Genomic Insights into the Molecular Basis of Broad Host Adaptability of the Entomopathogenic Fungus Conidiobolus coronatus (Entomophthoromycotina)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Culture and DNA Extraction
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Phylogenetics, Conserved Motifs, and Domain Analysis
2.4. Exploration of Terpene Metabolites in C. coronatus
3. Results
3.1. Genomic Characteristics of C. coronatus
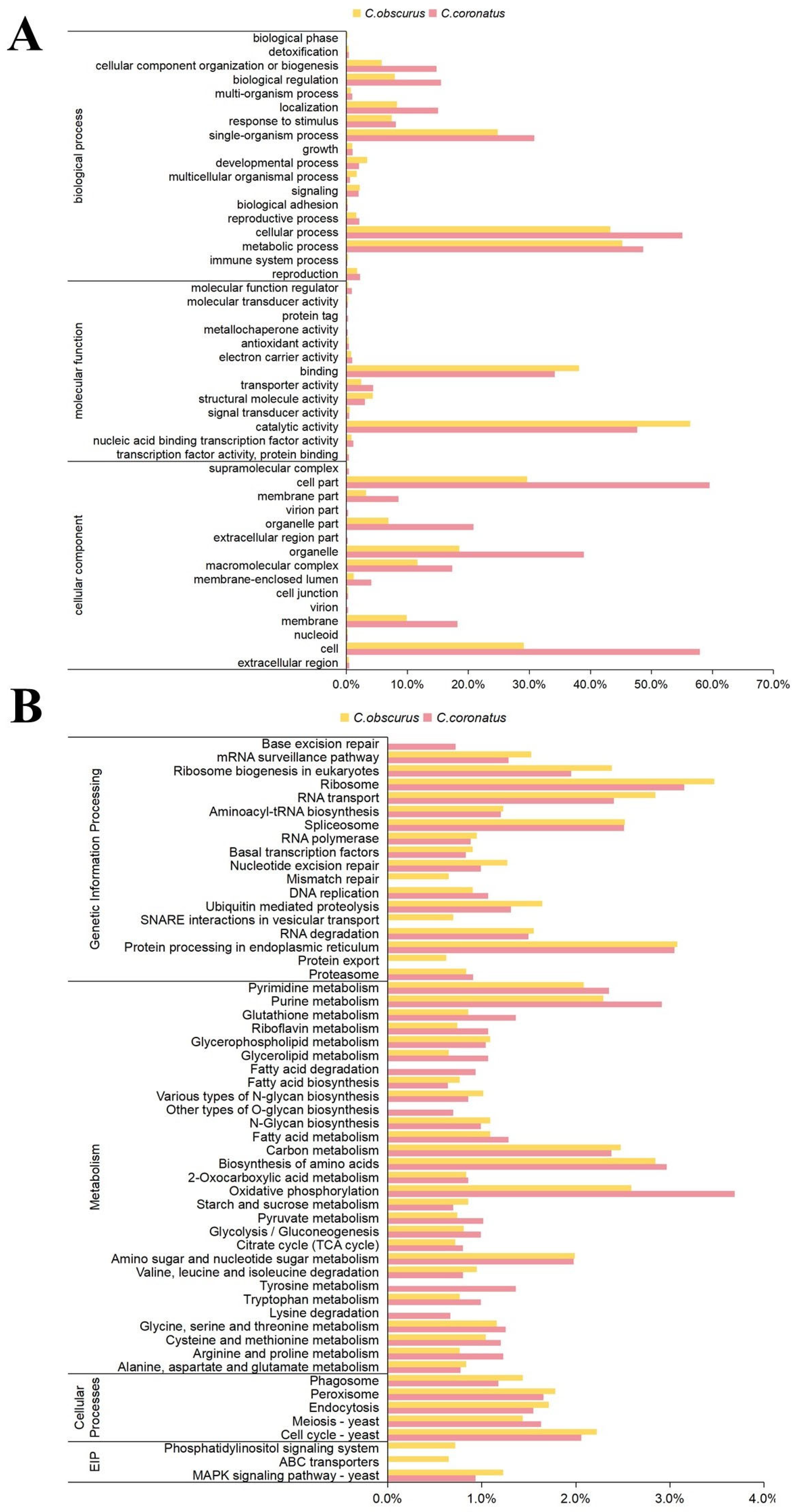
3.2. General Functional Annotation of C. coronatus Genes
3.3. Special Functional Annotation of C. coronatus Genes
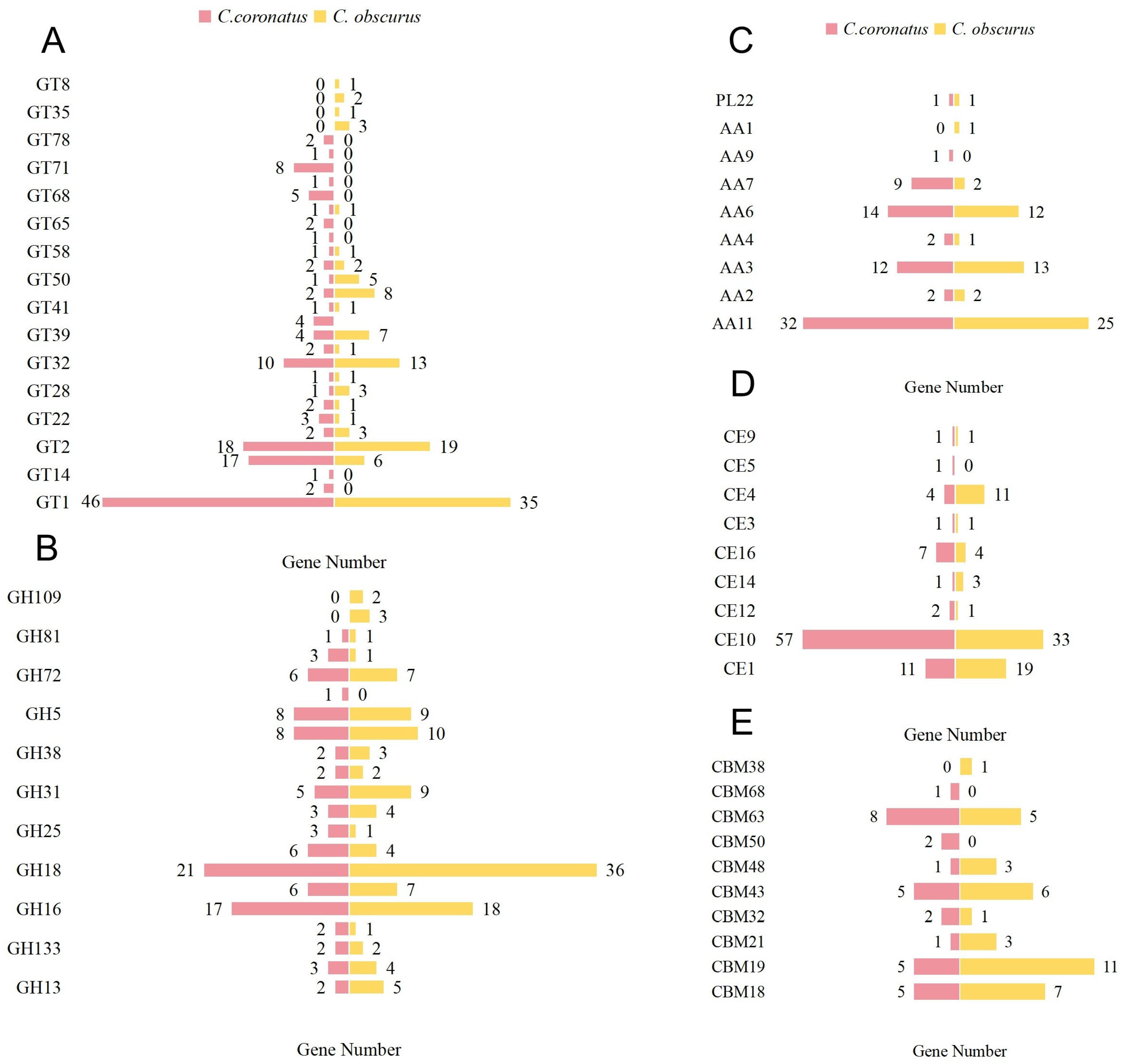
3.3.1. CAZy Database Annotation
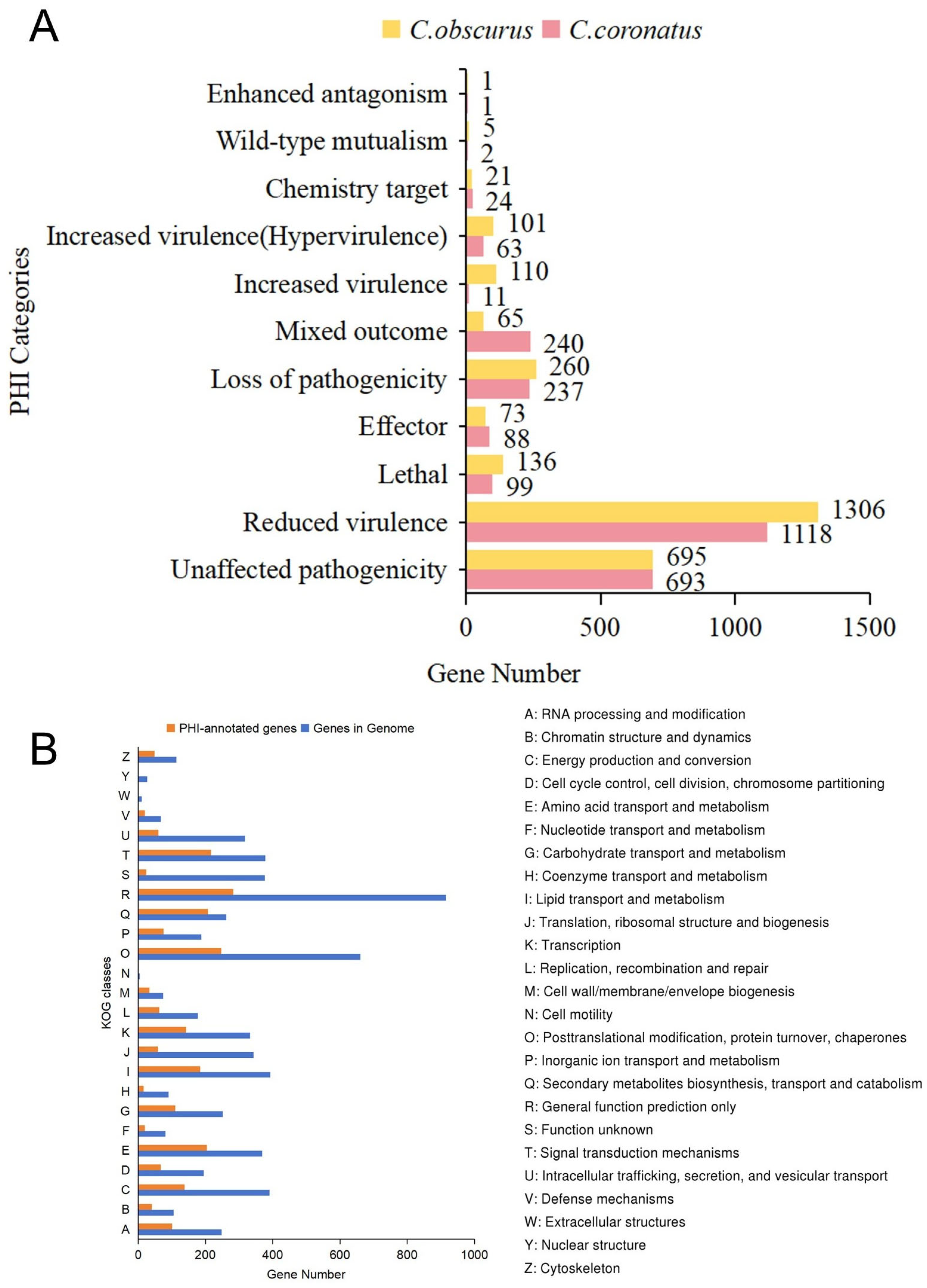
3.3.2. PHI-Base Annotations
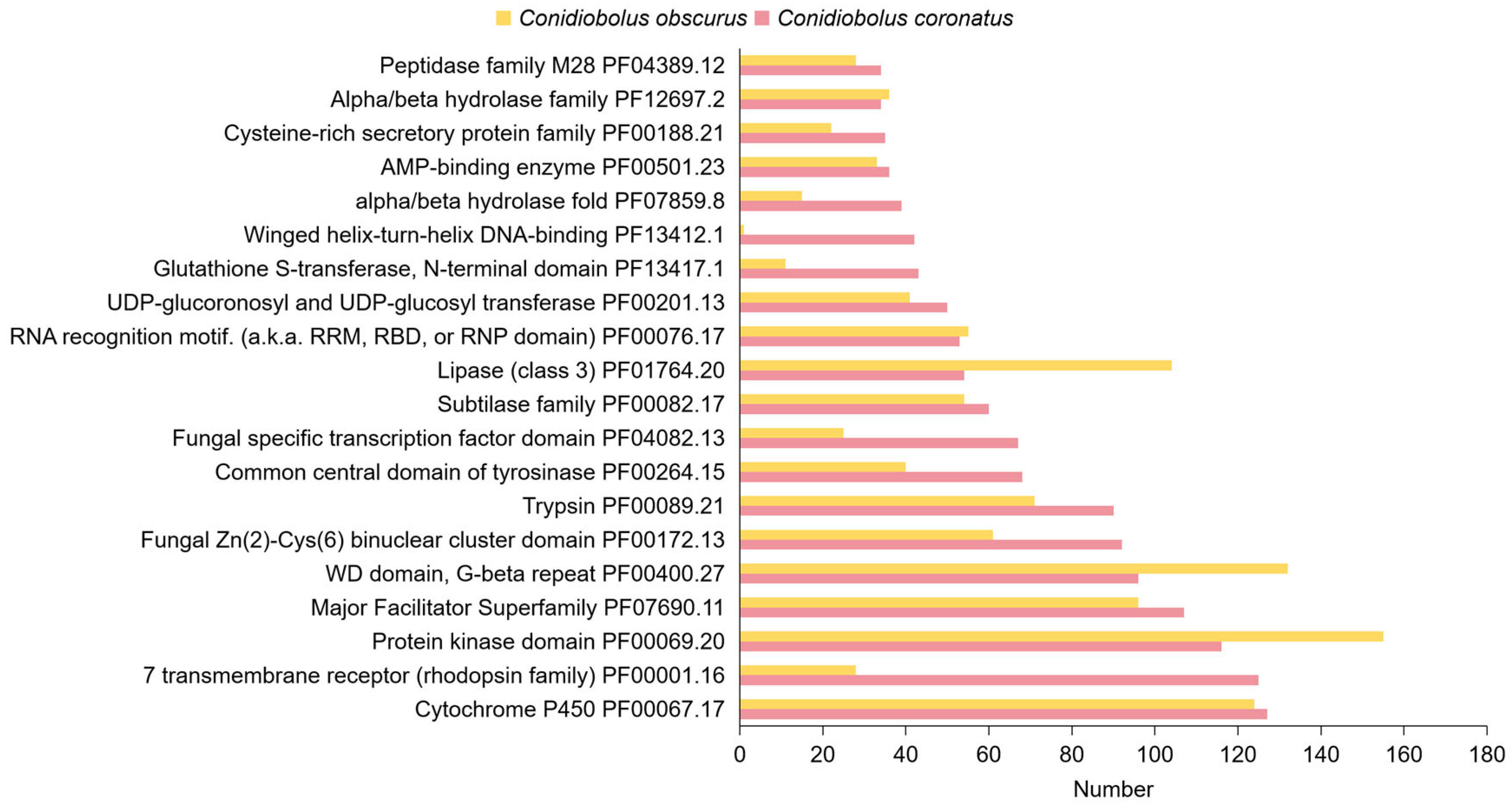
3.3.3. Protein Family (Pfam) Domain Annotation
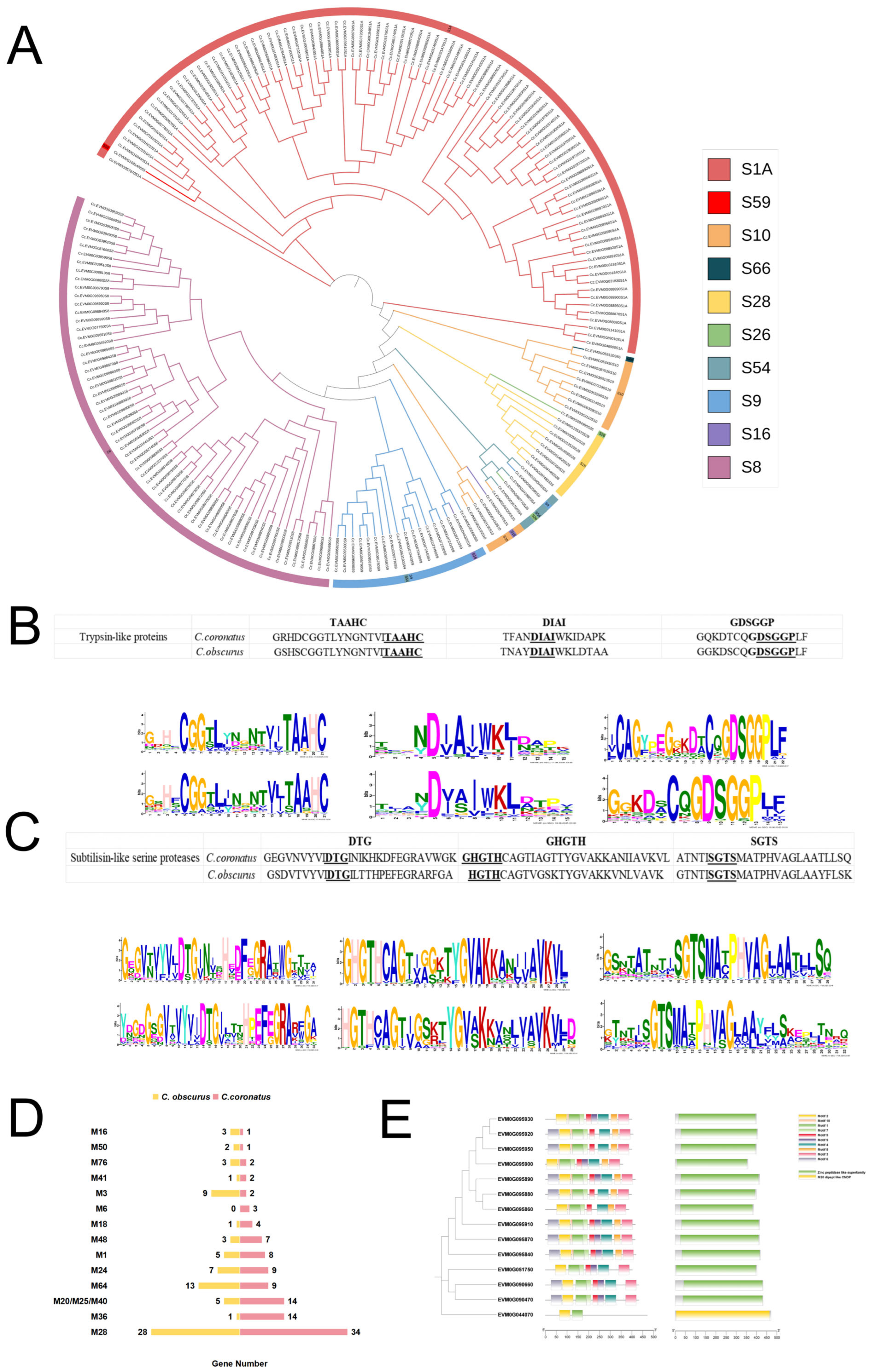
3.4. Abundant Serine Proteases and Metallopeptidases in C. coronatus
3.5. Expansion of G-Protein-Coupled Receptors in C. coronatus
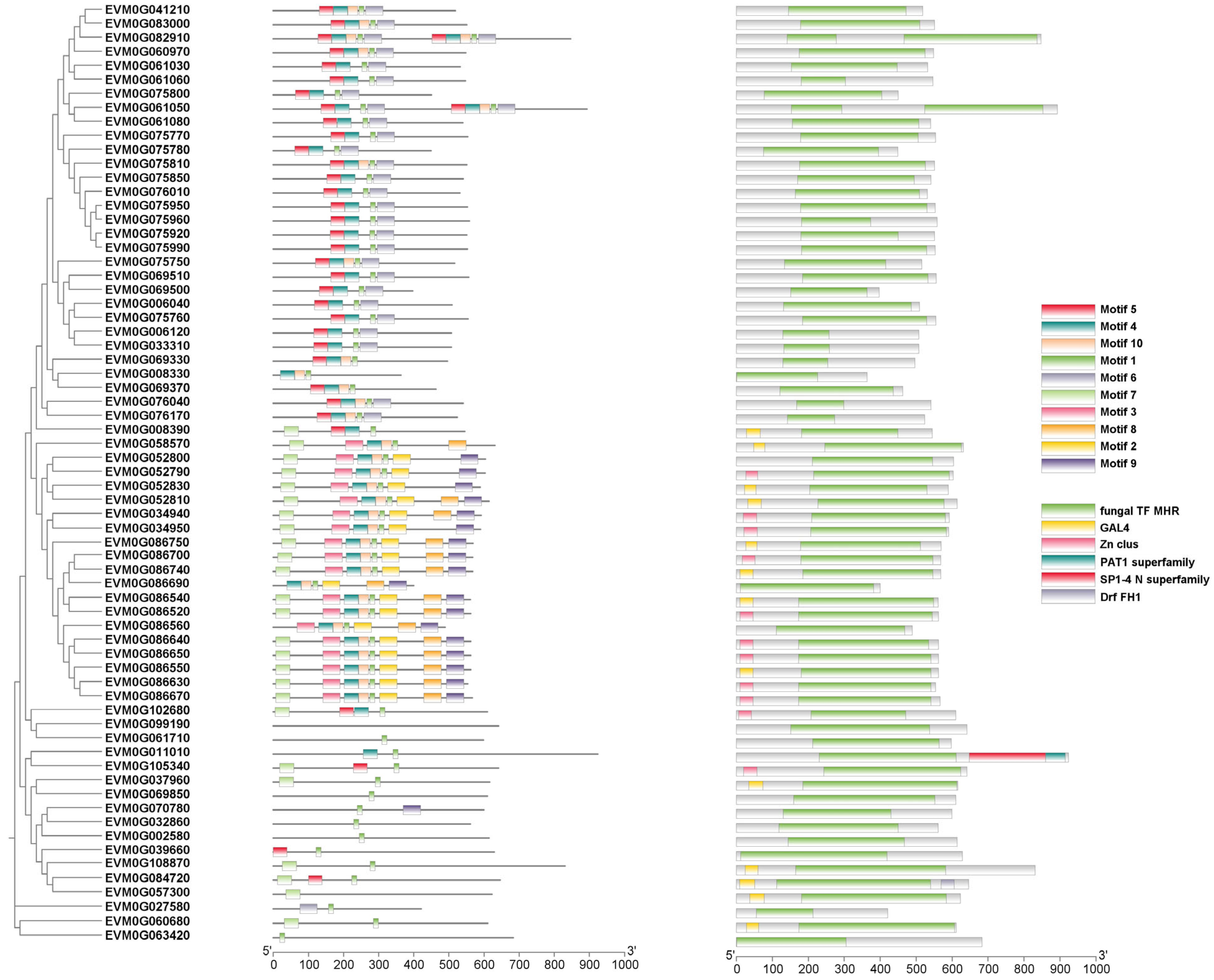
3.6. Expansion of Transcription Factors in C. coronatus
3.7. Metabolomic Profiling Reveals Enhanced Terpenoid Biosynthesis in C. coronatus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Bekker, C.; Beckerson, W.C.; Elya, C. Mechanisms behind the madness: How do zombie-making fungal entomopathogens affect host behavior to increase transmission? mBio 2021, 12, e0187221. [Google Scholar] [CrossRef]
- Wang, C.S.; Feng, M.G. Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol. Control 2014, 68, 129–135. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef]
- Lovett, B.; St Leger, R.J. Genetically engineering better fungal biopesticides. Pest Manag. Sci. 2018, 74, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Feng, M.G. Molecular basis and regulatory mechanisms underlying fungal insecticides’ resistance to solar ultraviolet irradiation. Pest Manag. Sci. 2022, 78, 30–42. [Google Scholar] [CrossRef]
