Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Activation and Sequencing of Fungal Pathogens
2.2. Screening for Fungicide Sensitivity
2.3. Transcriptome Sequencing After Different Fungicide Treatments
2.4. Determination of Physiological Indicators
3. Results
3.1. Fungicide Sensitivity Screening and EC50 Calculation
3.2. Transcriptome Sequencing and Assembly
3.3. Identification and Analysis of Differentially Expressed Genes (DEGs)
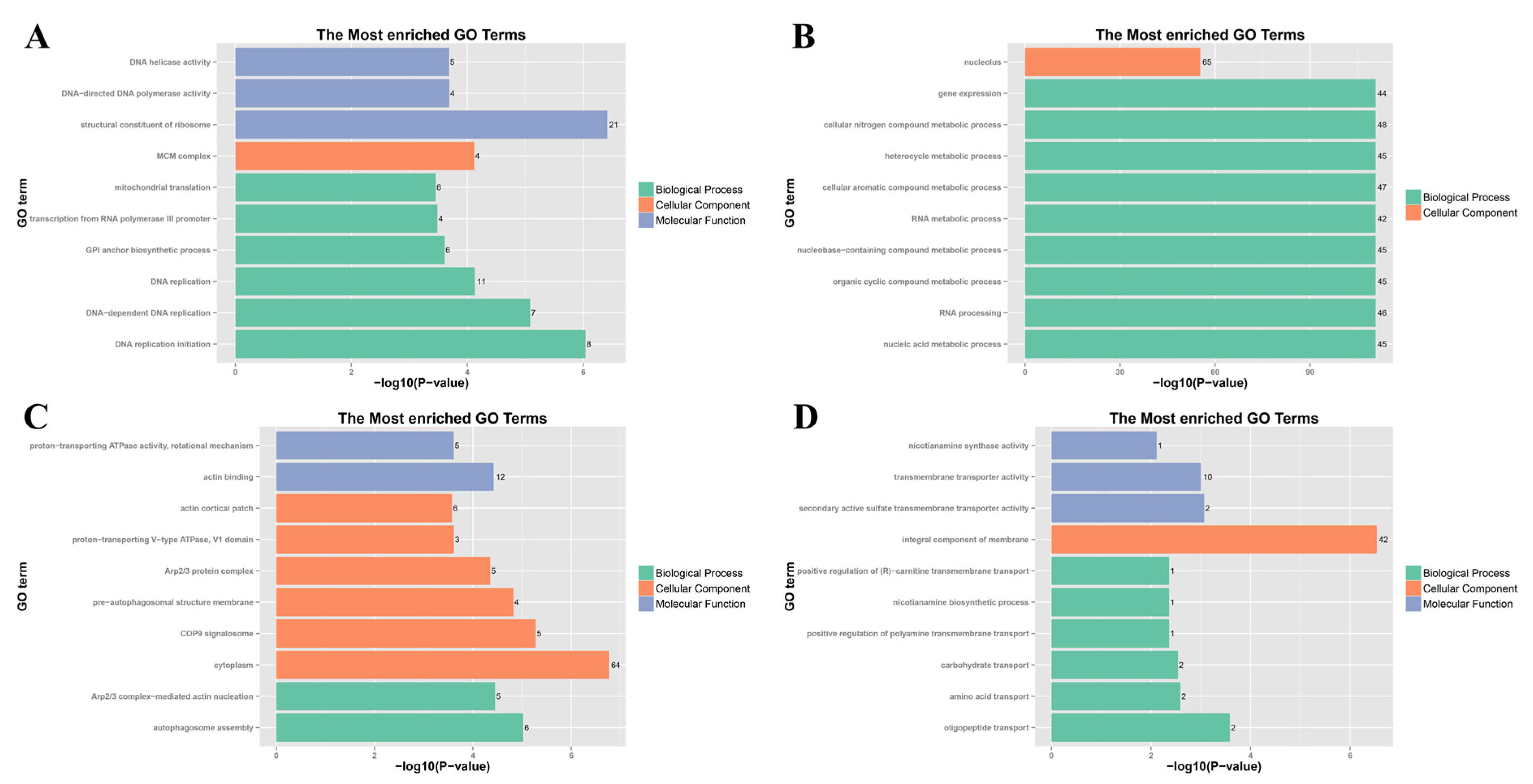
3.4. GO and KEGG Enrichment Analysis of DEGs
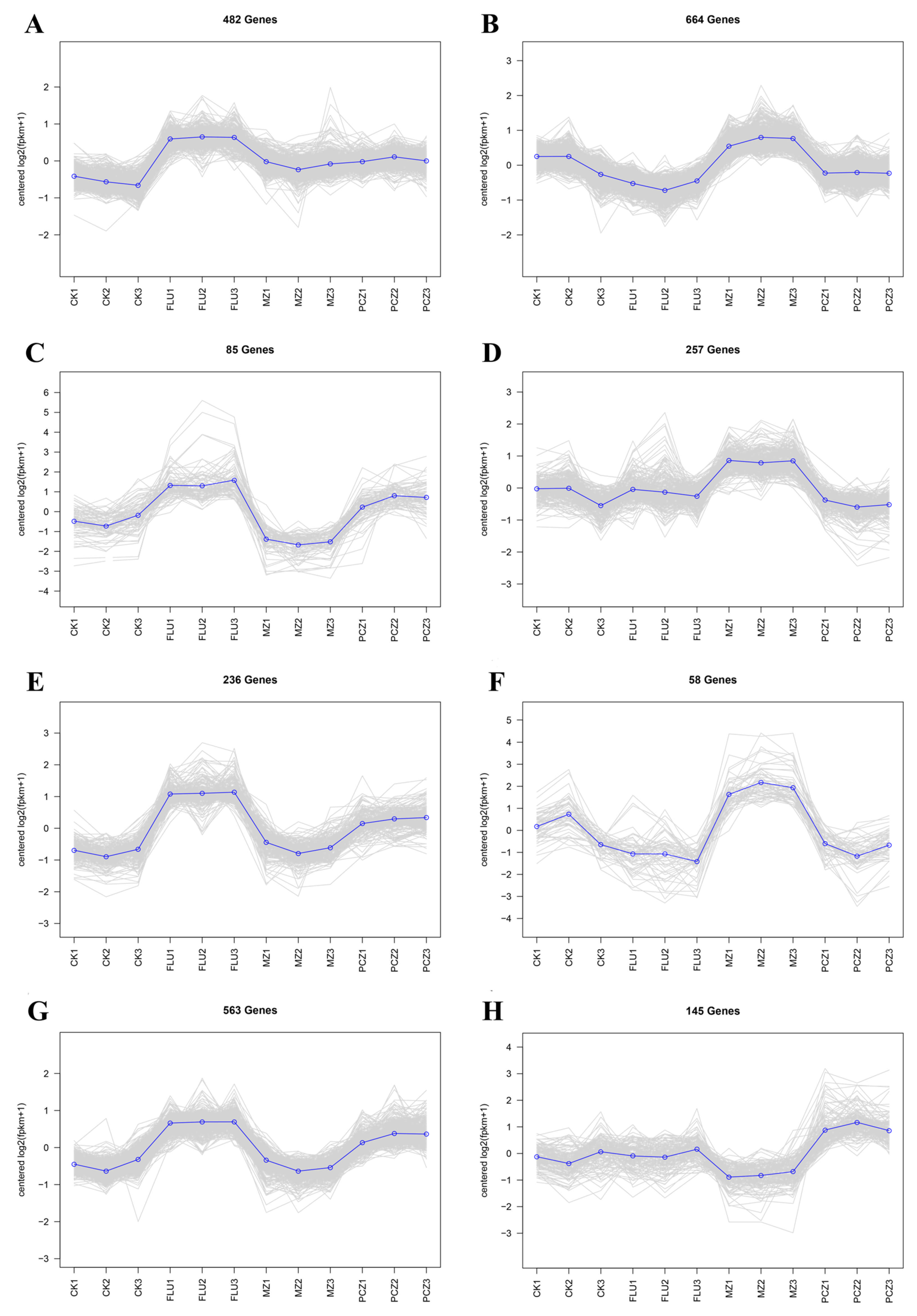
3.5. Collinear Trend Analysis Based on DEGs
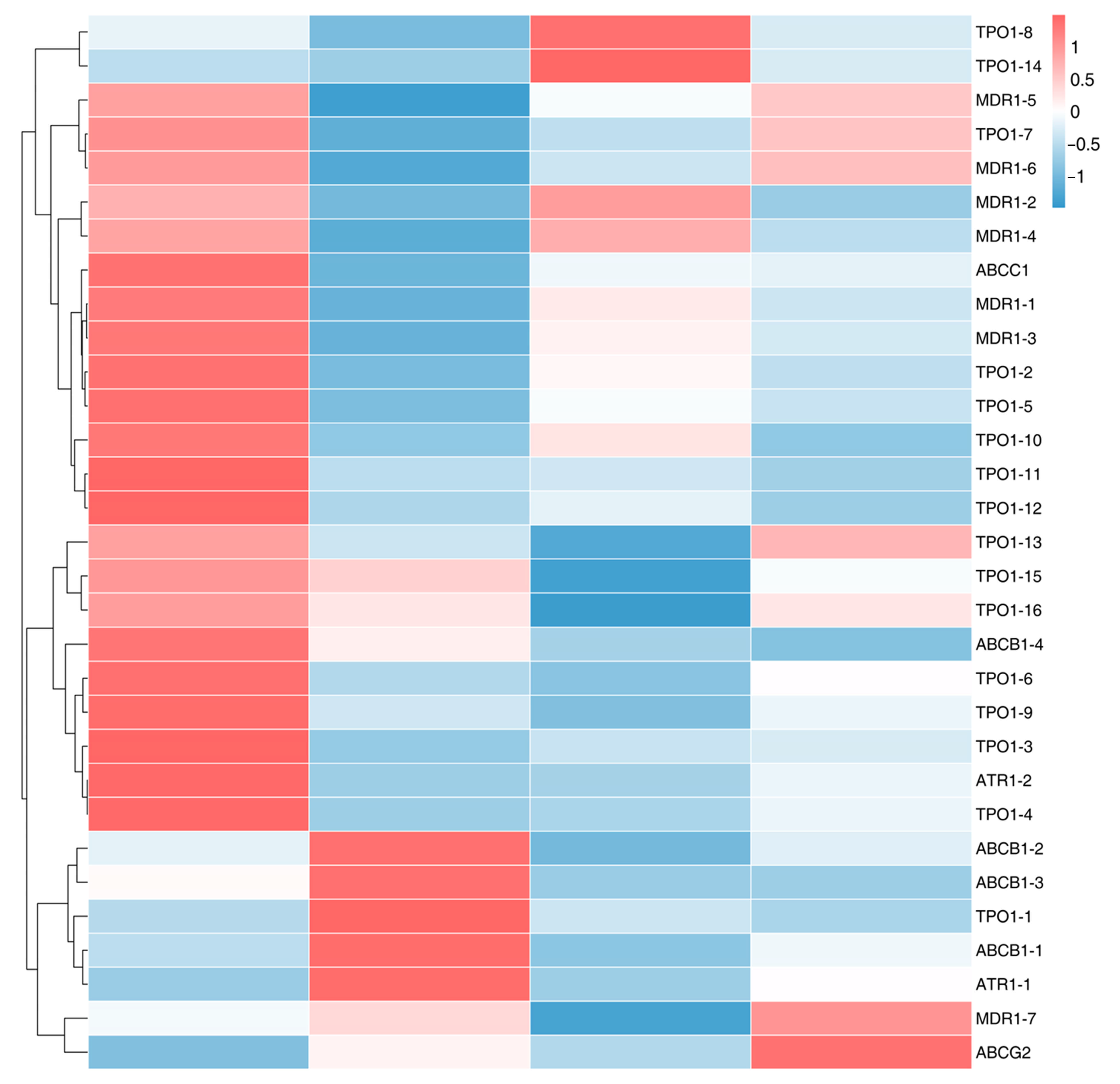
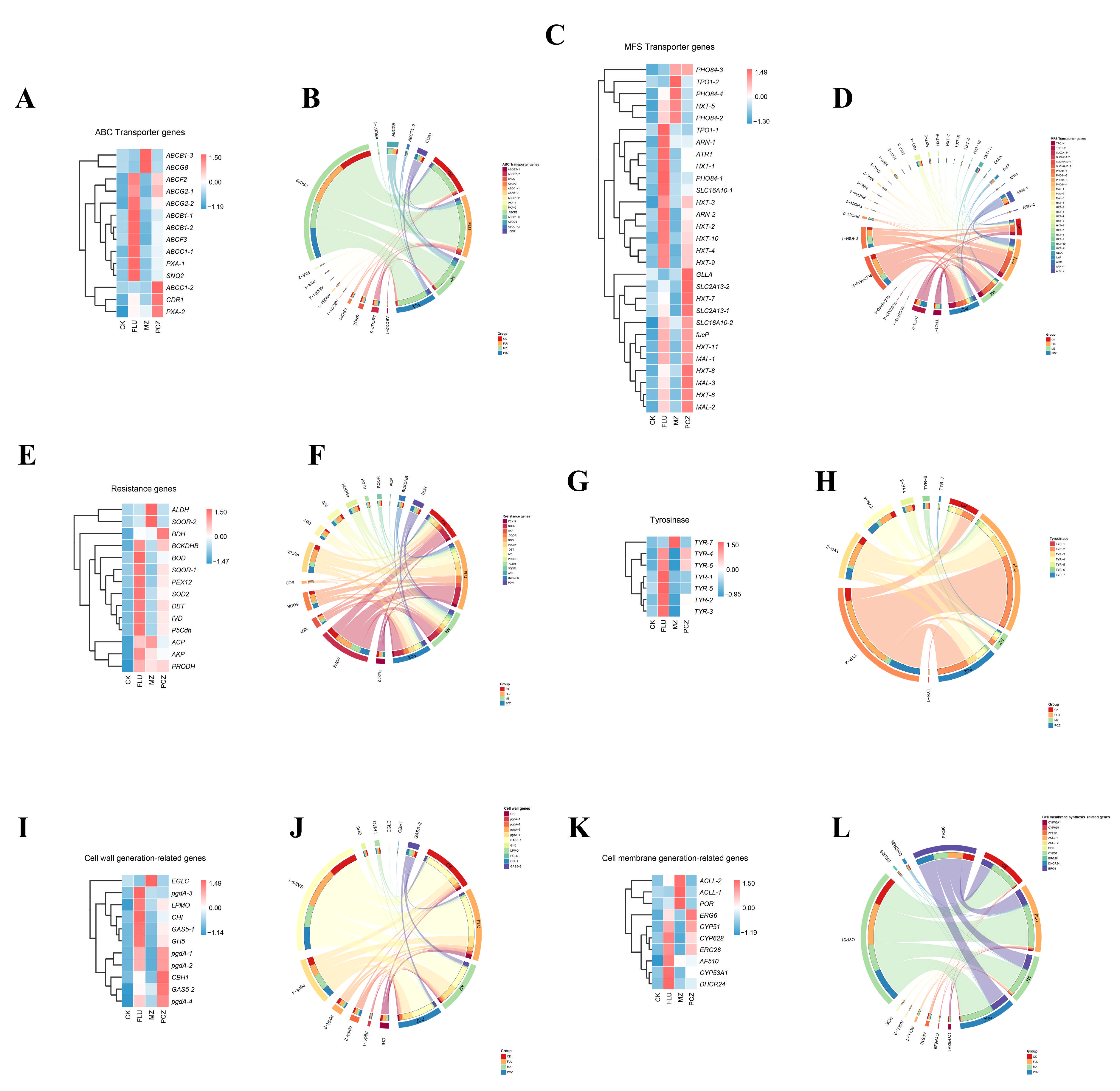
3.6. Analysis of Fungicide Resistance-Related Genes Based on DEGs
3.7. Analysis of Fungicide Resistance-Related Pathways Based on DEGs
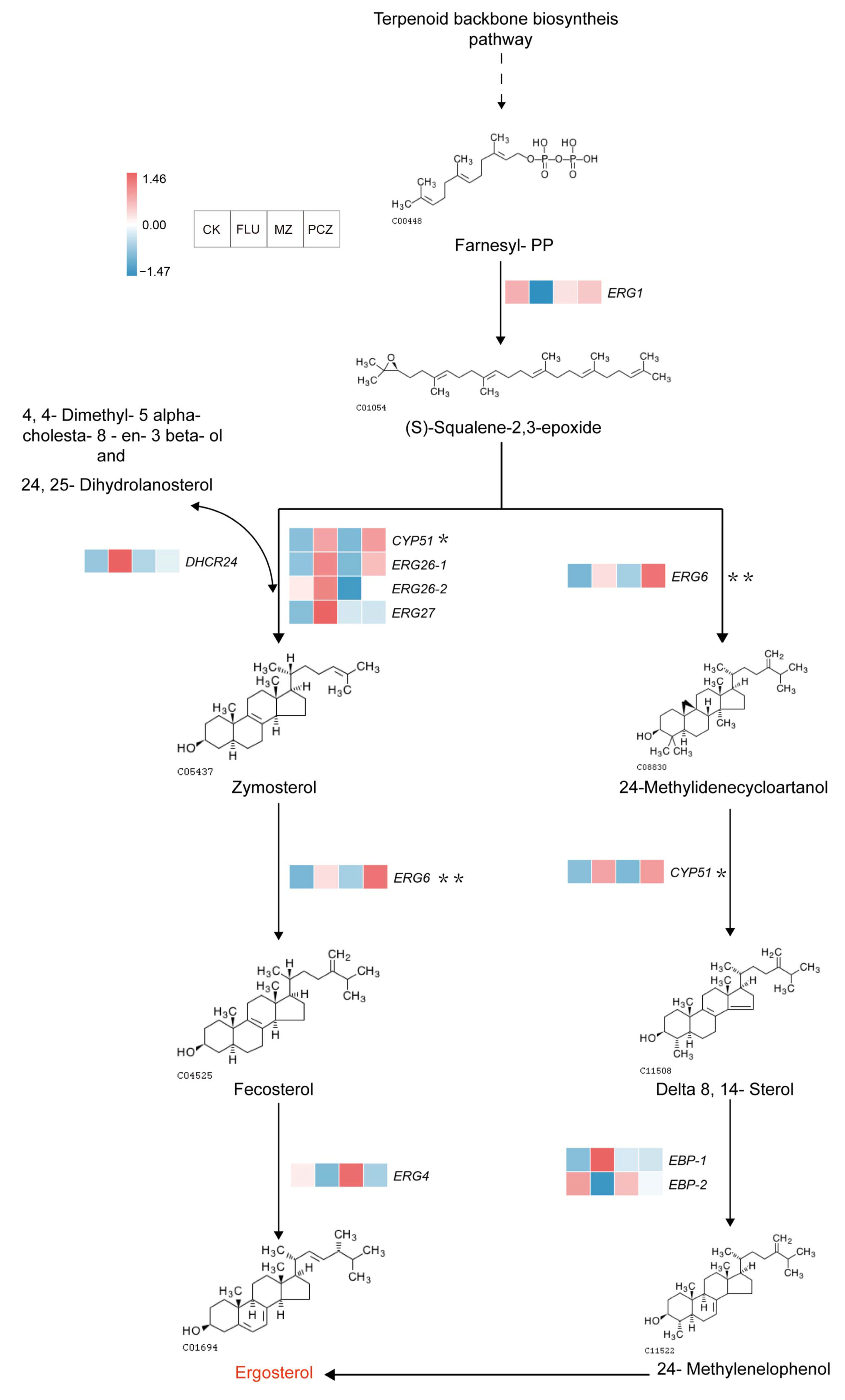
3.7.1. Ergosterol Biosynthesis Pathway
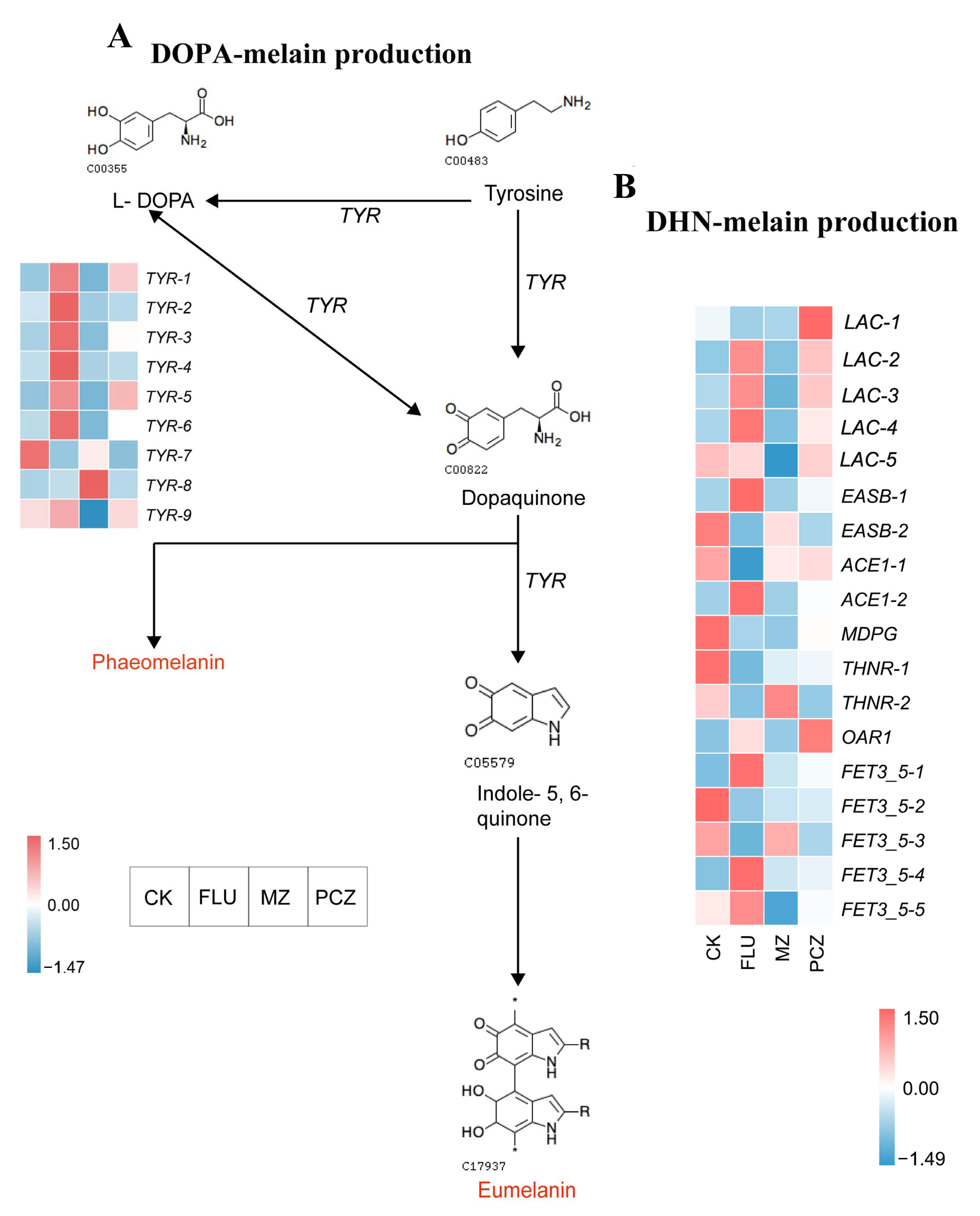
3.7.2. Melanin Biosynthesis Pathway
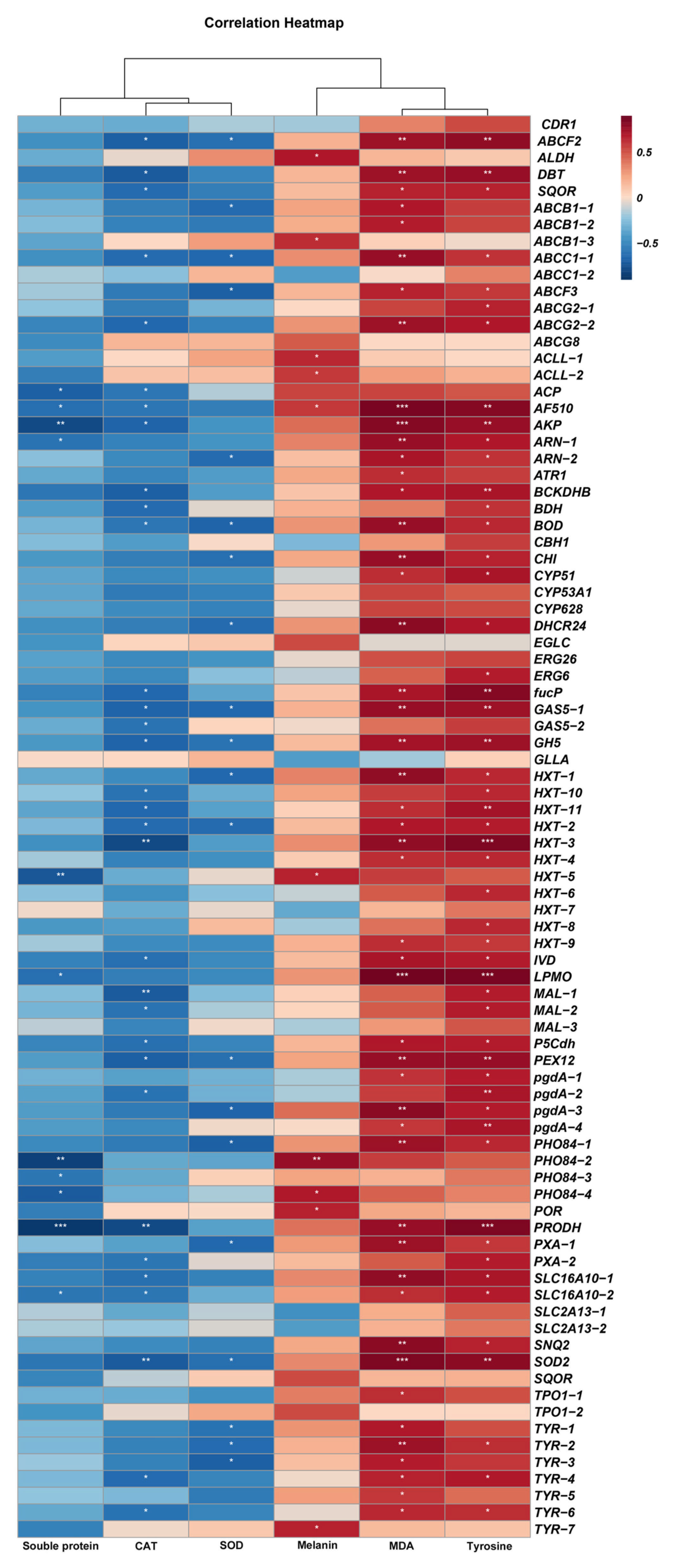
3.8. Correlation Between Some Physiological Indexes of A. alternata and Potential Fungicide Resistance-Related Genes
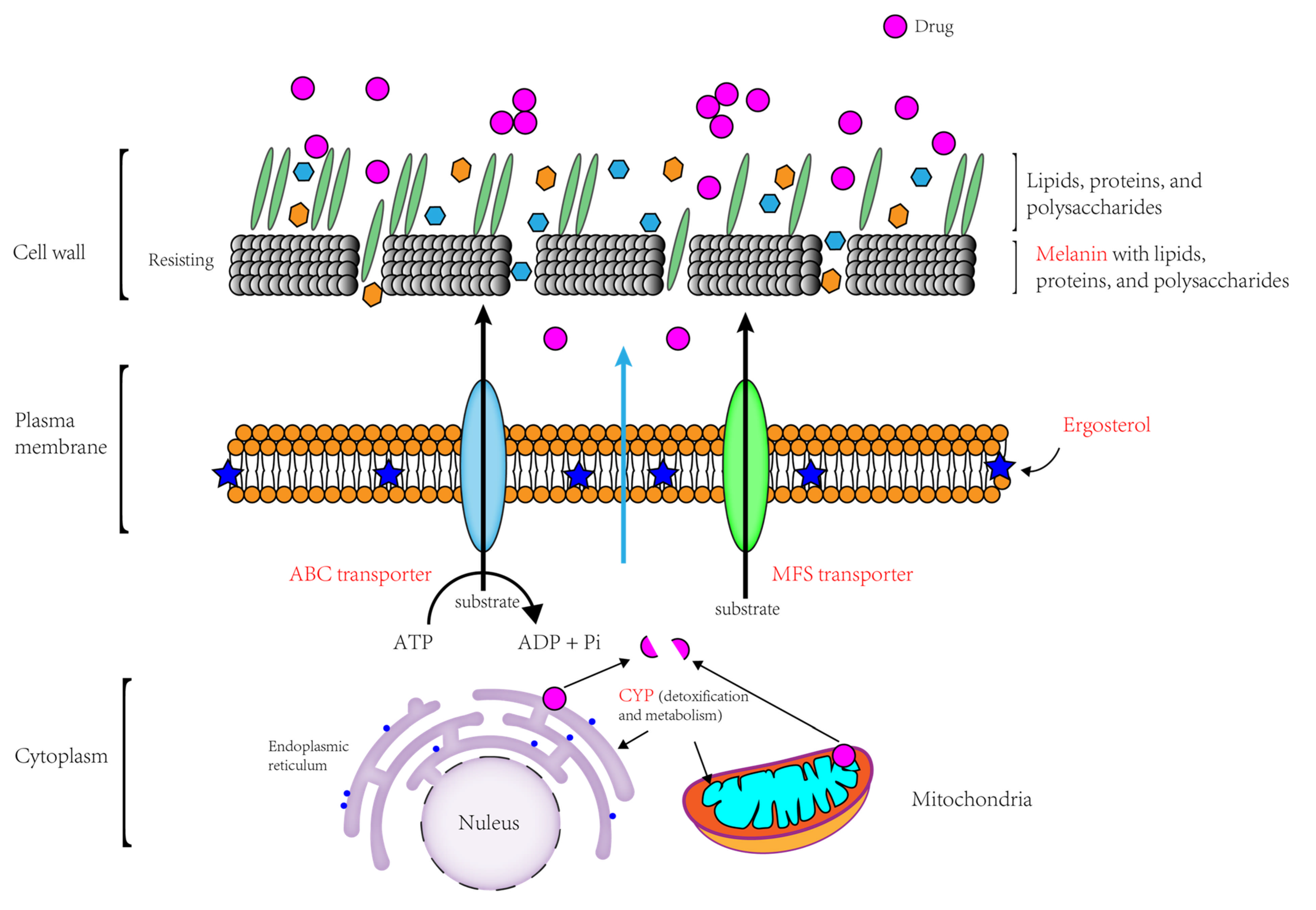
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FST | Fungicide sensitivity tests |
| MBC | Methyl benzimidazoles |
| DMIs | demethylation inhibitors |
| QoIs | Quinone outside Inhibitors |
| SDHI | Succinate dehydrogenase inhibitor |
| PPs | Phenylpyrroles |
| EC50 | 50% effective concentration |
| MDR | Multidrug resistance |
| ABC | ATP-binding cassette |
| MFS | Major facilitator superfamily |
| FLU | Fludioxonil |
| MZ | Mancozeb |
| PCZ | Prochloraz |
| PDA | Potato dextrose agar |
| FDR | False discovery rate |
| DEG | Differentially expressed gene |
| Log2FC | Log2 fold change |
| FPKM | Fragment Per Kilobase of exon model per Million mapped fragments |
| GO | Gene Ontology |
| CC | Cellular Component |
| MF | Molecular Function |
| FAD | Flavin adenine dinucleotide |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDA | Malondialdehyde |
| CAT | Catalase |
| SOD | Superoxide dismutase |
| MATE | Multidrug and toxic compound extrusion family |
| GST | Glutathione S-transferase |
| ROS | Reactive oxygen species |
Appendix A
| Fungicide Name and Active Ingredient | Functional Type | Physical Formulation | Manufacturer | Mode of Action Type | Primary Mechanism of Action |
|---|---|---|---|---|---|
| 50% Difenoconazole | Protective and Therapeutic | Suspension Concentrate | Shandong Libang Agrochemicals Co., Ltd., Qingzhou, China. | DMIs | Inhibits cytochrome P450-dependent 14α-demethylation, blocking ergosterol biosynthesis. |
| 450 g/L Prochloraz | Protective and Eradicant | Emulsifiable Concentrate | Shanghai Hulian Biochemical Co., Ltd., Shanghai, China. | DMIs | Inhibits ergosterol biosynthesis in fungi. |
| 40% Myclobutanil | Protective and Therapeutic | Suspension Concentrate | Jiangsu Yunnong Chemical Co., Ltd., Zhengjiang, China. | DMIs | Inhibits ergosterol biosynthesis in fungi. |
| 50% Carbendazim | Therapeutic | Wettable Powder | Leke Pesticide Co., Ltd., Yingkou, China. | Methyl Benzimidazole Carbamates (MBC) | Prevents cell division by inhibiting tubulin polymerization. |
| 50% Thiophanate-Methyl | Protective | Wettable Powder | Sichuan Runnong Technology Co., Ltd., Jianyang, China. | Methyl Benzimidazole MBC | After plant absorption, decomposes into MBC to disrupt mitotic spindle formation in fungal cells. |
| 75% Chlorothalonil | Protective | Wettable Powder | Chifeng Zhongnongda Biochemistry Technology Co., Ltd., Chifeng, China. | Multi-Site | Disrupts glyceraldehyde-3-phosphate dehydrogenase activity in cells. |
| 80% Mancozeb | Protective | Wettable Powder | Liming Chemical Co., Ltd., Xinyi, China. | Multi-Site | Inhibits pyruvate oxidation in fungi. |
| 80% Zineb | Protective | Wettable Powder | Shandong Xinxing Agrochemicals Co., Ltd., Qingzhou, China. | Multi-Site | Active ingredient oxidizes in water to isothiocyanate, inhibiting -SH group-containing enzymes in fungi. |
| 90% Phosethyl-Al | Therapeutic | Wettable Powder | Liming Chemical Co., Ltd., Xinyi, China. | Multi-Site | Effectively inhibits pyruvate oxidation in fungi. |
| 25 g/L Fludioxonil | Protective | Suspension Concentrate | Syngenta Nantong Crop Protection Co., Ltd., Nantong, China. | PPs | Inhibits glucose-phosphorylation-related transference. |
