| Rodents | 1 | 100% | 20.7–100 | 0 | 0% | - | 1 |
| Other mammals | 8 | 100% | 67.6–100 | 0 | 0% | - | 8 |
| Non-mammalian non-avian vertebrates | 11 | 100% | 67.6–100 | 0 | 0% | - | 11 |
| Total | 1011 | 95.6% | 94.0–96.6 | 48 | 4.5% | 3.4–6.0 | 1059 |
| Papiliotrema laurentii (Formerly C. laurentii ) | Filobasidium uniguttulatum (Formerly C. neoformans var. uniguttulatus) | Cryptococcus neoformans | Naganishia humicola (Formerly C. humicola) | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Breeds | n | % Within Breed | n | % Within Breed | n | % Within Breed | n | % Within Breed | n | |
| Canine | Boston Terrier | 0 | 0% | 0 | 0% | 2 | 100% | 0 | 0% | 2 |
| Dobermann | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 100% | 1 | |
| French Bulldog | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | |
| German Shepherd | 0 | 0% | 0 | 0% | 1 | 100% | 0 | 0% | 1 | |
| Saint Bernard | 0 | 0% | 0 | 0% | 1 | 100% | 0 | 0% | 1 | |
| Siberian Husky | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | |
| Yorkshire Terrier | 0 | 0% | 0 | 0% | 1 | 100% | 0 | 0% | 1 | |
| Mixed Breed | 1 | 14.3% | 1 | 14.3% | 5 | 71.4% | 0 | 0% | 7 | |
| Feline | European Shorthair | 0 | 0% | 0 | 0% | 4 | 100% | 0 | 0% | 4 |
| Total | 3 | 15.8% | 1 | 5.3% | 14 | 73.7% | 1 | 5.3% | 19 | |
| Papiliotrema laurentii (Formerly C. laurentii ) | Filobasidium uniguttulatum (Formerly C. neoformans var. uniguttulatus) | Cryptococcus neoformans | Solicoccozyma terreus (Formerly C. terreus) | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Avian Species | n | % Within Species | n | % Within Species | n | % Within Species | n | % Within Species | n |
| African grey parrot (Psittacus erithacus) | 1 | 50% | 0 | 0% | 0 | 0% | 1 | 50% | 2 |
| Atlantic canary (Serinus canaria) | 5 | 83.3% | 1 | 16.7% | 0 | 0% | 0 | 0% | 6 |
| Blue-and-yellow macaw (Ara ararauna) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Budgerigar (Melopsittacus undulatus) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Chestnut-billed toucanet (Aulacorhynchus castaneotis) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Cockatiel (Nymphicus hollandicus) | 0 | 0% | 0 | 0% | 1 | 100% | 0 | 0% | 1 |
| Eclectus parrot (Eclectus roratus) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Emerald toucanet (Aulacorhynchus prasinus) | 3 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 3 |
| European goldfinch (Carduelis carduelis) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Lovebird (Agapornis spp.) | 3 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 3 |
| Parakeet (Cyanoramphus spp.) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Red lory (Lorius lory erythrothorax) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Red-crowned parakeet (Cyanoramphus novaezelandiae) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Ring neck (Psittacula krameri) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Rock pigeon (Columba livia) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Society finch (Lonchura striata) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Stella’s lory (Lorius stella) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Toco toucan (Ramphastos toco) | 1 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | 1 |
| Tucumán amazon (Amazona tucumana) | 0 | 0% | 0 | 0% | 1 | 100% | 0 | 0% | 1 |
| Total | 25 | 86.2% | 1 | 3.5% | 2 | 6.9% | 1 | 3.5% | 29 |