- Faria, M.R.; Wraight, S.P. Mycoinsecticides and mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Brodeur, J. Host specificity in biological control: Insights from opportunistic pathogens. Evol. Appl. 2012, 5, 470–480. [Google Scholar] [CrossRef]
- Sacco, N.E.; Hajek, A.E. Diversity and breadth of host specificity among arthropod pathogens in the Entomophthoromycotina. Microorganisms 2023, 11, 1658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Winter prevalence of obligate aphid pathogen Pandora neoaphidis mycosis in the host Myzus persicae populations in southern China: Modeling description and biocontrol implication. Braz. J. Microbiol. 2012, 43, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Elya, C.; Lok, T.C.; Spencer, Q.E.; McCausland, H.; Martinez, C.C.; Eisen, M. Robust manipulation of the behavior of Drosophila melanogaster by a fungal pathogen in the laboratory. eLife 2018, 7, e34414. [Google Scholar] [CrossRef]
- Gryganskyi, A.P.; Nie, Y.; Hajek, A.E.; Hodge, K.T.; Liu, X.Y.; Aadland, K.; Voigt, K.; Anishchenko, I.M.; Kutovenko, V.B.; Kava, L.; et al. The early terrestrial fungal lineage of Conidiobolus-transition from saprotroph to parasitic lifestyle. J. Fungi 2022, 8, 789. [Google Scholar] [CrossRef] [PubMed]
- Boguś, M.I.; Kazek, M.; Drozdowski, M.; Kaczmarek, A.; Wrońska, A.K. The entomopathogenic fungus Conidiobolus coronatus has similar effects on the cuticular free fatty acid profile of sensitive and resistant insects. Insects 2023, 14, 895. [Google Scholar] [CrossRef] [PubMed]
- Torres Acosta, R.I.; Humber, R.A.; Sánchez-Peña, S.R. Zoophthora radicans (Entomophthorales), a fungal pathogen of Bagrada hilaris and Bactericera cockerelli (Hemiptera: Pentatomidae and Triozidae): Prevalence, pathogenicity, and interplay of environmental influence, morphology, and sequence data on fungal identification. J. Invertebr. Pathol. 2016, 139, 82–91. [Google Scholar] [CrossRef]
- De Fine Licht, H.H.; Hajek, A.E.; Eilenberg, J.; Jensen, A.B. Utilizing genomics to study entomopathogenicity in the fungal phylum Entomophthoromycota: A review of current genetic resources. Adv. Genet. 2016, 94, 41–65. [Google Scholar]