| 250 g/L Azoxystrobin | Therapeutic | Suspension Concentrate | Syngenta Nantong Crop Protection Co., Ltd., Nantong, China. | QoIs | Binds to the Qo site of the cytochrome bc1 complex, obstructing respiration and leading to fungal pathogen death. |
| 15% Triadimefon | Protective | Wettable Powder | Sichuan Guoguang Agrochemical Co., Ltd., Jianyang, China. | Conazoles | Inhibits ergosterol biosynthesis, thereby impeding appressorium and haustorium development, hyphal growth, and spore formation. |

| Fungicide Type | Fungicide Name | Fungicide Concentration | Inhibition Rate (%) | |||
|---|---|---|---|---|---|---|
| 2d | 3d | 4d | 5d | |||
| DMIs | Difenoconazole | 0.5 | 65.16 ± 1.53% i | 48.29 ± 0.79% i | 58.21 ± 0.12% i | 54.90 ± 0.61% k |
| 5.0 | 69.68 ± 1.15% g | 54.86 ± 0.27% h | 63.66 ± 0.12% h | 66.67 ± 0.25% i | ||
| 10.0 | 71.45 ± 0.58% f | 63.60 ± 0.31% f | 71.07 ± 0.21% f | 69.87 ± 0.20% h | ||
| 50.0 | 100 ± 0.00% a | 100 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| 100.0 | 100 ± 0.00% a | 100 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| Prochloraz | 0.5 | 67.30 ± 0.22% h | 63.69 ± 2.03% f | 63.66 ± 0.12% h | 65.88 ± 0.17% j | |
| 5.0 | 87.80 ± 0.21% d | 79.27 ± 0.79% d | 77.08 ± 0.53% e | 74.32 ± 0.34% f | ||
| 10.0 | 89.18 ± 0.95% c | 83.46 ± 0.53% c | 81.41 ± 1.16% d | 79.56 ± 0.00% d | ||
| 50.0 | 100 ± 0.00% a | 100 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| 100.0 | 100 ± 0.00% a | 100 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| Myclobutanil | 0.5 | 53.71 ± 0.33% j | 61.94 ± 0.53% g | 57.93 ± 0.32% i | 54.90 ± 0.17% k | |
| 5.0 | 69.31 ± 0.12% g | 66.76 ± 0.15% e | 69.67 ± 0.12% g | 71.85 ± 0.26% g | ||
| 10.0 | 75.00 ± 0.00% e | 78.84 ± 0.26% d | 76.59 ± 0.32% e | 74.94 ± 0.10% e | ||
| 50.0 | 91.82 ± 0.50% b | 91.51 ± 0.30% b | 92.38 ± 0.95% c | 86.43 ± 0.10% c | ||
| 100.0 | 100 ± 0.00% a | 100 ± 0.00% a | 97.55 ± 0.12% b | 96.45 ± 0.00% b | ||
| MBC | Carbendazim | 0.5 | 7.17 ± 0.38% g | 2.45 ± 0.15% f | 1.82 ± 0.12% g | 7.77 ± 0.17% ef |
| 5.0 | 9.94 ± 0.22% f | 6.56 ± 0.26% de | 8.04 ± 0.12% e | 10.19 ± 0.09% d | ||
| 10.0 | 12.83 ± 0.38% e | 8.66 ± 0.26% c | 11.60 ± 0.12% d | 16.95 ± 0.35% c | ||
| 50.0 | 22.39 ± 0.22% b | 10.94 ± 0.31% b | 14.05 ± 042% c | 22.30 ± 0.34% b | ||
| 100.0 | 27.42 ± 0.22% a | 15.58 ± 0.15% a | 15.79 ± 0.12% b | 24.32 ± 0.17% a | ||
| Thiophanate-Methyl | 0.5 | 4.53 ± 0.38% h | 5.69 ± 1.24% e | 2.10 ± 0.21% g | 2.42 ± 0.26% i | |
| 5.0 | 10.94 ± 0.76% ef | 7.44 ± 0.30% d | 3.98 ± 0.21% f | 4.00 ± 0.09% h | ||
| 10.0 | 17.86 ± 0.58% d | 11.37 ± 0.85% b | 13.56 ± 0.53% c | 5.41 ± 0.61% gh | ||
| 50.0 | 20.25 ± 0.58% c | 14.96 ± 0.26% a | 18.66 ± 0.00% a | 6.36 ± 2.03% fg | ||
| 100.0 | 25.91 ± 2.65% a | 15.83 ± 0.55% a | 18.73 ± 0.44% a | 9.18 ± 0.85% de | ||
| Multi-site | Chlorothalonil | 0.5 | 3.40 ± 0.38% n | 9.45 ± 0.26% n | 9.22 ± 0.21% n | 12.44 ± 0.35% k |
| 5.0 | 19.37 ± 0.21% k | 13.74 ± 1.6% l | 14.12 ± 0.53% l | 14.75 ± 0.68% j | ||
| 10.0 | 31.70 ± 1.51% g | 23.88 ± 0.69% i | 20.34 ± 0.55% j | 25.06 ± 1.70% g | ||
| 50.0 | 46.29 ± 0.95% d | 39.28 ± 0.80% e | 43.26 ± 0.12% c | 41.27 ± 0.68% c | ||
| 100.0 | 51.57 ± 1.53% c | 45.06 ± 0.66% c | 46.33 ± 0.63% b | 42.63 ± 0.35% b | ||
| Mancozeb | 0.5 | 3.77 ± 0.65% n | 17.41 ± 0.66% j | 20.96 ± 1.26% j | 8.61 ± 0.17% l | |
| 5.0 | 21.51 ± 0.76% j | 43.57 ± 0.26% d | 29.07 ± 0.95% f | 18.07 ± 0.29% i | ||
| 10.0 | 31.32 ± 0.38% g | 49.25 ± 0.15% b | 43.82 ± 0.21% c | 41.38 ± 1.03% bc | ||
| 50.0 | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| 100.0 | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| Zineb | 0.5 | 19.37 ± 0.58% k | 17.76 ± 0.15% j | 22.08 ± 0.12% i | 22.64 ± 0.17% h | |
| 5.0 | 21.26 ± 0.22% j | 27.56 ± 0.26% h | 24.95 ± 0.21% h | 25.00 ± 0.17% g | ||
| 10.0 | 25.66 ± 0.38% i | 30.54 ± 0.15% g | 26.97 ± 0.12% g | 26.97 ± 0.10% f | ||
| 50.0 | 35.85 ± 0.38% f | 31.76 ± 1.46% g | 30.40 ± 0.21% e | 36.38 ± 0.10% e | ||
| 100.0 | 59.75 ± 0.22% b | 43.83 ± 0.26% cd | 39.41 ± 0.42% d | 37.84 ± 0.17% d | ||
| Phosethy-Al | 0.5 | 11.83 ± 0.22% m | 9.19 ± 0.27% n | 2.52 ± 0.42% p | 0.56 ± 0.25% o | |
| 5.0 | 16.60 ± 0.38% l | 11.29 ± 0.27% m | 7.97 ± 0.42% o | 1.52 ± 1.27% o | ||
| 10.0 | 19.75 ± 0.43% k | 15.40 ± 0.40% k | 11.53 ± 0.21% m | 4.00 ± 0.09% n | ||
