Exploring the Biodiversity and Antibacterial Potential of the Culturable Soil Fungi in Nyingchi, Tibet
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Isolation and Identification of Fungi
2.3. Phylogenetic Analysis
2.4. Screening of Antibacterial Fungi by Agar Diffusion Assay
2.5. Fermentation and Extraction
2.6. Antibacterial Activity Tests by Filter Paper Method and 96-Well Plate Method
3. Results and Analysis
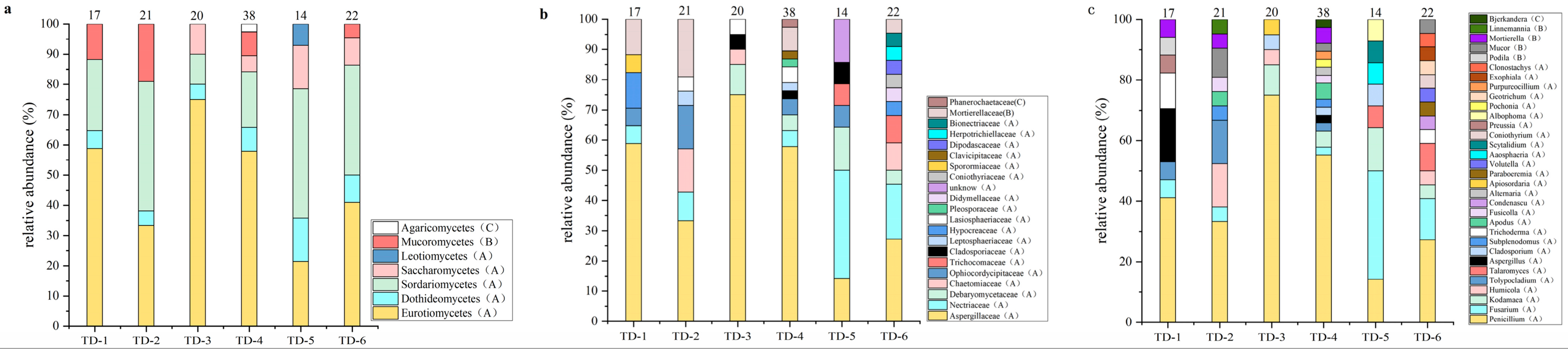
3.1. Soil Fungal Diversity in Nyingchi, Tibet
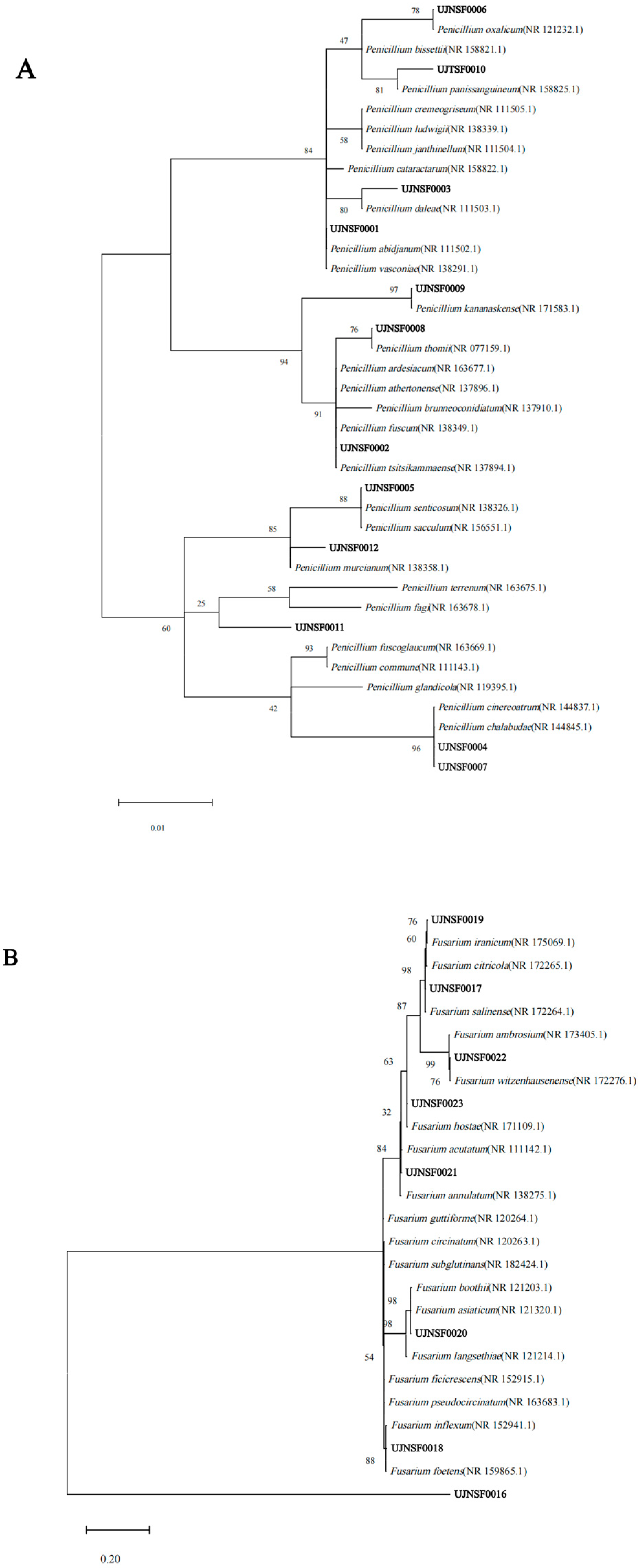
3.2. Phylogenetic Analysis of the Isolated Culturable Fungi
3.3. Screening of Antibacterial Fungi
3.4. The Antibacterial Activities of Crude Extracts of Fungi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11, 1215. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. World Health Organization 2017. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 12 February 2025).
- Jesudason, T. WHO publishes updated list of bacterial priority pathogens. Lancet Microbe 2024, 5, 100940. [Google Scholar] [PubMed]
- Gigante, V.; Sati, H.; Beyer, P. Recent advances and challenges in antibacterial drug development. Admet Dmpk 2022, 10, 147–151. [Google Scholar] [CrossRef]
- A Financial Model for an Impact Investment Fund for the Development of Antibacterial Treatments and Diagnostics: A User Guide; World Health Organization: Geneva, Switzerland, 2020.
- Zhang, D.; Li, S.; Fan, M.; Zhao, C. The Novel Compounds with Biological Activity Derived from Soil Fungi in the Past Decade. Drug Des. Devel. Ther. 2022, 16, 3493–3555. [Google Scholar]
- Yang, H.; Xu, Y.; Wang, L.J. Evaluation method and empirical application of construction land suitability and arable land suitability in alpine-gorge region of Qinghai-Tibet Plateau: A case study of Nyingchi city. J. Nat. Resour. 2023, 38, 1283–1299. [Google Scholar]
- Qin, Y.; Bobrov, A.; Puppe, D.; Li, H.; Man, B.; Gong, J.; Wang, J.; Cui, Y.; Gu, Y.; Herzschuh, U.; et al. Testate amoebae (Protozoa) in lakes of the Qinghai-Tibet Plateau: Biodiversity, community structures, and protozoic biosilicification in relation to environmental properties and climate warming. Sci. Total Environ. 2024, 913, 169661. [Google Scholar] [PubMed]
- Cao, P.-x.; Liu, Y.; Ma, H.-m.; Zhao, N.; Chen, S.-t.; Xu, G.-Q.; Liu, X. Fungal Diversity in the Soil of the Oxytropis glacialis Root System on the Qinghai-Tibet Plateau. Front. Microbiol. 2022, 13, 831783. [Google Scholar]