- St Leger, R.J. The evolution of complex Metarhizium-insect-plant interactions. Fungal Biol. 2024, 128, 2513–2528. [Google Scholar] [CrossRef]
- Stajich, J.E.; Lovett, B.; Lee, E.; Macias, A.M.; Hajek, A.E.; de Bivort, B.L.; Kasson, M.T.; De Fine Licht, H.H.; Elya, C. Signatures of transposon-mediated genome inflation, host specialization, and photoentrainment in Entomophthora muscae and allied entomophthoralean fungi. eLife 2024, 12, RP92863. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, Y.; Yu, D.; Xie, X.; Qin, L.; Yang, Y.; Huang, B. Genome-wide study of saprotrophy-related genes in the basal fungus Conidiobolus heterosporus. Appl. Microbiol. Biotechnol. 2020, 104, 6261–6272. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, T.; Yu, W.; Wang, X.; Zhou, X.; Zhou, X. Genome-wide study of conidiation-related genes in the aphid-obligate fungal pathogen Conidiobolus obscurus (Entomophthoromycotina). J. Fungi 2022, 8, 389. [Google Scholar] [CrossRef]
- Chen, E.C.H.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Krüger, M.; et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018, 220, 1161–1171. [Google Scholar] [CrossRef]
- Hane, J.K.; Anderson, J.P.; Williams, A.H.; Sperschneider, J.; Singh, K.B. Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 2014, 10, e1004281. [Google Scholar] [CrossRef] [PubMed]
- Lang, D.; Zhang, S.; Ren, P.; Liang, F.; Sun, Z.; Meng, G.; Tan, Y.; Li, X.; Lai, Q.; Han, L.; et al. Comparison of the two up-to-date sequencing technologies for genome assembly: HiFi reads of Pacific Biosciences Sequel II system and ultralong reads of Oxford Nanopore. Gigascience 2020, 9, giaa123. [Google Scholar] [CrossRef] [PubMed]
- Congrains, C.; Bremer, F.; Dupuis, J.R.; Barr, N.B.; Garzón-Orduña, I.J.; Rubinoff, D.; Doorenweerd, C.; Jose, M.S.; Morris, K.; Kauwe, A.; et al. CCS-Consensuser: A haplotype-aware consensus generator for PacBio amplicon sequences. Mol. Ecol. Resour. 2025, 4, e14113. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wessler, S.R. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010, 38, e199. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef]