| 50.0 | 29.06 ± 0.38% h | 17.59 ± 0.27% j | 14.47 ± 0.21% l | 6.31 ± 0.10% m | ||
| 100.0 | 37.49 ± 0.44% e | 36.22 ± 0.53% f | 19.22 ± 0.32% k | 8.95 ± 0.68% l | ||
| PPs | Azoxystrobin | 0.5 | 66.54 ± 0.43% b | 59.84 ± 0.26% d | 56.81 ± 0.21% c | 48.99 ± 0.17% c |
| 5.0 | 100.00 ± 0.00% a | 94.84 ± 0.40% c | 94.69 ± 0.12% b | 92.17 ± 0.10% b | ||
| 10.0 | 100.00 ± 0.00% a | 96.98 ± 0.15% b | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| 50.0 | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| 100.0 | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | 100.00 ± 0.00% a | ||
| QoIs | Azoxystrobin | 0.5 | 35.47 ± 1.13% e | 27.30 ± 0.53% e | 27.81 ± 2.45% e | 15.48 ± 1.61% e |
| 5.0 | 39.50 ± 1.53% d | 34.73 ± 1.45% d | 32.77 ± 0.12% d | 17.34 ± 0.35% d | ||
| 10.0 | 42.26 ± 0.38% c | 38.06 ± 0.00% c | 34.94 ± 0.12% c | 23.14 ± 1.50% c | ||
| 50.0 | 48.30 ± 0.65% b | 40.07 ± 0.15% b | 40.18 ± 0.12% b | 25.40 ± 0.10% b | ||
| 100.0 | 52.08 ± 0.38% a | 41.30 ± 0.15% a | 42.00 ± 0.12% a | 39.42 ± 0.10% a | ||
| Conazoles | Triadimefon | 0.5 | 7.30 ± 0.22% e | 0.88 ± 0.15% e | 2.73 ± 0.21% e | 11.54 ± 0.52% e |
| 5.0 | 29.43 ± 0.38% d | 28.70 ± 0.30% d | 26.21 ± 0.21% d | 28.78 ± 0.10% d | ||
| 10.0 | 37.36 ± 0.38% c | 31.41 ± 0.40% c | 32.42 ± 0.12% c | 35.58 ± 0.10% c | ||
| 50.0 | 50.69 ± 1.16% b | 42.34 ± 0.40% b | 39.13 ± 0.74% b | 41.55 ± 0.17% b | ||
| 100.0 | 56.73 ± 2.79% a | 51.53 ± 0.16% a | 43.75 ± 0.24% a | 45.67 ± 0.10% a 1 | ||
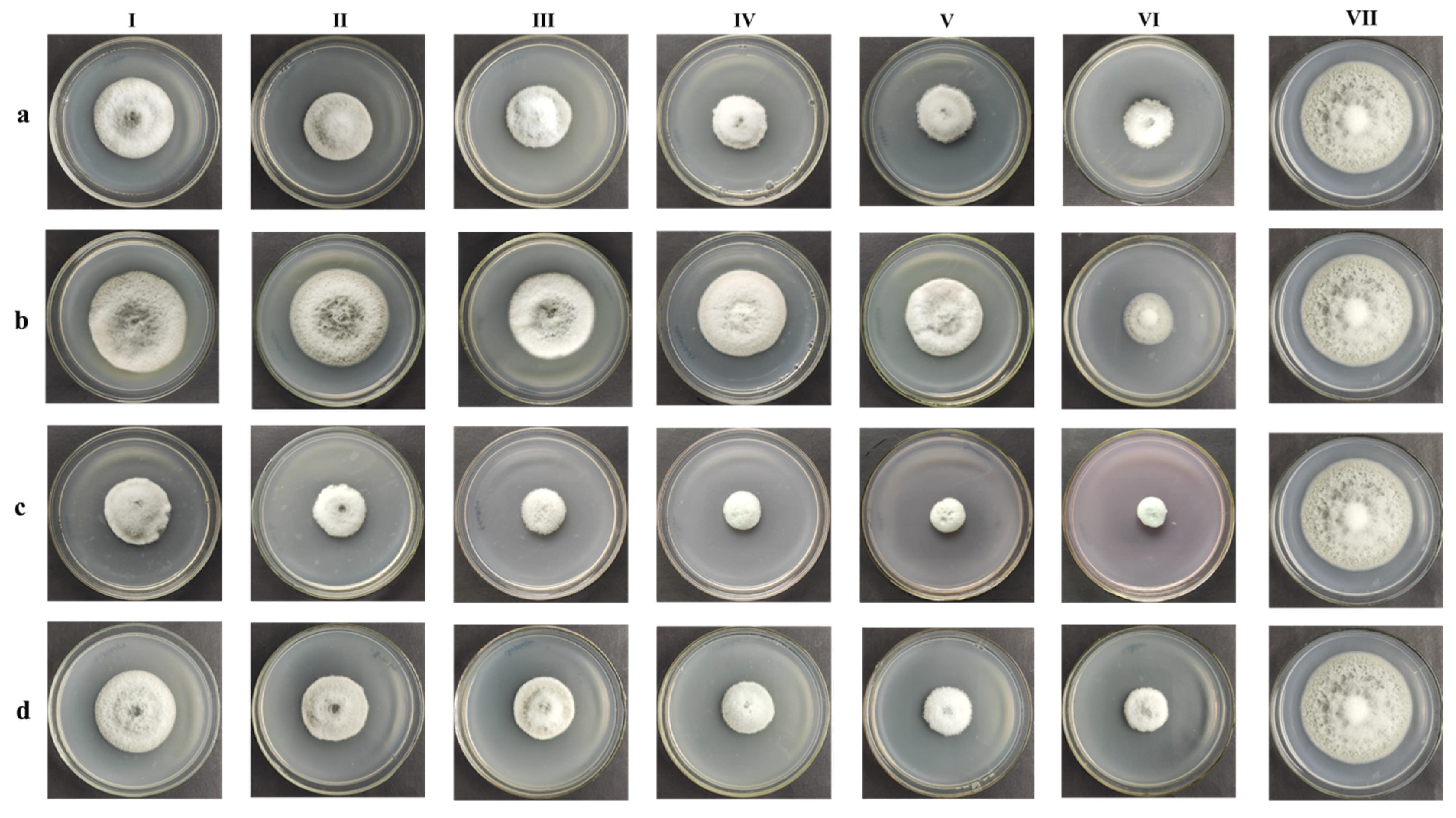
References
- Li, Z.; Li, H.; Zhang, J.; Zhang, S.; Zhao, Q.; Cheng, C.; Zhang, Y. Characterization of fungal species isolated from cankered apple barks demonstrates the Alternaria alternata causing apple canker disease. J. Fungi 2024, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- DeMers, M. Alternaria alternata as endophyte and pathogen. Microbiology 2022, 168, 001153. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Hamada, M.S.; Hahn, M.; Li, G.Q.; Luo, C.X. Fungicide resistance of Botrytis cinerea from strawberry to procymidone and zoxamide in Hubei, China. Phytopathol. Res. 2019, 1, 17. [Google Scholar] [CrossRef]
- Samaras, A.; Ntasiou, P.; Myresiotis, C.; Karaoglanidis, G. Multidrug resistance of Penicillium expansum to fungicides: Whole transcriptome analysis of MDR strains reveals overexpression of efflux transporter genes. Int. J. Food Microbiol. 2020, 335, 108896. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, F.; Yin, L.; Yin, W.; Lv, L.; Yang, X.; Chang, X.; Zhang, S.; Luo, C. Transcriptomic analysis of resistant and wild-type isolates revealed fludioxonil as a candidate for controlling the emerging isoprothiolane resistant populations of Magnaporthe oryzae. Front. Microbiol. 2022, 13, 874497. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, X.; Wang, Q.; Li, Y.; He, C.; Luo, H.; An, B. Integrated application of transcriptomics and metabolomics provides insights into the antifungal activity of α-phenylcinnamic acid against Colletotrichum gloeosporioides. Postharvest Biol. Technol. 2022, 186, 111834. [Google Scholar] [CrossRef]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month 2020, 66, 100971. [Google Scholar] [CrossRef]
- Song, G.; Du, S.; Sun, H.; Liang, Q.; Wang, H.; Yan, M.; Zhang, J. Antifungal mechanism of (E)-2-hexenal against Botrytis cinerea growth revealed by transcriptome analysis. Front. Microbiol. 2022, 13, 951751. [Google Scholar] [CrossRef]
- Maggiolini, F.A.M.; Prencipe, A.; Bergamini, C.; Marsico, A.D.; Vendemia, M.; Santamaria, M.; Giannandrea, M.A.; D’Amico, M.; Forleo, L.R.; Perniola, R. A comparative transcriptomic study reveals temporal and genotype-specific defense responses to Botrytis cinerea in Grapevine. J. Fungi 2025, 11, 124. [Google Scholar] [CrossRef]
- Alassimone, J.; Praz, C.; Lorrain, C.; De Francesco, A.; Carrasco-López, C.; Faino, L.; Shen, Z.; Meile, L.; Sánchez-Vallet, A. The Zymoseptoria tritici avirulence factor AvrStb6 accumulates in hyphae close to stomata and triggers a wheat defense response hindering fungal penetration. Mol. Plant-Microbe Interact. 2024, 37, 432–444. [Google Scholar] [CrossRef]