- Ren, Z.; Gao, H. Abundant and rare soil fungi exhibit distinct succession patterns in the forefield of Dongkemadi glacier on the central Qinghai-Tibet Plateau. Sci. Total Environ. 2022, 828, 154563. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Wang, H.; Ouyang, S.; Liu, X.; Lu, J. Pattern and drivers of soil fungal community along elevation gradient in the Abies georgei forests of Segila mountains, Southeast Tibet. Glob. Ecol. Conserv. 2022, 39, e02291. [Google Scholar]
- Xing, R.; Gao, Q.-b.; Zhang, F.-q.; Wang, J.-l.; Chen, S.-l. Environmental filtering affects fungal communities more than dispersal limitation in a high-elevation hyperarid basin on Qinghai-Tibet Plateau. FEMS Microbiol. Lett. 2021, 368, fnab033. [Google Scholar]
- Zaferanloo, B.; Pepper, S.A.; Coulthard, S.A.; Redfern, C.P.F.; Palombo, E.A. Metabolites of endophytic fungi from Australian native plants as potential anticancer agents. FEMS Microbiol. Lett. 2018, 365, fny078. [Google Scholar]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar]
- Jigjiddorj, E.-A.; Maidarjav, A.; Byambasuren, B.; Nyamgerel, D. Identification, Antimicrobial and Plant Growth Promoting Activities of Endophytic Fungi Associated with Cynomorium songaricum Rupr., a Traditional Medicinal Plant in Mongolia. Diversity 2024, 16, 122. [Google Scholar] [CrossRef]
- Fu, C.; Lan, X.; Yuan, J.; Li, C.; Li, L.; Yu, Z.; Tan, T.; Yuan, M.; Du, F. Research on the optimization, key chemical constituents and antibacterial activity of the essential oil extraction process of Thuja koraiensis Nakai. J. Microbiol. Methods 2022, 194, 106435. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Zhai, H.; Zhu, K.; Yu, J.-H.; Zhang, Y.; Wang, Y.; Jiang, C.-S.; Zhang, X.; Zhang, Y.; Zhang, H. Bioactive Pyridone Alkaloids from a Deep-Sea-Derived Fungus Arthrinium sp. UJNMF0008. Mar. Drugs 2018, 16, 174. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Wu, J.H.; Sun, Y.X.; Zhang, Y.Y.; Zhao, Y.F.; Huang, Z.; Duan, W.H. Climate and geochemistry at different altitudes influence soil fungal community aggregation patterns in alpine grasslands. Sci. Total Environ. 2023, 881, 163375. [Google Scholar] [CrossRef]
- Li, X.; Qu, Z.; Zhang, Y.; Ge, Y.; Sun, H. Soil Fungal Community and Potential Function in Different Forest Ecosystems. Diversity 2022, 14, 520. [Google Scholar] [CrossRef]
- Shi, X.; Wang, J.; Lucas-Borja, M.E.; Wang, Z.; Li, X.; Huang, Z. Microbial diversity regulates ecosystem multifunctionality during natural secondary succession. J. Appl. Ecol. 2021, 58, 2833–2842. [Google Scholar] [CrossRef]