- Edgar, R.C.; Myers, E.W. PILER: Identification and classification of genomic repeats. Bioinformatics 2005, 21, i152–i158. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017, 45, D604–D610. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Sirim, D.; Wagner, F.; Lisitsa, A.; Pleiss, J. The cytochrome P450 engineering database: Integration of biochemical properties. BMC Biochem. 2009, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Lara-Martínez, D.; Tristán-Flores, F.E.; Cervantes-Montelongo, J.A.; Silva-Martínez, G.A. Fungal stress responses and the importance of GPCRs. J. Fungi 2025, 11, 213. [Google Scholar] [CrossRef]
- Vilela, R.; Mendoza, L. Human pathogenic entomophthorales. Clin. Microbiol. Rev. 2018, 31, e00014–e00018. [Google Scholar] [CrossRef]
- Hong, S.; Shang, J.; Sun, Y.; Tang, G.; Wang, C. Fungal infection of insects: Molecular insights and prospects. Trends Microbiol. 2024, 32, 302–316. [Google Scholar] [CrossRef]
- Zhou, X.; Montalva, C.; Arismendi, N.; Hong, F. Neozygites linanensis sp. nov. a fungal pathogen infecting bamboo aphids in southeast China. Mycotaxon 2017, 132, 305–315. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Guo, K.; Zhang, X.; Lin, H.; Montalva, C. Transcriptomic insight into pathogenicity-associated factors of Conidiobolus obscurus, an obligate aphid-pathogenic fungus belonging to Entomopthoromycota. Pest. Manag. Sci. 2018, 74, 1677–1686. [Google Scholar] [CrossRef]
- Xue, C.; Hsueh, Y.P.; Heitman, J. Magnificent seven: Roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 2008, 32, 1010–1032. [Google Scholar] [CrossRef]
- Ying, S.H.; Feng, M.G.; Keyhani, N.O. A carbon responsive G-protein coupled receptor modulates broad developmental and genetic networks in the entomopathogenic fungus, Beauveria bassiana. Environ. Microbiol. 2013, 15, 2902–2921. [Google Scholar] [CrossRef]