- Zhang, M.; Ge, J.; Yu, X. Transcriptome analysis reveals the mechanism of fungicidal of thymol against Fusarium oxysporum f. sp. niveum. Curr. Microbiol. 2018, 75, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; He, K.; Li, M.; Zhang, Y.; Jiang, J.; Qian, L.; Gao, X.; Zhang, C.; Liu, S. Comparative transcriptome analysis of Fusarium graminearum challenged with distinct fungicides and functional analysis of FgICL gene. Genomics 2024, 116, 110869. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, K.; Yang, P.; Hou, F.; Rajput, S.A.; Qi, D.; Wang, S. Transcriptomics reveals the effect of thymol on the growth and toxin production of Fusarium graminearum. Toxins 2022, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Miao, J.; Shao, W.; Liu, X.; Zhao, Y.; Ma, Z. Fungicide resistance: Progress in understanding mechanism, monitoring, and management. Phytopathology® 2023, 113, 707–718. [Google Scholar] [CrossRef]
- Vicentini, S.N.C.; Moreira, S.I.; da Silva, A.G.; de Oliveira, T.Y.K.; Silva, T.C.; Assis Junior, F.G.; Krug, L.D.; de Paiva Custódio, A.A.; Leite Júnior, R.P.; Teodoro, P.E. Efflux pumps and multidrug-resistance in Pyricularia oryzae Triticum lineage. Agronomy 2022, 12, 2068. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, S.; Sun, M.; Li, S.; Hu, Y.; Song, H. Transcriptomic response of Clonostachys rosea mycoparasitizing Rhizoctonia solani. J. Fungi 2023, 9, 818. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef]
- Fan, F.; Zhu, Y.; Wu, M.; Yin, W.; Li, G.; Hahn, M.; Hamada, M.S.; Luo, C. Mitochondrial inner membrane ABC transporter Bcmdl1 is involved in conidial germination, virulence, and resistance to anilinopyrimidine fungicides in Botrytis cinerea. Microbiol. Spectr. 2023, 11, e00108–e00123. [Google Scholar] [CrossRef]
- Pasqua, M.; Bonaccorsi di Patti, M.C.; Fanelli, G.; Utsumi, R.; Eguchi, Y.; Trirocco, R.; Prosseda, G.; Grossi, M.; Colonna, B. Host-bacterial pathogen communication: The wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 2021, 8, 723274. [Google Scholar] [CrossRef]
- Dwibedi, V.; Rath, S.K. New insights into and updates on antimicrobial agents. In Emerging Modalities in Mitigation of Antimicrobial Resistance; Springer: Berlin/Heidelberg, Germany, 2022; pp. 179–217. [Google Scholar]
- Boyce, K.J. The microevolution of antifungal drug resistance in pathogenic fungi. Microorganisms 2023, 11, 2757. [Google Scholar] [CrossRef]
- Keighley, C.; Gall, M.; Halliday, C.L.; Chaw, K.; Newton, P.; Sintchenko, V.; Chen, S.C. Breakthrough Candida albicans bloodstream infection associated with in vivo development of pan-azole resistance related to ERG3 gene deletion. Pathology 2024, 56, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Angiolella, L. Virulence regulation and drug-resistance mechanism of fungal infection. Microorganisms 2022, 10, 409. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.; Konings, M.; Parel, F.; Eadie, K.; Strepis, N.; Fahal, A.; Verbon, A.; van de Sande, W.W. Inhibiting DHN-and DOPA-melanin biosynthesis pathway increased the therapeutic value of itraconazole in Madurella mycetomatis infected Galleria mellonella. Med. Mycol. 2022, 60, myac003. [Google Scholar] [CrossRef] [PubMed]
- Ma, X. Screening of Chemical Fungicides and Potential Drug Resistance Mechanism of Poplar Leaf Blight. Master’s Thesis, Northeast Forestry University, Harbin, China, 2023. [Google Scholar]
- Pineda-Rodriguez, Y.Y.; Pompelli, M.F.; Jarma-Orozco, A.; Rodríguez, N.V.; Rodriguez-Paez, L.A. A new and profitable protocol to DNA extraction in Limnospira maxima. Methods Protoc. 2023, 6, 62. [Google Scholar] [CrossRef]
- Li, P.; Zou, J.; Dong, Y.; Jiang, J.; Liang, W.; Li, D. Tetrandrine, a potent antifungal agent, inhibits mycelial growth and virulence of Botrytis cinerea. Phytopathology® 2021, 111, 1152–1157. [Google Scholar] [CrossRef]
- Budde-Rodriguez, S.; Pasche, J.S.; Mallik, I.; Gudmestad, N.C. Sensitivity of Alternaria spp. from potato to pyrimethanil, cyprodinil, and fludioxonil. Crop Prot. 2022, 152, 105855. [Google Scholar] [CrossRef]
