Solving the Enigma of the Identity of Laccaria laccata
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphology
2.2. Molecular Analysis
2.3. Sequence Alignment, Dataset Assembly and Phylogenetic Analysis
2.4. Estimation of the Distribution of Laccaria laccata Using the nrITS Region
3. Results
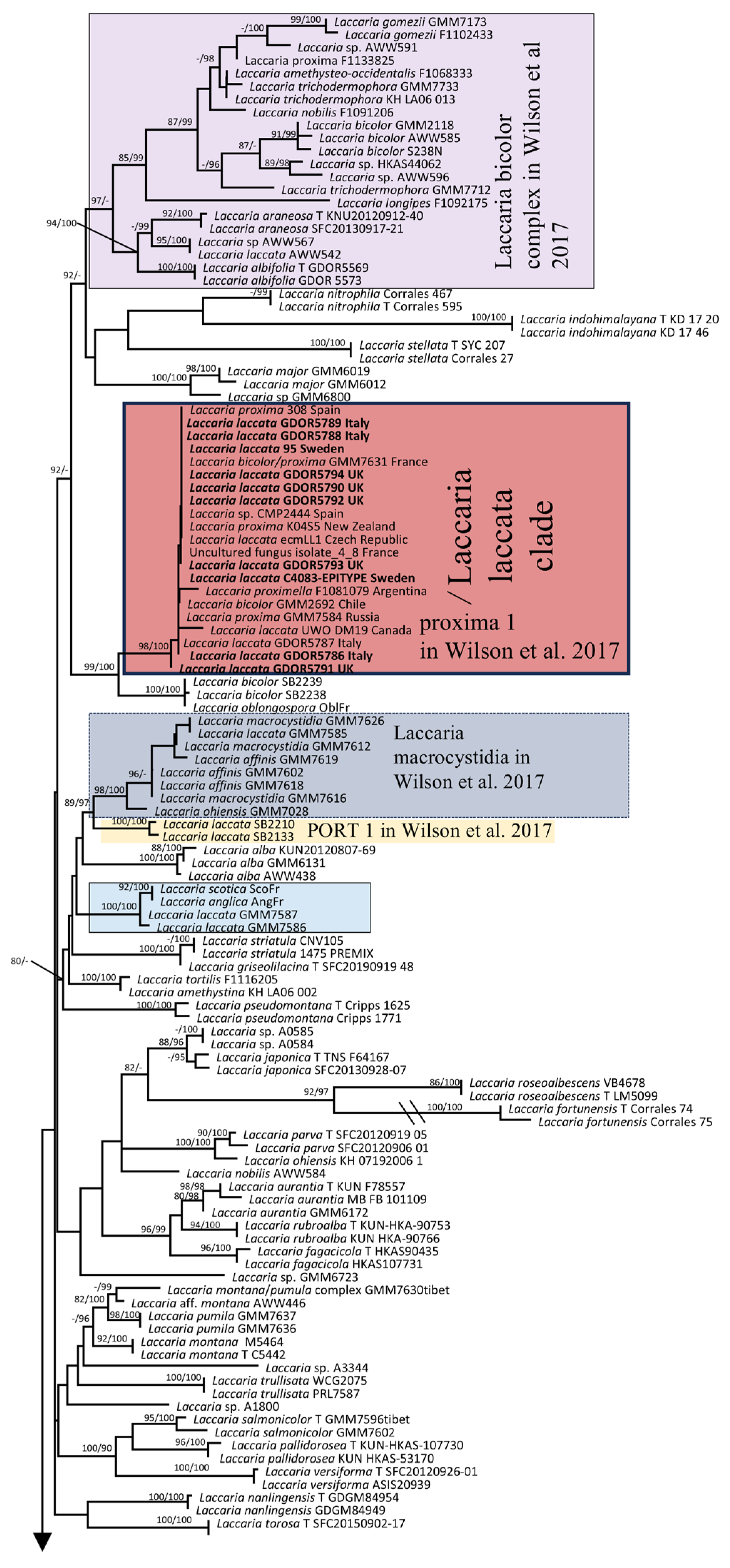
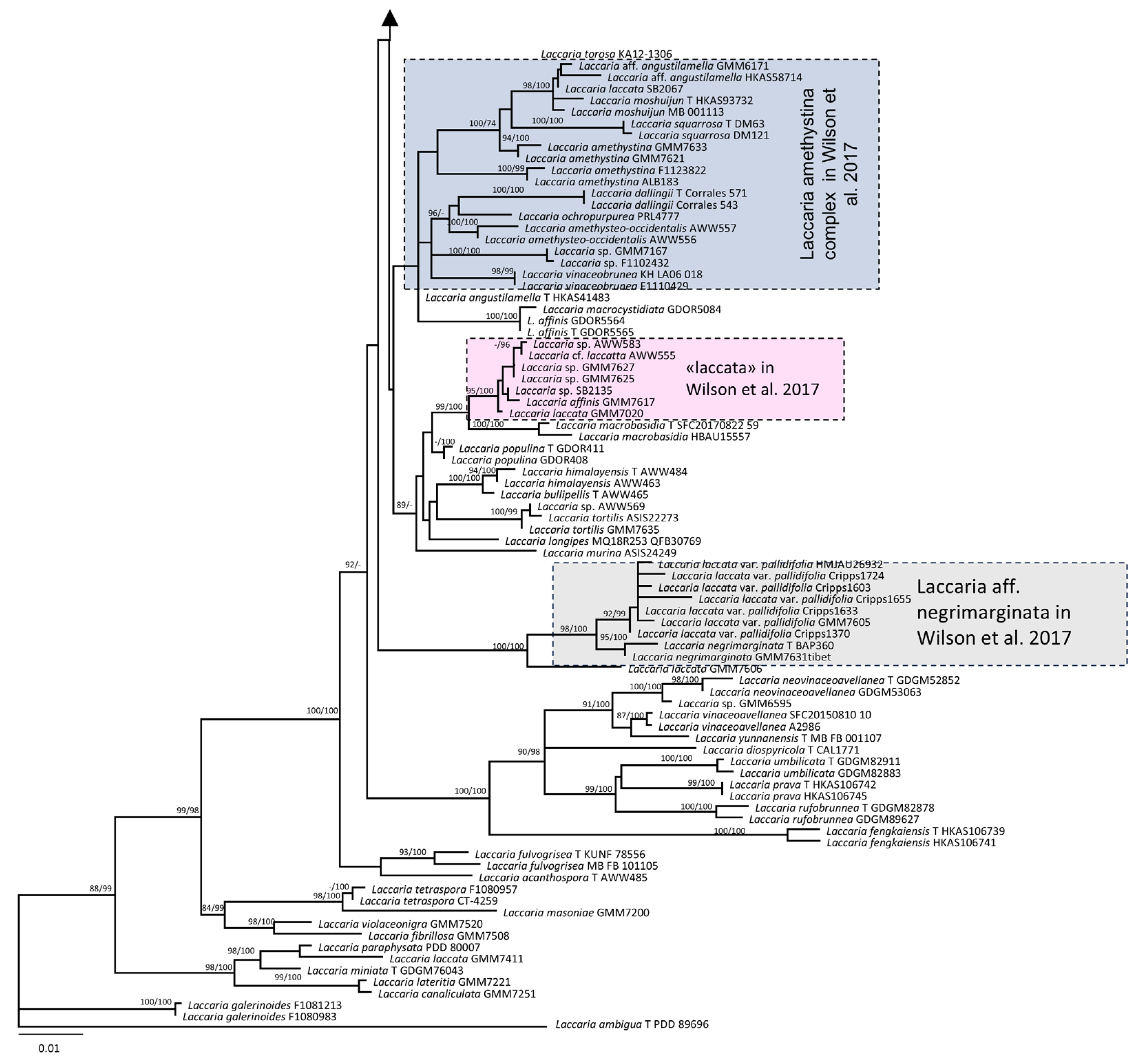
3.1. Molecular and Phylogeny
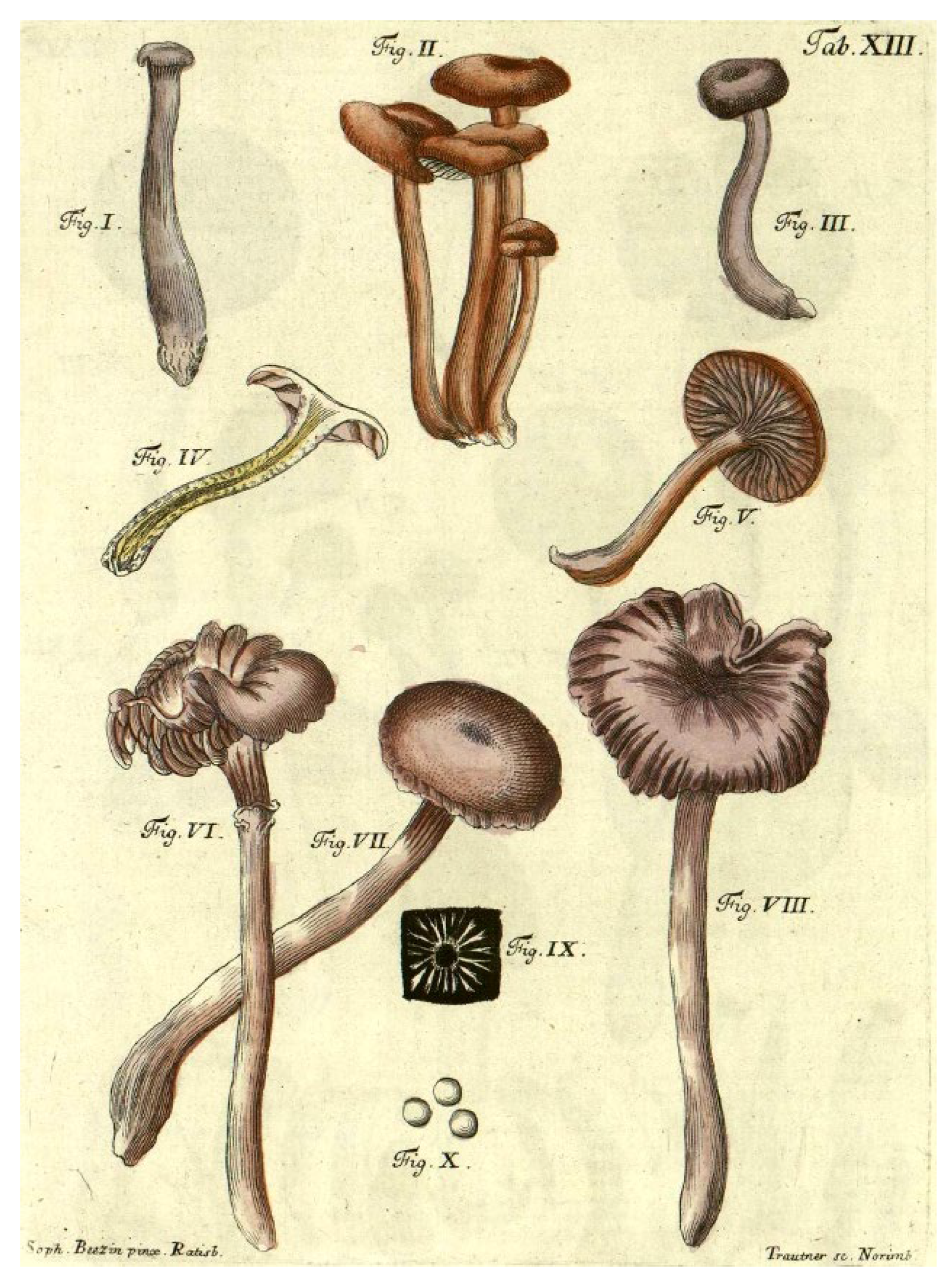
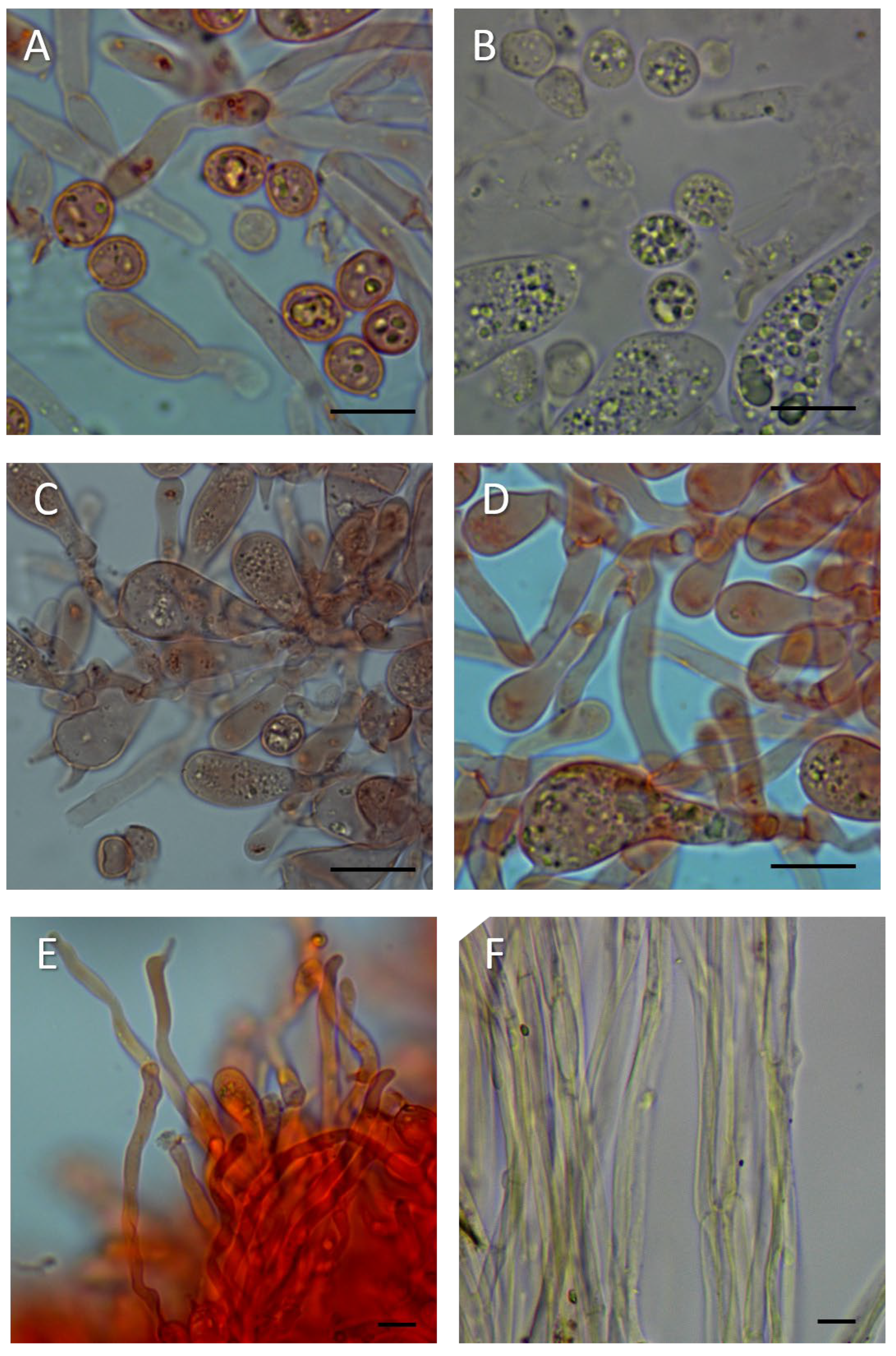
3.2. Morphology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, A.W.; Hosaka, K.; Mueller, G. Evolution of ectomycorrhizas as a driver of diversification and biogeographic patterns in the model mycorrhizal mushroom genus Laccaria. New Phytol. 2017, 213, 1862–1873. [Google Scholar] [CrossRef]
- Wilson, A.W.; May, T.W.; Mueller, G.M. Biogeography of the ectomycorrhizal mushroom genus Laccaria. In Biogeography of Mycorrhizal Symbiosis; Ecological Studies (Analysis and Synthesis); Tedersoo, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 230, pp. 273–297. [Google Scholar] [CrossRef]
- Mueller, G.M. Systematics of Laccaria (Agaricales) in the continental United States and Canada, with discussions on extralimital taxa and descriptions of extant types. Fieldiana Bot. 1992, 30, 1–158. [Google Scholar]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2010; ISBN 978-0-08-055934-6. [Google Scholar]
- Branzanti, M.B.; Rocca, E.; Pisi, A. Effect of ectomycorrhizal fungi on chestnut ink disease. Mycorrhiza 1999, 9, 103–109. [Google Scholar] [CrossRef]
- Singer, R.; Moser, M. Forest Mycology and forest communities in South America. Mycopathol. Mycol. Appl. 1965, 26, 129–191. [Google Scholar] [CrossRef]
- Landeweert, R.; Hoffland, E.; Finlay, R.D.; Kuyper, T.W.; Van Breemen, N. Linking Plants to Rocks: Ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol. Evol. 2001, 16, 248–254. [Google Scholar] [CrossRef]