| Papiliotrema laurentii (Formerly C. laurentii ) | Filobasidium uniguttulatum (Formerly C. neoformans var. uniguttulatus ) | Cryptococcus neoformans | Naganishia humicola (Formerly C. humicola) | Solicoccozyma terreus (Formerly C. terreus ) | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Region(NUTS2) | n | % Within Region | n | % Within Region | n | % Within Region | n | % Within Region | n | % Within Region | n |
| North | 3 | 10.7% | 1 | 50% | 10 | 62.5% | 1 | 100% | 0 | 0% | 15 |
| Centre | 25 | 89.3% | 1 | 50% | 3 | 18.8% | 0 | 0% | 1 | 100% | 30 |
| Algarve | 0 | 0% | 0 | 0% | 3 | 18.8% | 0 | 0% | 0 | 0% | 3 |
| Total | 28 | 100% | 2 | 100% | 16 | 100% | 1 | 100% | 1 | 100% | 48 |
| Species | Year | Location | Cryptococcus spp. (Strain) | Clinical Manifestation | Outcome | Ref. |
|---|---|---|---|---|---|---|
| HIV/AIDS human patients | 2000s | Lisbon | C. neoformans (VNI, VNII, VNIII, VNIV) | Meningoencephalitis | Some recovery, some death | [37] |
| Middle-aged man, male | 2007 | Lisbon | C. gattii (VGII) | Pulmonary and systemic | Recovery | [34] |
| Domestic ferret (Mustela putorius furo) | 2014 | V. N. Milfontes | C. gattii VGIII (AFLP6) | Pulmonary cryptococcosis (nodular masses in the lung) | Euthanised | [38] |
| Goat (Bravia breed) | 2014 | Lisbon | C. neoformans | Cryptococcal meningitis (post mortem diagnosis) | Death | [39] |
| Domestic cat (Felis catus) | 2015 | Vila Real | C. neoformans | Ocular cryptococcosis (blepharitis) | Recovery | [36] |
| Adult man, male | 2019 | Azores | C. deuterogattii (VGII) | Lung cryptococcomas, CNS lesions | Recovery | [40] |
| African grey parrot (Psittacus erithacus) | 2021 | Almada | C. bacillisporus VGIII (AFLP5) | Rhinothecal cryptococcoma | Recovery | [41] |
| Region | Year | Sample Source | Cryptococcus spp. (Strain) | Ref. |
|---|---|---|---|---|
| Lisbon | 2001 | Pigeons’ roost droppings | C. neoformans | [44] |
| Mediterranean region | 2012–2015 | Tree hollows, trunks, bark, and soil | C. neoformans | [35] |
| Vila Real | 2014 | Pigeon droppings; eucalyptus tree detritus | C. neoformans (VNIV, VNI, VNIII) | [43] |
| Various sites in Portugal | 2016 | Tree hollows | C. neoformans (VNI) | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, R.; Garcês, A.; Carvalho, H.L.d.; Silva, V.; Sampaio, F.; Fernandes, C.; Barros, G.; Brito, A.S.d.; Silva, A.R.; Duarte, E.L.; et al. What Do We Know About Cryptococcus spp. in Portugal? One Health Systematic Review in a Comprehensive 13-Year Retrospective Study (2013–2025). J. Fungi 2025, 11, 672. https://doi.org/10.3390/jof11090672
Lopes R, Garcês A, Carvalho HLd, Silva V, Sampaio F, Fernandes C, Barros G, Brito ASd, Silva AR, Duarte EL, et al. What Do We Know About Cryptococcus spp. in Portugal? One Health Systematic Review in a Comprehensive 13-Year Retrospective Study (2013–2025). Journal of Fungi. 2025; 11(9):672. https://doi.org/10.3390/jof11090672
Chicago/Turabian StyleLopes, Ricardo, Andreia Garcês, Hugo Lima de Carvalho, Vanessa Silva, Filipe Sampaio, Cátia Fernandes, Gonçalo Barros, Alexandre Sardinha de Brito, Ana Rita Silva, Elsa Leclerc Duarte, and et al. 2025. "What Do We Know About Cryptococcus spp. in Portugal? One Health Systematic Review in a Comprehensive 13-Year Retrospective Study (2013–2025)" Journal of Fungi 11, no. 9: 672. https://doi.org/10.3390/jof11090672
APA StyleLopes, R., Garcês, A., Carvalho, H. L. d., Silva, V., Sampaio, F., Fernandes, C., Barros, G., Brito, A. S. d., Silva, A. R., Duarte, E. L., Cardoso, L., & Coelho, A. C. (2025). What Do We Know About Cryptococcus spp. in Portugal? One Health Systematic Review in a Comprehensive 13-Year Retrospective Study (2013–2025). Journal of Fungi, 11(9), 672. https://doi.org/10.3390/jof11090672