- Yao, X.-M.; Lv, G.-Z.; Yang, H.; Zhao, Z.-H.; Chen, R. Studies of Fungal Flora in Forest Soil of Changbai Mountains. J. Fungal Res. 2007, 5, 43–46. [Google Scholar]
- Feng, Y.; Zhang, J.; Berdugo, M.; Guirado, E.; Guerra, C.A.; Egidi, E.; Wang, J.; Singh, B.K.; Delgado-Baquerizo, M. Temperature thresholds drive the global distribution of soil fungal decomposers. Glob. Change Biol. 2022, 28, 2779–2789. [Google Scholar]
- Liu, S.; Garcia-Palacios, P.; Tedersoo, L.; Guirado, E.; van der Heijden, M.G.A.; Wagg, C.; Chen, D.; Wang, Q.; Wang, J.; Singh, B.K.; et al. Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nat. Ecol. Evol. 2022, 6, 900–909. [Google Scholar]
- King, A.J.; Farrer, E.C.; Suding, K.N.; Schmidt, S.K. Co-occurrence patterns of plants and soil bacteria in the high-alpine subnival zone track environmental harshness. Front. Microbiol. 2012, 3, 347. [Google Scholar]
- Braga, A.F.V.C.; do Rosario, M.S.; Gomes, J.B.N.; de A Monteiro, C.; Farias, F.A.C.; Filho, E.R.; Filho, A.J.C. Antimicrobial Potential of Soil/Sediment Mangrove Associated Fungi: A Review. J. Braz. Chem. Soc. 2024, 35, e-20240032. [Google Scholar]
- Wang, X.; Sun, K.; Wang, B. Bioactive Pimarane Diterpenes from the Arctic Fungus Eutypella sp. D-1. Chem. Biodivers. 2018, 15, e1700501. [Google Scholar]
- Ning, Y.; Qu, Y.; Fu, Y.; Zhang, S.; Xu, Y.; Jiao, B.; Lu, X. Discovery of Bioactive Terpenes Derived from a Polar Fungus. Chem. Biodivers. 2024, 21, e202401750. [Google Scholar]
- Xu, L.-L.; Cao, F.; Tian, S.-S.; Zhu, H.-J. Alkaloids and Polyketides from the Soil Fungus Aspergillus terreus and Their Antibacterial Activities. Chem. Nat. Compd. 2017, 53, 1212–1215. [Google Scholar]
- Liu, J.H.; Guo, J.N.; Lu, H.; Lin, J. Activity-Based Screening of Soil Samples from Nyingchi, Tibet, for Amylase-Producing Bacteria and Other Multifunctional Enzyme Capacities. Int. J. Microbiol. 2022, 2022, 2401766. [Google Scholar]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Don Duy, N.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar]

| Samples | Altitude (m) | Features | Sample Environment | Soil Depth (cm) |
|---|---|---|---|---|
| TD-1 | 4089.2 | Yellow sand granular | meadow | 0–10 |
| TD-2 | 4466.7 | Black clumps | meadow | 10–20 |
| TD-3 | 2936.6 | Brown sand blocks | bush | 10–20 |
| TD-4 | 2956.2 | Black sand block | forest | 10–20 |
| TD-5 | 3138.9 | Black mass | forest | 20–30 |
| TD-6 | 2913.4 | Black clumps | forest | 10–20 |