- Martin, F.; Aerts, A.; Ahrén, D.; Brun, A.; Danchin, E.; Duchaussoy, F.; Gibon, J.; Kohler, A.; Lindquist, E.; Pereda, V. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 2008, 452, 88–92. [Google Scholar] [CrossRef] [PubMed]
- He, X.L.; Peng, W.H.; Wang, D. An Illustration of Important Wild Edible Fungi in Sichuan; Science Press: Beijing, China, 2021; pp. 1–206. [Google Scholar]
- Wu, F.; Zhou, L.-W.; Yang, Z.-L.; Bau, T.; Li, T.-H.; Dai, Y.-C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- He, M.Q.; Zhao, R.L.; Hyde, K.D.; Begerow, D.; Kemler, M.; Yurkov, A.; McKenzie, E.H.C.; Raspé, O.; Kakishima, M.; Sánchez- Ramírez, S.; et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019, 99, 105–367. [Google Scholar] [CrossRef]
- Scopoli, G.A. Flora Carniolica Tom. 2.; Ioannis Pauli Krauss, Bibliopolae Vindobonensis: Vienna, Austria, 1772; pp. 1–496. [Google Scholar]
- Fries, E. Systema Mycologicum; Lundae: Ex Officina Berlingiana: Uppsala, Sweden, 1821; Volume 1. [Google Scholar]
- Schaeffer, J.C. Fungorum Qui in Bavaria et Palatinatu Circa Ratisbonam Nascuntur Icones; Typis Zunkelianis: Regensburg, Germany, 1762. [Google Scholar]
- Persoon, C.H. Commentarius D. Jac. Christ. Schaefferi … Fungorum Bavariae Indigenorum Icones Pictas Differentiis Specificis, Synonymis et Observationibus Selectis Illustrans; Ioannem Iacobum Palmium: Erlangae, Germany, 1800; pp. 1–130. [Google Scholar]
- Singer, R. Notes sur le genre Laccaria. Bull. Trimest. Société Mycol. Fr. 1967, 83, 104–123. [Google Scholar]
- Lanjouw, J.; Baehni, C.; Robyns, W.; Ross, R.; Rousseau, J.; Schopf, J.M.; Schulze, G.M.; Smith, A.C.; Vilmorin, R.d.; Stafleu, F.A. International Code of Botanical Nomenclature: Adopted by the Ninth International Botanical Congress, Montreal, August 1959, Regnum Vegetabile 23 ed.; International Bureau for Plant Taxonomy and Nomenclature of the International Association for Plant Taxonomy: Utrecht, The Netherlands, 1961; 372p. [Google Scholar]