- Hu, X.; Xiao, G.; Zheng, P.; Shang, Y.; Su, Y.; Zhang, X.; Liu, X.; Zhan, S.; St Leger, R.J.; Wang, C. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, 16796–16801. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, S.; Sugiura, R.; Lu, Y.; Maeda, T.; Kawagishi, K.; Yokoyama, M.; Tohda, H.; Giga-Hama, Y.; Shuntoh, H.; Kuno, T. Zinc finger protein Prz1 regulates Ca2+ but not Cl− homeostasis in fission yeast. Identification of distinct branches of calcineurin signaling pathway in fission yeast. J. Biol. Chem. 2003, 278, 18078–18084. [Google Scholar] [CrossRef]
- Braun, B.R.; Head, W.S.; Wang, M.X.; Johnson, A.D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 2000, 156, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Finkel, J.S.; Xu, W.; Huang, D.; Hill, E.M.; Desai, J.V.; Woolford, C.A.; Nett, J.E.; Taff, H.; Norice, C.T.; Andes, D.R.; et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012, 8, e1002525. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Bürckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000, 19, 2569–2579. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.F.; Zhang, L.H.; Zhou, X. Long noncoding RNAs are potentially involved in the degeneration of virulence in an aphid-obligate pathogen, Conidiobolus obscurus (Entomophthoromycotina). Virulence 2021, 21, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Włóka, E.; Boguś, M.I.; Wrońska, A.K.; Drozdowski, M.; Kaczmarek, A.; Sobich, J.; Gołębiowski, M. Insect cuticular compounds affect Conidiobolus coronatus (Entomopthorales) sporulation and the activity of enzymes involved in fungal infection. Sci. Rep. 2022, 12, 13641. [Google Scholar] [CrossRef]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterase. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Gao, B.J.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. Subtilisin-like Pr1 proteases marking the evolution of pathogenicity in a wide-spectrum insect-pathogenic fungus. Virulence 2020, 11, 365–380. [Google Scholar] [CrossRef]
- Ngwenya, M.L.; Chen, W.; Basson, A.K.; Shandu, J.S.; Yu, J.H.; Nelson, D.R.; Syed, K. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. 2018, 19, 1711. [Google Scholar] [CrossRef]