- Hou, Y.; Guo, Y.; Cheng, Z.; Liu, S.; Yang, Y.; Li, Y.; Liu, S.; Hou, X.; Xu, J. Inhibitory activities of five fungicides on Alternaria suffruticosae and their field control efficacy against tree peony black spot. Plant Dis. 2024, 108, 2830–2837. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, M.; Kumar, R.S.; Basavarajappa, D.S.; Bhat, M.P.; Nagaraja, S.K.; Almansour, A.I.; Perumal, K.; Nayaka, S. Penicillium citrinum NP4 mediated production, extraction, physicochemical characterization of the melanin, and its anticancer, apoptotic, photoprotection properties. Int. J. Biol. Macromol. 2023, 245, 125547. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, R.L.; Ishaq, M.; Saleemuddin, M. Protein assay by Coomassie brilliant blue G-250-binding method is unsuitable for plant tissues rich in phenols and phenolases. Anal. Biochem. 1987, 163, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, J.; Zhang, Y.; Wang, H.; Wei, S.; Zhang, X.; Zhang, D.; Ma, H.; Ding, Q.; Ma, L. Morphophysiological, proteomic and metabolomic analyses reveal cadmium tolerance mechanism in common wheat (Triticum aestivum L.). J. Hazard. Mater. 2023, 445, 130499. [Google Scholar] [CrossRef]
- Wang, L.; Lu, H.; Zhang, X.; He, Y.; Zhang, J.; Guo, X.; Fu, H.; Ye, G.; Shu, Q. Disruption of serotonin biosynthesis increases resistance to striped stem borer without changing innate defense response in rice. J. Pineal Res. 2023, 75, e12895. [Google Scholar] [CrossRef]
- Strycker, B.D.; Han, Z.H.; Bahari, A.; Pham, T.; Lin, X.R.; Shaw, B.D.; Sokolov, A.V.; Scully, M.O. Raman characterization of fungal DHN and DOPA melanin biosynthesis pathways. J. Fungi 2021, 7, 841. [Google Scholar] [CrossRef]
- Camiletti, B.X.; Lichtemberg, P.S.F.; Paredes, J.A.; Carraro, T.A.; Velascos, J.; Michailides, T.J. Characterization, pathogenicity, and fungicide sensitivity of Alternaria isolates associated with preharvest fruit drop in California citrus. Fungal Biol. 2022, 126, 277–289. [Google Scholar] [CrossRef]
- Noh, J.S.; Hwang, S.H.; Maung, C.E.H.; Cho, J.Y.; Kim, K.Y. Enhanced control efficacy of Bacillus subtilis NM4 via integration of chlorothalonil on potato early blight caused by Alternaria solani. Microb. Pathog. 2024, 190, 106604. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Wang, P.; Zhao, L.; Zhang, H.; Qu, H.; Xu, F. Inhibitory effect and mechanism of carvacrol against black mold disease agent Alternaria alternata in Goji Berries. J. Fungi 2024, 10, 402. [Google Scholar] [CrossRef]
- Nottensteiner, M.; Absmeier, C.; Zellner, M. QoI fungicide resistance mutations in Alternaria solani and Alternaria alternata are fully established in potato growing areas in bavaria and dual resistance against SDHI fungicides is upcoming. Gesunde Pflanz. 2019, 71, 155–164. [Google Scholar] [CrossRef]
- Ivanovic, Z.; Blagojevic, J. Distribution of the F129L mutation conferring resistance to strobilurins in Alternaria solani populations in Serbia. Ann. Appl. Biol. 2022, 181, 117–126. [Google Scholar] [CrossRef]
- Budde-Rodriguez, S.; Celoy, R.M.; Mallik, I.; Pasche, J.S.; Gudmestad, N.C. Impact of SDH mutations in Alternaria solani on recently developed SDHI fungicides adepidyn and solatenol. Plant Dis. 2021, 105, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Förster, H.; Luo, Y.; Hou, L.; Adaskaveg, J.E. Mutations in Sdh gene subunits confer different cross-resistance patterns to SDHI fungicides in Alternaria alternata causing Alternaria leaf spot of Almond in California. Plant Dis. 2022, 106, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Saito, S.; Xiao, C. Fungicide resistance of Alternaria alternata and A. arborescens isolates from mandarin fruit and its influence on control of postharvest Alternaria rot. Plant Dis. 2023, 107, 1538–1543. [Google Scholar] [CrossRef]
- Sofianos, G.; Piombo, E.; Dubey, M.; Karlsson, M.; Karaoglanidis, G.; Tzelepis, G. Transcriptomic and functional analyses on a Botrytis cinerea multidrug-resistant (MDR) strain provides new insights into the potential molecular mechanisms of MDR and fitness. Mol. Plant Pathol. 2024, 25, e70004. [Google Scholar] [CrossRef]
- Child, H.T.; Deeks, M.J.; Rudd, J.J.; Bates, S. Comparison of the impact of two key fungal signalling pathways on Zymoseptoria tritici infection reveals divergent contribution to invasive growth through distinct regulation of infection-associated genes. Mol. Plant Pathol. 2023, 24, 1220–1237. [Google Scholar] [CrossRef]
- Ding, Y.; Gardiner, D.M.; Kazan, K. Transcriptome analysis reveals infection strategies employed by Fusarium graminearum as a root pathogen. Microbiol. Res. 2022, 256, 126951. [Google Scholar] [CrossRef]