- Mueller, G.M.; Vellinga, E.C. Taxonomic and nomenclatural notes on Laccaria B. & Br. Laccaria amethystea, L. fraterna, L. laccata, L. pumila, and their synonyms. Persoonia 1986, 13, 27–43. [Google Scholar]
- Voss, E.G.; Burdet, H.M.; Chaloner, W.G.; Demoulin, V.; Hiepko, P.; McNeill, J.; Meikle, R.D.; Nicolson, D.H.; Rollins, R.C.; Silva, P.C.; et al. (Eds.) International Code of Botanical Nomenclature Adopted by the Thirteenth International Botanical Congress, Sydney, August 1981; Bohn, Scheltema & Holkema: Utrecht, The Netherlands, 1983; Volume 111. [Google Scholar]
- McNeill, J.; Barrie, F.; Buck, W.R.; Demoulin, V.; Greuter, W.; Hawksworth, D.J.; Herendeen, P.S.; Knapp, S.; Marhold, K.; Prado, J. (Eds.) International Code of Nomenclature for Algae, Fungi and Plants (Melbourne Code); Koeltz Scientific Books: Königstein, Germany, 2012; Volume 154. [Google Scholar]
- Parra, L.A.; Zamora, J.C.; Hawksworth, D.L.; Hibbett, D.S.; Kirk, P.M.; Lücking, R.; Rambold, G.; Bensch, K.; Yao, Y.J.; Robert, V.; et al. Proposals for Consideration at IMC11 to Modify Provisions Related Solely to Fungi in the International Code of Nomenclature for Algae, Fungi, and Plants. IMA Fungus 2018, 9, i–vii. [Google Scholar] [CrossRef]
- May, T.W.; Redhead, S.A.; Bensch, K.; Hawksworth, D.L.; Lendemer, J.; Lombard, L.; Turland, N.J. Chapter F of the International Code of Nomenclature for algae, fungi, and plants as approved by the 11th International Mycological Congress, San Juan, Puerto Rico, July 2018. IMA Fungus 2019, 10, 21. [Google Scholar] [CrossRef]
- Dovana, F.; Para, R.; Moreno, G.; Scali, E.; Garbelotto, M.; Lechner, B.E.; Forte, L. Description of the New Species Laccaria albifolia (Hydnangiaceae, Basidiomycota) and a Reassessment of Laccaria affinis Based on Morphological and Phylogenetic Analyses. J. Fungi 2025, 11, 11. [Google Scholar] [CrossRef]