| Name | Formulas (L) |
|---|---|
| YPD | Yeast extract 5.0 g, peptone 10.0 g, glucose 20.0 g, agar 20.0 g |
| PDA | Potato 200.0 g, boiled to get filtrate, glucose 20.0 g, agar 20.0 g |
| MEA | Malt extract 20.0 g, peptone 1.0 g, agar 20.0 g |
| CSA | Corn syrup 7.0 g, agar 20.0 g |
| SSA | Soluble starch 10.0 g, peptone 1.0 g, glucose 10.0 g, K2HPO4 1.0 g, MgSO4·7H2O 1.0 g, agar 20.0 |
| RBA | Peptone 5.0 g, glucose 10.0 g, KH2PO4 1.0 g, MgSO4·7H2O 0.5 g, rose bengal red 0.03 g, agar 20.0 g |
| CDA | NaNO3 3.0 g, K2HPO4 1.0 g, MgSO4·7H2O 0.5 g, KC1 0.5 g, FeSO4·7H2O 0.01 g, sucrose 30.0 g, agar 20.0 g |
| Names | Formulas |
|---|---|
| 1# (solid) | Rice 80.0 g, yeast extract 0.1 g, dextrose 0.1 g, distilled water 120 mL |
| 2# (solid) | Oat 80.0 g, yeast extract 0.1 g, dextrose 0.1 g, distilled water 120 mL |
| 3# (liquid) | Glucose 10.0 g, mannitol 20.0 g, maltose 20.0 g, corn syrup 1.0 g, distilled water 1 L |
| 4# (liquid) | Yeast extract 3.0 g, malt extract 3.0 g, peptone 1.0 g, glucose 2.0 g, distilled water 1 L |
| Fungal Family | Fungal Genus | Number of Isolates (Isolation Frequency (%)) | |||||
|---|---|---|---|---|---|---|---|
| TD-1 | TD-2 | TD-3 | TD-4 | TD-5 | TD-6 | ||
| Aspergillaceae | Penicillium | 7 (41.2) | 7 (33.3) | 15 (75.0) | 21 (55.3) | 2 (14.3) | 6 (27.3) |
| Aspergillus | 3 (17.6) | 0 (0) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) | |
| Nectriaceae | Fusarium | 1 (5.9) | 1 (4.8) | 0 (0) | 1 (2.6) | 5 (35.7) | 3 (13.6) |
| Fusicolla | 0 (0) | 1 (4.8) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) | |
| Volutella | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) | |
| Debaryomycetaceae | Kodamaea | 0 (0) | 0 (0) | 2 (10.0) | 2 (5.3) | 2 (14.3) | 1 (4.5) |
| Chaetomiaceae | Humicola | 0 (0) | 3 (14.3) | 1 (5.0) | 0 (0) | 0 (0) | 1 (4.5) |
| Condenascu | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) | |
| Ophiocordycipitaceae | Tolypocladium | 1 (5.9) | 3 (14.3) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) |
| Albophoma | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (7.1) | 0 (0) | |
| Purpureocillium | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | |
| Trichocomaceae | Talaromyces | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (7.1) | 2 (9.1) |
| Cladosporiaceae | Cladosporium | 0 (0) | 0 (0) | 1 (5.0) | 1 (2.6) | 1 (7.1) | 0 (0) |
| Leptosphaeriaceae | Subplenodomus | 0 (0) | 1 (4.8) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) |