- Seo, S.Y.; Sharma, V.K.; Sharma, N. Mushroom tyrosinase: Recent prospects. Agric. Food Chem. 2003, 51, 2837–2853. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhang, Y.; Guo, H.; Gan, P.; Cai, M.; Kang, Z.; Cheng, Y. Identification and functional analysis of CAP genes from the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. J. Fungi 2023, 9, 734. [Google Scholar] [CrossRef]
- Boguś, M.I.; Wieloch, W.; Ligęza-Żuber, M. Coronatin-2 from the entomopathogenic fungus Conidiobolus coronatus kills Galleria mellonella larvae and incapacitates hemocytes. B. Entomol. Res. 2017, 107, 66–76. [Google Scholar] [CrossRef]
- Wieloch, W.; Boguś, M.I.; Ligęza, M.; Koszela-Piotrowska, I.; Szewczyk, A. Coronatin-1 isolated from entomopathogenic fungus Conidiobolus coronatus kills Galleria mellonella hemocytes in vitro and forms potassium channels in planar lipid membrane. Toxicon 2011, 58, 369–379. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Wang, J.; Zhou, X. Characterization of a cytolytic-like gene from the aphid-obligate fungal pathogen Conidiobolus obscurus. J. Invertebr. Pathol. 2020, 173, 107366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.H.; Yang, T.; Siu, X.; Zhang, X.Q.; Zhou, X. Debilitation of Galleria mellonella hemocytes using CytCo a cytolytic-like protein derived from the entomopathogen Conidiobolus obscurus. Pestic. Biochem. Phys. 2023, 193, 105418. [Google Scholar] [CrossRef]
- Liu, X.; Liu, X.; Chen, S.; Chen, Y.; Su, X.; Zhang, X.; Guo, K.; Zhou, X. Calcium leakage involved in nematotoxic effects of the Conidiobolus obscurus CytCo protein on the pine wood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 2024, 80, 6366–6374. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, Y.; Zhang, L.; Yang, T.; Zhou, X. Transcriptomic profiling of bean aphid Megoura crassicauda upon exposure to the aphid-obligate entomopathogen Conidiobolus obscurus (Entomophthoromycotina) and screening of CytCo-binding aphid proteins through a pull-down assay. Insects 2024, 15, 388. [Google Scholar] [CrossRef]