- Moreno, G.; Castillo, A.; Thüs, H. Critical revision of type material of Stemonitales (Myxogastria) at the Natural History Museum London (BM). Phytotaxa 2018, 344, 149–159. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Eyre, C.A.; Hayden, K.M.; Dhillon, J.; Garbelotto, M.M. Back to basics: An evaluation of NaOH and alternative rapid DNA extraction protocols for DNA barcoding, genotyping, and disease diagnostics from fungal and oomycete samples. Mol. Ecol. Resour. 2013, 13, 66–74. [Google Scholar] [CrossRef]
- Del Visco, C.L.; Scali, E. A Modified CTAB Method for the SimultaneousExtraction of Host and Pathogen Total RNA of Inoculated Conifer Plants. Protocols.io. 2025. Available online: https://dx.doi.org/10.17504/protocols.io.rm7vz9mr5gx1/v1 (accessed on 1 May 2025).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based genome alignment and genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Mol. Phylogenet Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E.A. Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Sheedy, E.M.; Van de Wouw, A.P.; Howlett, B.J.; May, T.W. Multigene sequence data reveal morphologically cryptic phylogenetic species within the genus Laccaria in southern Australia. Mycologia 2013, 105, 547–563. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Cripps, C.L.; Mueller, G.M. Morphological and molecular systematics of Rocky Mountain alpine Laccaria. Mycologia 2005, 97, 949–972. [Google Scholar] [CrossRef]
- Walbert, K.; Ramsfield, T.D.; Ridgway, H.J.; Jones, E.E. Ectomycorrhizal species associated with Pinus radiata in New Zealand including novel associations determined by molecular analysis. Mycorrhiza 2010, 20, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Vincenot, L.; Nara, K.; Sthultz, C.; Labbé, J.; Dubois, M.P.; Tedersoo, L.; Martin, F.; Selosse, M.A. Extensive