- Zhang, T.; Cao, Q.; Li, N.; Liu, D.; Yuan, Y. Transcriptome analysis of fungicide-responsive gene expression profiles in two Penicillium italicum strains with different response to the sterol demethylation inhibitor (DMI) fungicide prochloraz. BMC Genom. 2020, 21, 156. [Google Scholar] [CrossRef]
- Liu, J.; Wang, S.; Qin, T.; Li, N.; Niu, Y.; Li, D.; Yuan, Y.; Geng, H.; Xiong, L.; Liu, D.L. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genom. 2015, 16, 855. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Lu, L. Fungal cytochrome P450 protein Cyp51: What we can learn from its evolution, regulons and Cyp51-based azole resistance. Fungal Biol. Rev. 2018, 32, 131–142. [Google Scholar] [CrossRef]
- Pütsepp, R.; Mäe, A.; Põllumaa, L.; Andresen, L.; Kiiker, R. Fungicide sensitivity profile of Pyrenophora teres f. teres in field population. J. Fungi 2024, 10, 260. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Carvalhaes, C.G.; Deshpande, L.M.; Rhomberg, P.R.; Castanheira, M. In vitro activity of isavuconazole and other mould-active azoles against Aspergillus fumigatus with and without CYP51 alterations. J. Fungi 2023, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, P.; Gruez, A.; Babin, A.-L.; Frippiat, J.-P.; Machouart, M.; Debourgogne, A. CYP51 mutations in the Fusarium solani species complex: First clue to understand the low susceptibility to azoles of the genus Fusarium. J. Fungi 2022, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Van Rhijn, N.; Bromley, M.; Richardson, M.; Bowyer, P. CYP51 paralogue structure is associated with intrinsic azole resistance in fungi. mBio 2021, 12, e0194521. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Q.; Tian, P.; Li, Y.; Duan, G.; Li, D.; Zhan, J.; Chen, F. Induced expression of CYP51 associated with difenoconazole resistance in the pathogenic Alternaria sect. on potato in China. Pest Manag. Sci. 2020, 76, 1751–1760. [Google Scholar] [CrossRef]
- Li, S.; Ying, Z.; Xue, Y.; Sun, Z.; Liu, J.; Liu, Q. Effects of different drugs and hormone treatment on Toxoplasma gondii glutathione S-transferase 2. Parasites Vectors 2022, 15, 461. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- Michael, H.S.R.; Subiramanian, S.R.; Thyagarajan, D.; Mohammed, N.B.; Saravanakumar, V.K.; Govindaraj, M.; Maheswari, K.M.; Karthikeyan, N.; Ramesh Kumar, C. Melanin biopolymers from microbial world with future perspectives—A review. Arch. Microbiol. 2023, 205, 306. [Google Scholar] [CrossRef]
- Surendirakumar, K.; Pandey, R.R.; Muthukumar, T.; Sathiyaseelan, A.; Loushambam, S.; Seth, A. Characterization and biological activities of melanin pigment from root endophytic fungus, Phoma sp. RDSE17. Arch. Microbiol. 2022, 204, 171. [Google Scholar] [CrossRef]
- Xu, X.; Cao, R.; Li, K.; Wan, Q.; Wu, G.; Lin, Y.; Huang, T.; Wen, G. The protective role and mechanism of melanin for Aspergillus niger and Aspergillus flavus against chlorine-based disinfectants. Water Res. 2022, 223, 119039. [Google Scholar] [CrossRef]
- Sanglard, D.; Coste, A.; Ferrari, S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009, 9, 1029–1050. [Google Scholar] [CrossRef]
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal cytochrome P450s and the P450 complement (CYPome) of Fusarium graminearum. Toxins 2018, 10, 112. [Google Scholar] [CrossRef]
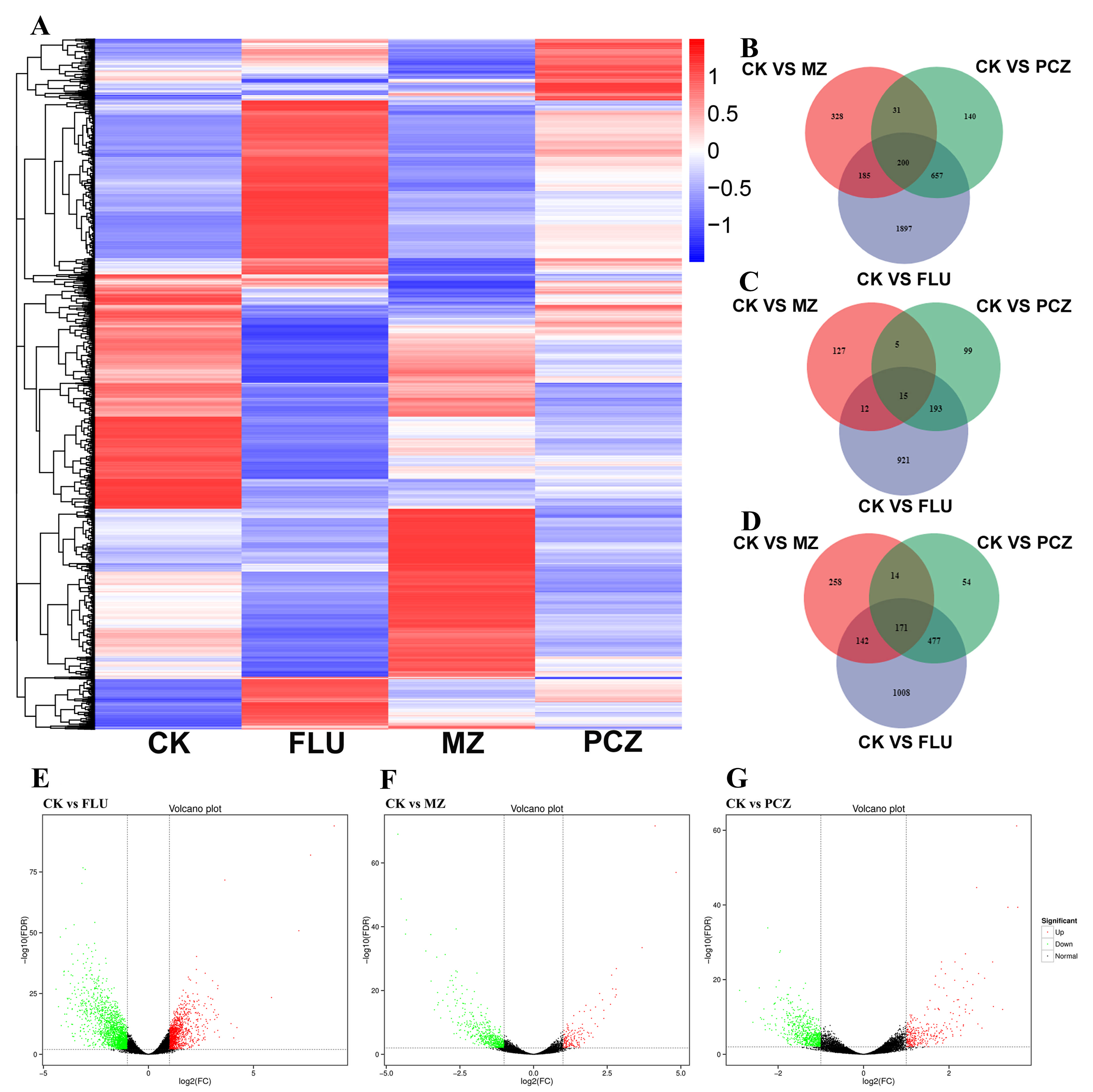
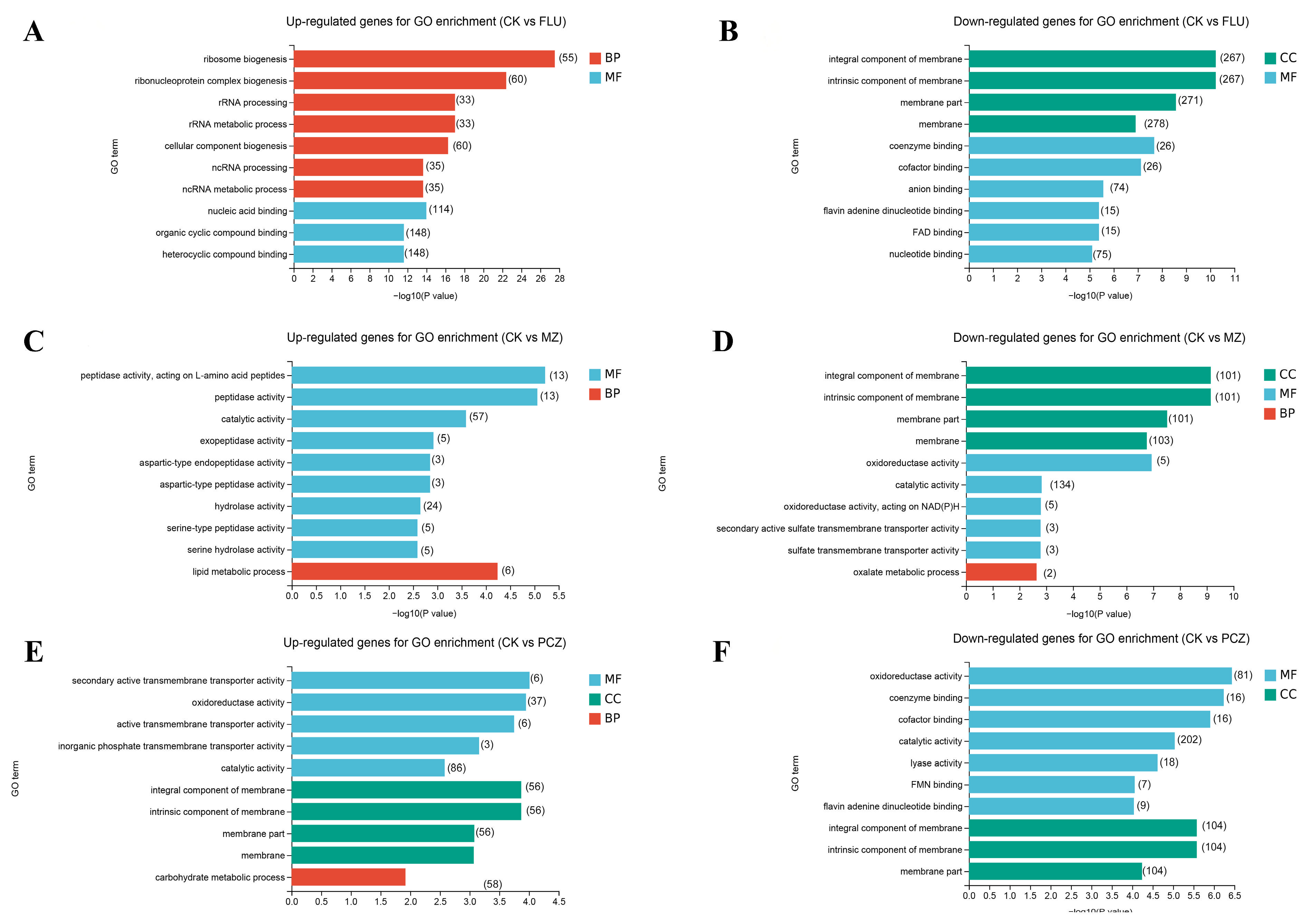
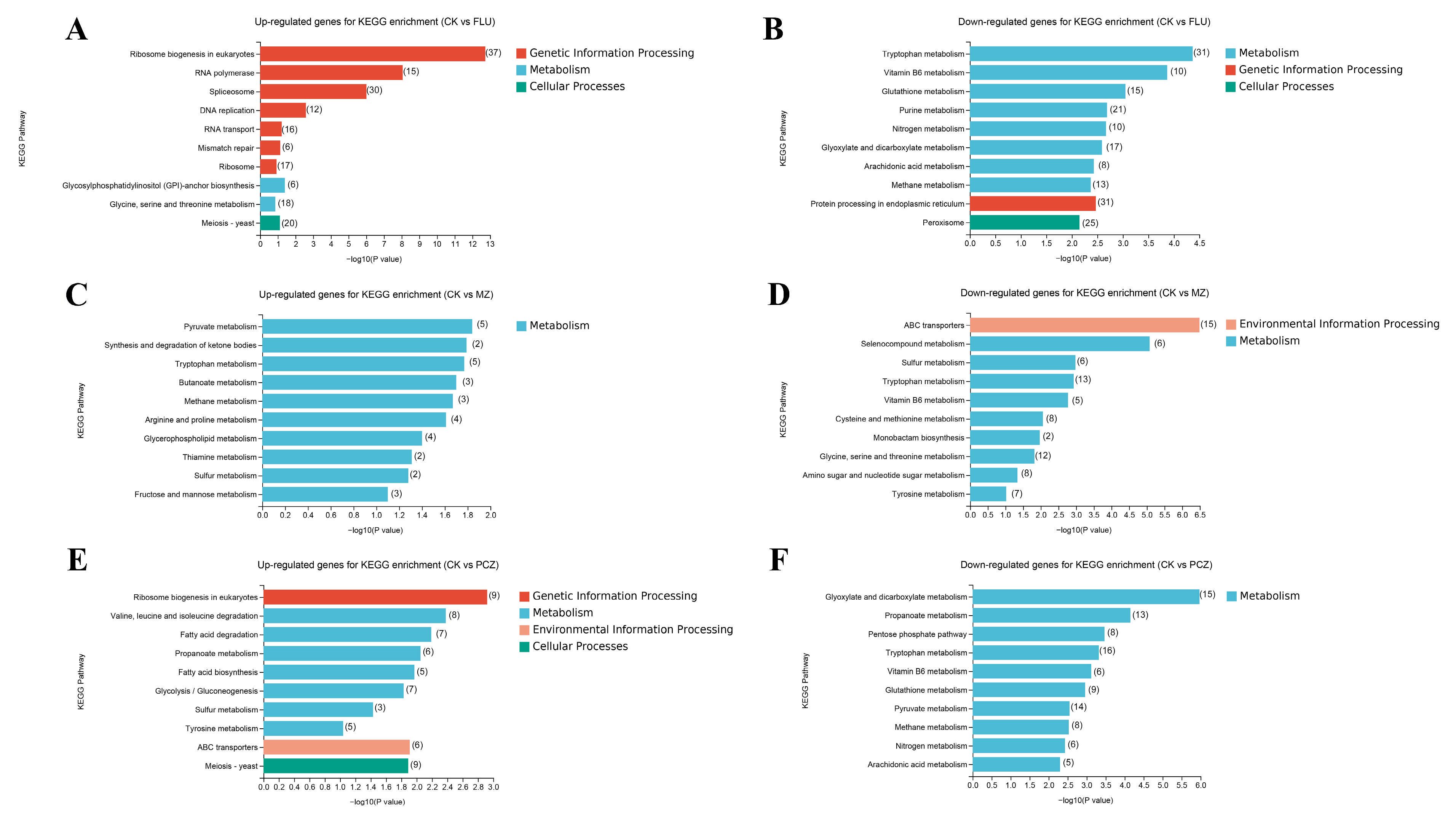
| Fungicide Name | Fungicide Concentration (μg/mL) | 2d | 3d | 4d | 5d |
|---|---|---|---|---|---|
| Mancozeb | 0.1 | 8.30 ± 0.54% f | 3.49 ± 1.50% f | 3.13 ± 0.21% f | 0.48 ± 0.37% f |
| 0.5 | 12.98 ± 1.27% e | 5.82 ± 0.56% e | 8.70 ± 0.84% e | 1.19 ± 0.45% e | |
| 1 | 15.44 ± 2.19% d | 9.41 ± 0.54% d | 20.60 ± 0.53% d | 21.45 ± 0.27% d | |
| 5 | 17.89 ± 0.35% c | 20.61 ± 0.31% c | 34.86 ± 0.21% c | 26.38 ± 0.36% c | |
| 10 | 22.11 ± 0.35% b | 33.15 ± 0.94% b | 36.74 ± 0.42% b | 40.88 ± 0.37% b | |
| 15 | 72.87 ± 0.73% a | 60.30 ± 0.41% a | 52.61 ± 0.42% a | 46.11 ± 0.27% a | |
| Prochloraz | 0.1 | 21.40 ± 3.86% e | 16.31 ± 0.16% f | 25.96 ± 0.53% f | 23.35 ± 0.36% e |
| 0.2 | 36.26 ± 1.58% d | 34.77 ± 0.41% e | 35.56 ± 0.12% e | 36.13 ± 0.10% d | |
| 0.4 | 40.81 ± 1.42% c | 38.80 ± 0.68% d | 38.48 ± 0.32% d | 36.60 ± 0.27% d | |
| 0.8 | 48.66 ± 0.20% b | 49.10 ± 0.41% c | 50.73 ± 0.42% c | 49.20 ± 0.36% c | |
| 2 | 54.97 ± 0.41% a | 52.42 ± 0.54% b | 54.56 ± 0.32% b | 53.24 ± 0.27% b | |