- Wrońska, A.K.; Boguś, M.I. Harman and norharman, metabolites of the entomopathogenic fungus Conidiobolus coronatus (Entomophthorales), affect the serotonin levels and phagocytic activity of hemocytes, insect immunocompetent cells, in Galleria mellonella (Lepidoptera). Cell Biosci. 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed]
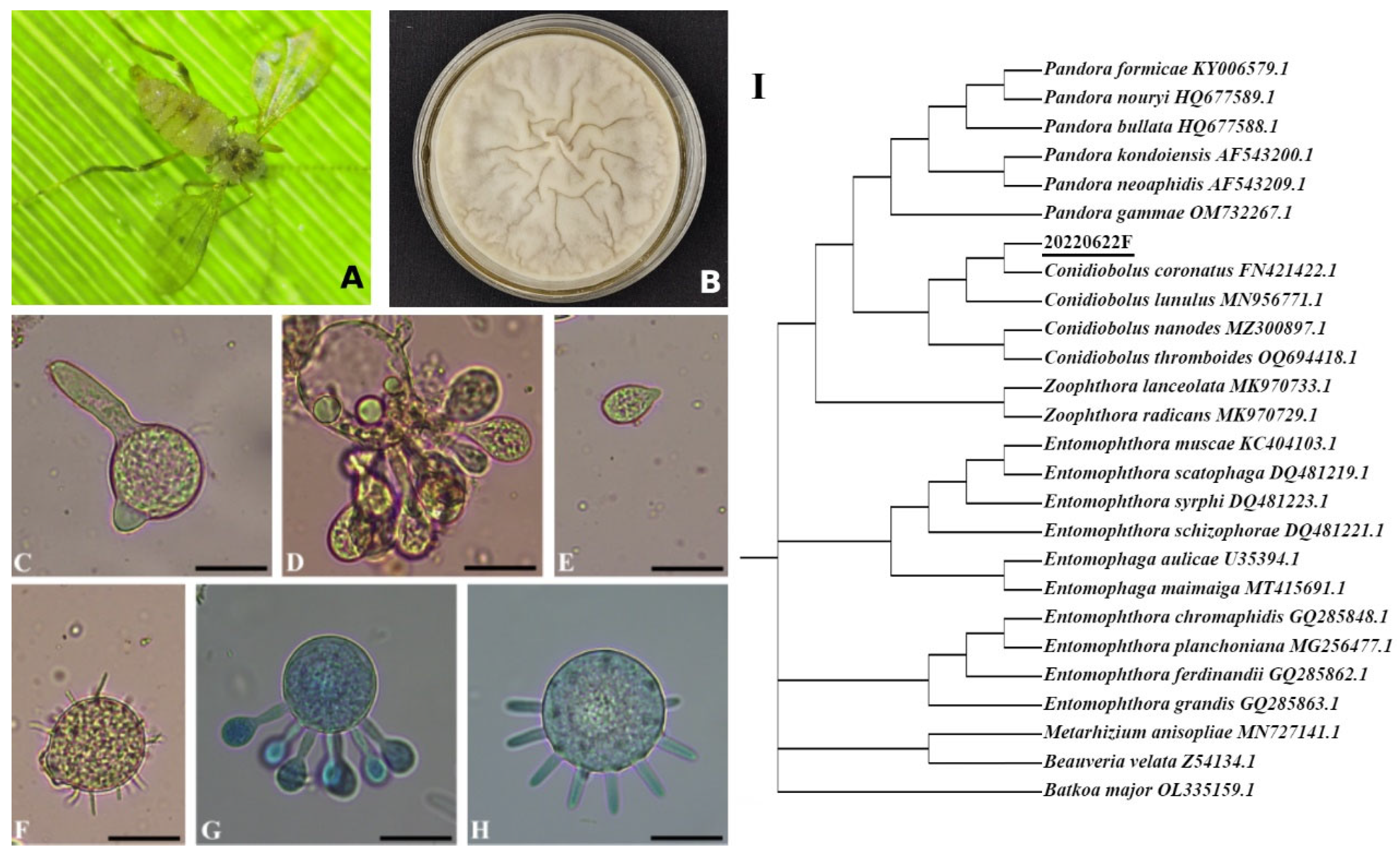

| Species | C. coronatus |
|---|---|
| Genome assembly (Mb) | 44.21 |
| Coverage (×) | 82.44 |
| Scaffold number | 77 |
| Scaffold N50 length (bp) | 1,415,773 |
| Scaffold N90 length (bp) | 755,109 |
| G + C content (%) | 27.65 |
| Protein-encoding genes | 11,128 |
| Average number of exons per gene | 2.85 |
| Repeat sequences (bp) | 7,745,087 |
| rRNA, family number | 294, 4 |
| tRNA, family number | 1294, 46 |
| Other ncRNA, family number | 43, 17 |
| Species [Ref.] | C. coronatus 20220622F | C. coronatus NRRL28638 [16] | C. heterosporus RCEF6331 [17] | C. obscurus ARSEF 7217 [18] | E. muscae ARSEF 13514 [16] |
|---|---|---|---|---|---|
| Genome size (Mb) | 44.21 | 39.9 | 33.4 | 37.6 | 1030 |
| Coverage (×) | 82.44 | 16.31 | 466 | 318 | 83.12 |
| Complete fungal BUSCOs | 93.45% | ~80% | / | 80.0% | 81.3% |
| Scaffolds | 77 | 1050 | 96 | 167 | 7810 |
| Scaffold N50 (bp) | 1,415,773 | 102,410 | 1,660,467 | 1,104,530 | 329,600 |
| Scaffold N90 (bp) | 755,109 | 19,422 | 86,181 | 75,751 | 79,400 |
| G + C content (%) | 27.65 | 27.5 | 36.88 | 26.46 | 41.03 |
| Repeat sequence (%) | 17.52 | / | 12.9 | 17.44 | 90.9% |
| Protein-coding genes | 11,128 | 10,568 | 10,857 | 10,262 | 38,917 |
| Compound Name | Annotation (LIPID MAPS/KEGG) | Relative Abundance | Log2(FC) | |
|---|---|---|---|---|
| C. coronatus | C. obscurus | |||
| Limonene-1,2-diol | C10 isoprenoids (monoterpenes) (PR0102) | 658.1 | 286.9 | 1.20 |
| (1S,4R)-1-Hydroxy-2-oxolimonene | 390.1 | 271.5 | 0.52 | |
| 6,10-Dimethyl-9-methylene-undec-5E-en-2-one | C15 isoprenoids (sesquiterpenes) (PR0103) | 881.7 | 1.1 | 9.71 |
| 5-(1-Oxopropan-2-yl)isolongifol-5-ene | 17,817.5 | 320.6 | 5.80 | |
| 5-(1-Hydroxypropan-2-yl)isolongifol-5-ene | 281.6 | 8.3 | 5.09 | |
| Dihydrophaseic acid | 1613.9 | 215.5 | 2.90 | |
| (2-trans,6-trans)-Farnesol | 1753.0 | 427.3 | 2.04 | |