gene flow over Europe and possible speciation over Eurasia in the ectomycorrhizal basidiomycete Laccaria amethystina complex. Mol. Ecol. 2012, 21, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.W.; Hosaka, K.; Perry, B.A.; Mueller, G.M. Laccaria (Agaricomycetes, Basidiomycota) from Tibet (Xizang Autonomous Region, China). Mycoscience 2013, 54, 406–419. [Google Scholar] [CrossRef]
- Popa, F.; Rexer, K.H.; Donges, K.; Yang, Z.L.; Kost, G. Three new Laccaria species from Southwest China (Yunnan). Mycol. Prog. 2014, 13, 1105–1117. [Google Scholar] [CrossRef]
- Montoya, L.; Bandala, V.M.; Baroni, T.J.; Horton, T.R. A new species of Laccaria in montane cloud forest from eastern Mexico. Mycoscience 2015, 56, 597–605. [Google Scholar] [CrossRef]
- Wilson, A.W.; Wickett, N.J.; Grabowski, P.; Fant, J.; Borevitz, J.; Mueller, G.M. Examining the efficacy of a genotyping-by-sequencing technique for population genetic analysis of the mushroom Laccaria bicolor and evaluating whether a reference genome is necessary to assess homology. Mycologia 2015, 107, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ye, L.; Chen, J.; Karunarathna, S.C.; Xu, J.C.; Hyde, K.D.; Mortimer, P.E. Laccaria rubroalba sp. nov. (Hydnangiaceae, Agaricales) from Southwestern China. Phytotaxa 2016, 284, 041–050. [Google Scholar] [CrossRef]
- Popa, F.; Jimenéz, S.Y.C.; Weisenborn, J.; Donges, K.; Rexer, K.H.; Piepenbring, M. A new Laccaria species from cloud forest of Fortuna, Panama. Mycol. Prog. 2016, 15, 19. [Google Scholar] [CrossRef]
- Ramos, A.; Bandala, V.M.; Montoya, L. A new species and a new record of Laccaria (Fungi, Basidiomycota) found in a relict forest of the endangered Fagus grandifolia var. mexicana. MycoKeys 2017, 27, 77–94. [Google Scholar] [CrossRef]