| 4 | 57.08 ± 0.20% a | 55.65 ± 0.27% a | 57.83 ± 0.55% a | 58.59 ± 0.27% a | |
| Fludioxonil | 0.1 | 29.82 ± 0.70% f | 37.10 ± 0.54% f | 41.13 ± 0.63% e | 34.52 ± 0.37% f |
| 0.2 | 32.51 ± 1.07% e | 40.50 ± 0.41% e | 48.50 ± 0.53% d | 50.09 ± 0.18% e | |
| 0.4 | 41.99 ± 0.54% d | 45.61 ± 0.41% d | 54.98 ± 0.32% cd | 55.08 ± 0.18% d | |
| 0.8 | 55.67 ± 1.42% c | 50.63 ± 0.41% c | 57.90 ± 0.12% bc | 62.98 ± 0.27% c | |
| 1 | 60.82 ± 0.54% b | 56.36 ± 0.41% b | 62.91 ± 0.32% ab | 64.77 ± 0.27% b | |
| 2 | 75.20 ± 0.88% a | 72.76 ± 0.41% a | 66.53 ± 8.08% a | 73.32 ± 0.27% a | |
| Difenoconazole | 0.1 | 17.19 ± 0.70% f | 15.50 ± 0.68% f | 20.18 ± 0.32% f | 19.25 ± 0.36% f |
| 0.2 | 24.44 ± 0.88% e | 27.78 ± 0.68% e | 30.13 ± 0.12% e | 32.44 ± 0.47% e | |
| 0.4 | 27.66 ± 1.23% d | 31.00 ± 0.68% d | 35.49 ± 0.21% d | 35.83 ± 0.36% d | |
| 0.8 | 41.29 ± 1.23% c | 37.28 ± 0.16% c | 43.42 ± 0.21% c | 42.78 ± 0.18% c | |
| 2 | 52.28 ± 0.35% b | 41.31 ± 0.31% b | 45.30 ± 0.21% b | 45.75 ± 0.21% b | |
| 4 | 61.75 ± 1.40% a | 53.85 ± 0.16% a | 52.61 ± 0.21% a | 54.90 ± 0.36% a 1 |
| Fungicide Name | Regression Equation | r2 | K | EC50 (µg/mL) |
|---|---|---|---|---|
| Mancozeb | y = 0.0290x + 0.0749 | R2 = 0.8389 | 0.0290 | 14.66 |
| Prochloraz | y = 0.0723x + 0.3381 | R2 = 0.6979 | 0.0723 | 2.24 |
| Fludioxonil | y = 0.1686x + 0.4415 | R2 = 0.7655 | 0.1686 | 0.35 |
| Difenoconazole | y = 0.0692x + 0.2984 | R2 = 0.7281 | 0.0692 | 2.91 |
| Samples | Clean Reads | Clean Bases | GC Content | % ≥Q30 |
|---|---|---|---|---|
| CK1 | 21,861,040 | 6,535,074,204 | 54.56% | 95.13% |
| CK2 | 20,913,041 | 6,257,953,670 | 54.38% | 95.10% |
| CK3 | 19,647,287 | 5,880,173,540 | 54.46% | 95.29% |
| Fludioxonil 1 | 20,434,717 | 6,113,480,574 | 54.72% | 95.24% |
| Fludioxonil 2 | 19,729,983 | 5,905,210,182 | 54.98% | 95.50% |
| Fludioxonil 3 | 19,955,369 | 5,970,527,032 | 54.85% | 95.08% |
| Mancozeb 1 | 21,962,043 | 6,573,537,506 | 54.39% | 95.02% |
| Mancozeb 2 | 23,142,253 | 6,924,447,538 | 54.48% | 95.09% |
| Mancozeb 3 | 20,167,429 | 6,034,657,224 | 54.63% | 94.99% |
| Prochloraz 1 | 20,672,993 | 6,186,553,534 | 54.37% | 95.20% |
| Prochloraz 2 | 20,387,397 | 6,101,144,570 | 54.70% | 94.87% |
| Prochloraz 3 | 22,234,030 | 6,649,083,082 | 54.72% | 95.19% |
| Sample | Total Reads | Mapped Reads | Uniquely Mapped Reads | Multi-Mapped Reads | Positive Strand Mapped Reads | Negative Strand Mapped Reads |
|---|---|---|---|---|---|---|
| CK1 | 43,722,080 | 42,296,656 (96.74%) | 42,206,36 (96.53%) | 90,295 (0.21%) | 21,184,372 (48.45%) | 21,210,242 (48.51%) |
| CK2 | 41,826,082 | 40,492,095 (96.81%) | 40,409,351 (96.61%) | 82,744 (0.20%) | 20,279,645 (48.49%) | 20,303,583 (48.54%) |
| CK3 | 39,294,574 | 38,062,886 (96.87%) | 37,988,848 (96.68%) | 74,038 (0.19%) | 19,059,135 (48.50%) | 19,083,660 (48.57%) |
| Fludioxonil 1 | 40,869,434 | 39,448,017 (96.52%) | 39,351,670 (96.29%) | 96,347 (0.24%) | 19,793,641 (48.43%) | 19,757,558 (48.34%) |
| Fludioxonil 2 | 39,459,966 | 38,159,320 (96.70%) | 38,063,855 (96.46%) | 95,465 (0.24%) | 19,123,192 (48.46%) | 19,138,605 (48.50%) |
| Fludioxonil 3 | 39,910,738 | 38,446,001 (96.33%) | 38,354,808 (96.10%) | 91,193 (0.23%) | 19,263,963 (48.27%) | 19,279,011 (48.31%) |
| Mancozeb 1 | 43,924,086 | 42,486,492 (96.73%) | 42,379,620 (96.48%) | 106,872 (0.24%) | 21,342,440 (48.59%) | 21,258,963 (48.40%) |
| Mancozeb 2 | 46,284,506 | 44,207,251 (95.51%) | 44,106,443 (95.29%) | 100,808 (0.22%) | 22,165,125 (47.89%) | 22,150,397 (47.86%) |
| Mancozeb 3 | 40,334,858 | 38,930,703 (96.52%) | 38,848,657 (96.32%) | 82,046 (0.20%) | 19,500,824 (48.35%) | 19,518,747 (48.39%) |
| Prochloraz 1 | 41,345,986 | 40,117,348 (97.03%) | 40,016,431 (96.78%) | 100,917 (0.24%) | 20,157,307 (48.75%) | 20,067,135 (48.53%) |
| Prochloraz 2 | 40,774,794 | 39,462,161 (96.78%) | 39,375,847 (96.57%) | 86,314 (0.21%) | 19,770,515 (48.49%) | 19,786,084 (48.53%) |
| Prochloraz 3 | 44,468,060 | 42,986,501 (96.67%) | 42,884,843 (96.44%) | 101,658 (0.23%) | 21,541,368 (48.44%) | 21,554,549 (48.47%) |
| Sample Comparison | Gene Expression Trend | ABC Transporters | MFS Transporters | |||
|---|---|---|---|---|---|---|
| Subfamily B | Subfamily C | Subfamily G | DHA1 Family | DHA2 Family | ||
| CK vs. FLU | UP | 1 | 0 | 0 | 1 | 1 |
| CK vs. FLU | DOWN | 0 | 1 | 0 | 17 | 1 |