| Gibberellin A8-catabolite | C20 isoprenoids (diterpenes) (PR0104) | 3087.4 | 61.9 | 5.64 |
| (-)-Reiswigin A | 1965.6 | 144.1 | 3.77 | |
| Gibberellin A34-catabolite | 2408.0 | 1187.8 | 1.02 | |
| (-)-2,7-Dolabelladiene-6beta,10alpha,18-triol | 12,478.5 | 6731.4 | 0.89 | |
| Gibberellin A36 | 32,494.9 | 26,459.1 | 0.30 | |
| Astaxanthin | C40 isoprenoids (tetraterpenes) (PR0107) | 2557.2 | 9.9 | 8.02 |
| Adonixanthin | 2844.9 | 110.6 | 4.69 | |
| 1′-Hydroxytorulene | 30,533.4 | 3964.0 | 2.95 | |
| Octaprenyl diphosphate | 3803.3 | 1120.3 | 1.76 | |
| Neurosporaxanthin | 1082.4 | 438.6 | 1.30 | |
| Echinenone | 260.3 | 132.9 | 0.97 | |
| Thiothece-474 | 22,722.8 | 12,189.4 | 0.90 | |
| Canthaxanthin | 841.7 | 499.2 | 0.75 | |
| Terpendole K | Indole diterpene alkaloid biosynthesis (ko00403) | 177.3 | 50.1 | 1.82 |
| Terpendole G | 2762.2 | 1725.8 | 0.68 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, F.; Yang, T.; Zhang, L.; Yang, J.; Chen, X.; Zhou, X. Genomic Insights into the Molecular Basis of Broad Host Adaptability of the Entomopathogenic Fungus Conidiobolus coronatus (Entomophthoromycotina). J. Fungi 2025, 11, 600. https://doi.org/10.3390/jof11080600
Bai F, Yang T, Zhang L, Yang J, Chen X, Zhou X. Genomic Insights into the Molecular Basis of Broad Host Adaptability of the Entomopathogenic Fungus Conidiobolus coronatus (Entomophthoromycotina). Journal of Fungi. 2025; 11(8):600. https://doi.org/10.3390/jof11080600
Chicago/Turabian StyleBai, Fan, Tian Yang, Lvhao Zhang, Jiaqi Yang, Xinyu Chen, and Xiang Zhou. 2025. "Genomic Insights into the Molecular Basis of Broad Host Adaptability of the Entomopathogenic Fungus Conidiobolus coronatus (Entomophthoromycotina)" Journal of Fungi 11, no. 8: 600. https://doi.org/10.3390/jof11080600
APA StyleBai, F., Yang, T., Zhang, L., Yang, J., Chen, X., & Zhou, X. (2025). Genomic Insights into the Molecular Basis of Broad Host Adaptability of the Entomopathogenic Fungus Conidiobolus coronatus (Entomophthoromycotina). Journal of Fungi, 11(8), 600. https://doi.org/10.3390/jof11080600