- Vincenot, L.; Popa, F.; Laso, F.; Donges, K.; Rexer, K.H.; Kost, G.; Yang, Z.L.; Nara, K.; Selosse, M.A. Out of Asia: Biogeography of fungal populations reveals Asian origin of diversification of the Laccaria amethystina complex, and two new species of violet Laccaria. Fungal Biol. 2017, 121, 939–955. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Park, M.S.; Lee, H.; Oh, S.Y.; Wilson, A.W.; Mueller, G.M.; Lim, Y.W. A systematic revision of the ectomycorrhizal genus Laccaria from Korea. Mycologia 2018, 110, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Hay, C.R.J.; Thorn, R.G.; Jacobs, C.R. Taxonomic survey of Agaricomycetes (Fungi: Basidiomycota) in Ontario tallgrass prairies determined by fruiting body and soil rDNA sampling. Can. Field Nat. 2019, 132, 407–424. [Google Scholar] [CrossRef]
- Wang, X.H.; Das, K.; Bera, I.; Chen, Y.H.; Bhatt, R.P.; Ghosh, A.; Hembrom, M.E.; Hofdtetter, V.; Parihar, A.; Vizzini, A.; et al. Fungal Biodiversity Profiles 81–90. Cryptogam. Mycol. 2019, 40, 57–95. [Google Scholar] [CrossRef]
- Cho, H.J.; Lee, H.; Park, M.S.; Park, K.H.; Park, J.H.; Cho, Y.; Kim, C.; Lim, Y.W. Two new species of Laccaria (Agaricales, Basidiomycota) from Korea. Mycobiology 2020, 48, 1786961. [Google Scholar] [CrossRef]
- Corrales, A.; Wilson, A.W.; Mueller, G.M.; Ovrebo, C. Novel Laccaria Species From Juglandaceae Forest in Panama With Notes on Their Ecology. Front. Microbiol. 2020, 11, 1597. [Google Scholar] [CrossRef]
- Li, F. Two new species of Laccaria from South China, with a note on Hodophilus glaberipes. Mycol. Prog. 2020, 19, 525–539. [Google Scholar] [CrossRef]
- Crous, P.W.; Osieck, E.R.; Jurjevi, Ž.; Boers, J.; Van Iperen, A.L.; Starink-Willemse, M.; Dima, B.; Balashov, S.; Bulgakov, T.S.; Johnston, P.R.; et al. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178–374. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.; Jones, G.E.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef]