| CK vs. MZ | UP | 0 | 0 | 0 | 1 | 0 |
| CK vs. MZ | DOWN | 3 | 0 | 0 | 9 | 1 |
| CK vs. PCZ | UP | 0 | 0 | 1 | 0 | 0 |
| CK vs. PCZ | DOWN | 1 | 0 | 0 | 13 | 1 |
| Gene ID | Gene Name | CK vs. FLU log2FC | CK vs. MZ log2FC | CK vs. PCZ log2FC | KEGG Annotation |
|---|---|---|---|---|---|
| scaffold29.g33 | ERG1 | −1.61 | \ | \ | squalene monooxygenase |
| scaffold30.g31 | DHCR24 | 1.21 | \ | \ | Delta24-sterol reductase |
| scaffold70.g28 | ERG4 | −1.21 | \ | \ | Delta24 (24 (1))-sterol reductase |
| scaffold221.g2 | ERG6 | 1.04 | \ | 1.55 | sterol 24-C-methyltransferase |
| scaffold31.g1 | CYP51 | 1.02 | \ | 1.05 | sterol 14alpha-demethylase |
| scaffold76.g38 | ERG26-1 | 2.65 | \ | 2.18 | sterol-4alpha-carboxylate 3-dehydrogenase |
| scaffold84.g8 | ERG26-2 | \ | −1.14 | \ | sterol-4alpha-carboxylate 3-dehydrogenase |
| scaffold40.g88 | ERG27 | 1.08 | \ | \ | 3-keto steroid reductase |
| scaffold73.g57 | EBP-1 | 1.47 | \ | \ | cholestenol Delta-isomerase |
| Alternaria_alternata_newGene_1524 | EBP-2 | −1.24 | \ | \ | cholestenol Delta-isomerase |
| Gene ID | Gene Name | CK vs. FLU log2 FC | CK vs. MZ log2 FC | CK vs. PCZ log2 FC | KEGG Annotation |
|---|---|---|---|---|---|
| scaffold1.g420 | TYR-1 | 1.20 | \ | \ | tyrosinase |
| scaffold17.g5 | TYR-2 | 1.21 | \ | \ | tyrosinase |
| scaffold3.g176 | TYR-3 | 2.29 | −1.33 | 1.11 | tyrosinase |
| scaffold39.g62 | TYR-4 | 3.27 | \ | \ | tyrosinase |
| scaffold44.g51 | TYR-5 | 1.23 | \ | 1.02 | tyrosinase |
| scaffold84.g9 | TYR-6 | 1.55 | −1.36 | \ | tyrosinase |
| scaffold99.g40 | TYR-7 | −1.08 | \ | −1.10 | tyrosinase |
| scaffold44.g82 | TYR-8 | \ | 3.68 | \ | tyrosinase |
| scaffold62.g73 | TYR-9 | \ | −1.84 | \ | tyrosinase |
| scaffold30.g2 | LAC-1 | −1.09 | \ | −1.24 | laccase |
| scaffold19.g123 | LAC-2 | 1.22 | \ | \ | laccase |
| scaffold137.g5 | LAC-3 | 1.30 | −1.29 | \ | laccase |
| scaffold20.g78 | LAC-4 | 1.70 | \ | \ | laccase |
| scaffold43.g9 | LAC-5 | \ | −2.26 | \ | laccase |
| scaffold3.g55 | EASB-1 | 1.90 | \ | \ | polyketide synthase |
| scaffold184.g4 | EASB-2 | −2.08 | \ | −1.52 | polyketide synthase |
| scaffold106.g31 | ACE1-1 | −1.78 | \ | \ | polyketide synthase |
| Alternaria_alternata_newGene_1437 | ACE1-2 | 1.49 | \ | \ | polyketide synthase |
| scaffold45.g101 | MDPG | −2.89 | −4.35 | \ | polyketide synthase |
| scaffold73.g47 | THNR-1 | −1.45 | \ | \ | tetrahydroxynaphthalene reductase |
| scaffold36.g38 | THNR-2 | −2.00 | \ | −1.63 | tetrahydroxynaphthalene reductase |
| scaffold78.g52 | OAR1 | \ | \ | 1.15 | tetrahydroxynaphthalene reductase |
| scaffold15.g120 | FET3_5-1 | 1.67 | \ | \ | multicopper oxidase |
| scaffold1.g5 | FET3_5-2 | −1.97 | −1.39 | −1.17 | multicopper oxidase |
| scaffold36.g36 | FET3_5-3 | −2.51 | \ | −1.51 | multicopper oxidase |
| scaffold132.g2 | FET3_5-4 | 1.33 | −1.67 | \ | multicopper oxidase |
| scaffold26.g100 | FET3_5-5 | \ | −1.56 | \ | multicopper oxidase |
| Sample | CK | FLU | MZ | PCZ |
|---|---|---|---|---|
| Soluble protein (mg·mL−1) | 24.35 ± 4.98 c | 58.34 ± 3.86 ab | 52.30 ± 2.75 b | 63.38 ± 1.50 a |
| Tyrosinase (U·g−1) | 253.61 ± 18.34 d | 598.69 ± 16.19 a | 465.21 ± 44.88 c | 552.34 ± 11.71 b |
| Melanin (g·L−1) | 0.32 ± 0.03 c | 0.53 ± 0.01 ab | 0.48 ± 0.04 b | 0.57 ± 0.06 a |
| MDA (µmol·g−1) | 4.67 ± 0.61 c | 23.79 ± 3.34 a | 14.46 ± 1.45 b | 15.88 ± 0.43 b |
| CAT (U·g−1) | 27.86 ± 0.48 a | 23.62 ± 1.69 b | 24.63 ± 1.39 b | 25.50 ± 2.08 ab |
| SOD (U·g−1) | 155.34 ± 29.73 a | 121.70 ± 5.47 a | 152.73 ± 15.70 a | 150.82 ± 16.08 a 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Q.; Ma, X.; Hayat, M.; Tang, Y.; Wang, Z. Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms. J. Fungi 2025, 11, 305. https://doi.org/10.3390/jof11040305
Bai Q, Ma X, Hayat M, Tang Y, Wang Z. Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms. Journal of Fungi. 2025; 11(4):305. https://doi.org/10.3390/jof11040305
Chicago/Turabian StyleBai, Qian, Xinbo Ma, Mansoor Hayat, Yuxin Tang, and Zhanbin Wang. 2025. "Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms" Journal of Fungi 11, no. 4: 305. https://doi.org/10.3390/jof11040305
APA StyleBai, Q., Ma, X., Hayat, M., Tang, Y., & Wang, Z. (2025). Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms. Journal of Fungi, 11(4), 305. https://doi.org/10.3390/jof11040305