- Cui, Y.Y.; Cai, Q.; Li, J.; Yang, Z.L. Two new Laccaria species from China based on molecular and morphological evidence. Mycol. Pro. 2021, 20, 567–576. [Google Scholar] [CrossRef]
- Dovana, F.; Moreno, G.; Para, R.; Lavorato, C.; Mucciarelli, M. Phylogenetic reappraisal and epitypification of Laccaria macrocystidiata (Hydnangiaceae, Basidiomycota). Phytotaxa 2021, 514, 129–139. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, X.L.; Mu, L.Q.; Deng, W.Q. Morphology and Molecular Phylogeny Reveal Five New Species of Laccaria (Hydnangiaceae, Agaricales) from Southern China. J. Fungi 2023, 9, 1179. [Google Scholar] [CrossRef] [PubMed]
- Armada, F.; Bellanger, J.M.; Moreau, P.-A. Champignons de la Zone Alpine; Fédération Mycologique et Botanique Dauphiné-Savoie: Annemasse, France, 2023; pp. 1–376. [Google Scholar]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.F.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Le Vinh, S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Bon, M. Tricholomataceae of France and Western Europe (Leucopaxilloideae); Éditions Bordas: Paris, France, 1978; pp. 1–456. [Google Scholar]
- Bon, M. Tricholomataceae de France et d’Europe occidentale; Clé Monogr. Doc. Mycol. 1983, 51, 1–51. [Google Scholar]
- Ballero, M.; Contu, M. Tassonomia ed ecologia del genere Laccaria Berk. & Br. (Basidiomycetes, Agaricales, Tricholomataceae) in Sardegna. Candollea 1987, 42, 601–611. [Google Scholar]
- Breitenbach, J.; Kränzlin, F. Champignons de Suisse, Tome 3. ed.; Mykologia: Lucerne, Switzerland, 1991; pp. 1–364. [Google Scholar]
- Pázmány, D. Zur Systematik der Gattung Laccaria Bk. et Br. Z. Mykol. 1994, 60, 5–12. [Google Scholar]
- Vellinga, E.C. Laccaria . In Flora Agaricina Neerlandica, Critical Monograph on the Families of Agarics and Boleti Occuring in The Netherlends; Tricholomataceae, Bas, C., Kuyper, T.W., Noordeloos, M.E., Vellinga, E.C., Eds.; A.A. Balkema: Rotterdam, The Netherlands, 1995; Volume 3, pp. 1–183. [Google Scholar]
- Consiglio, G.; Papetti, C. Photographic Atlas of the Mushrooms of Italy; Centro Studi Micologici “A.M.B.”: Vicenza, Italy, 2001; Volume 2, pp. 1–450. [Google Scholar]
- Contu, M. Il Genere Laccaria (Basidiomycotina, Agaricales) in Italia, con Note sulle Rimanenti Specie in Europa. Boll. Gr. Micol. Bresadola 2003, 46, 5–58. [Google Scholar]
- Vesterholt, J. Laccaria. In Funga Nordica, 1st ed.; Knudsen, H., Vesterholt, J., Eds.; Nordisk Forlag: Copenhagen, Denmark, 2009. [Google Scholar]
- Schaeffer, J.C.; Persoon, C.H. Fungorum Qui in Bavaria et Palatinatu Circa Ratisbonam Nascuntur Icons; Tom, I., Ed.; editio nova; Ioannem Iacobum Palmium: Erlangae, Germany, 1800; pp. 1–443. [Google Scholar]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dovana, F.; Scali, E.; Lopez Del Visco, C.; Moreno, G.; Para, R.; Lechner, B.E.; Garbelotto, M.; May, T.W. Solving the Enigma of the Identity of Laccaria laccata. J. Fungi 2025, 11, 575. https://doi.org/10.3390/jof11080575
Dovana F, Scali E, Lopez Del Visco C, Moreno G, Para R, Lechner BE, Garbelotto M, May TW. Solving the Enigma of the Identity of Laccaria laccata. Journal of Fungi. 2025; 11(8):575. https://doi.org/10.3390/jof11080575
Chicago/Turabian StyleDovana, Francesco, Edoardo Scali, Clarissa Lopez Del Visco, Gabriel Moreno, Roberto Para, Bernardo Ernesto Lechner, Matteo Garbelotto, and Tom W. May. 2025. "Solving the Enigma of the Identity of Laccaria laccata" Journal of Fungi 11, no. 8: 575. https://doi.org/10.3390/jof11080575
APA StyleDovana, F., Scali, E., Lopez Del Visco, C., Moreno, G., Para, R., Lechner, B. E., Garbelotto, M., & May, T. W. (2025). Solving the Enigma of the Identity of Laccaria laccata. Journal of Fungi, 11(8), 575. https://doi.org/10.3390/jof11080575