| Hypocreaceae | Trichoderma | 2 (11.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Lasiosphaeriaceae | Apodus | 0 (0) | 1 (4.8) | 0 (0) | 2 (5.2) | 0 (0) | 0 (0) |
| Apiosordaria | 0 (0) | 0 (0) | 1 (5.0) | 0 (0) | 0 (0) | 0 (0) | |
| Pleosporaceae | Alternaria | 0 (0) | 0 (0) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) |
| Didymellaceae | Paraboeremia | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Coniothyriaceae | Coniothyrium | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Sporormiaceae | Preussia | 1 (5.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Clavicipitaceae | Pochonia | 0 (0) | 0 (0) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) |
| Dipodascaceae | Geotrichum | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Herpotrichiellaceae | Exophiala | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Bionectriaceae | Clonostachys | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4.5) |
| Mortierellaceae | Linnemannia | 0 (0) | 1 (4.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Podila | 1 (5.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Mucor | 0 (0) | 2 (9.2) | 0 (0) | 1 (2.6) | 0 (0) | 1 (4.5) | |
| Mortierella | 1 (5.9) | 1 (4.8) | 0 (0) | 2 (5.2) | 0 (0) | 0 (0) | |
| Phanerochaetaceae | Bjerkandera | 0 (0) | 0 (0) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) |
| Unknow | Aaosphaeria | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (7.1) | 0 (0) |
| Scytalidium | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (7.1) | 0 (0) | |
| Total | 17 | 21 | 20 | 38 | 14 | 22 | |
| Fungal Class | Fungal Genus | Fungal Taxa | Representative Isolate (Accession Number in GenBank) | Sequence in Genbank | Identities | Number of Fungal Isolates | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TD-1 | TD-2 | TD-3 | TD-4 | TD-5 | TD-6 | ||||||
| Eurotiomycetes | Penicillium | P. vasconiae | UJNSF0001 (PQ152240) | NR 138291.1 | 100.00% | 1 | 0 | 4 | 0 | 0 | 1 |
| P. fuscum | UJNSF0002 (PQ152241) | NR 138349.1 | 100.00% | 2 | 0 | 1 | 2 | 0 | 0 | ||
| P. daleae | UJNSF0003 (PQ152242) | NR 111503.1 | 98.42% | 1 | 2 | 0 | 2 | 1 | 1 | ||
| P. chalabudae | UJNSF0004 (PQ152243) | NR 144845.1 | 99.47% | 1 | 0 | 10 | 14 | 0 | 0 | ||
| P.sacculum | UJNSF0005 (PQ152244) | NR 156551.1 | 99.80% | 1 | 0 | 0 | 0 | 0 | 0 | ||
| P.oxalicum | UJNSF0006 (PQ152245) | NR 121232.1 | 100.00% | 1 | 0 | 0 | 0 | 0 | 0 | ||
| P. cinereoatrum | UJNSF0007 (PQ152246) | NR 144837.1 | 99.60% | 0 | 2 | 0 | 2 | 0 | 0 | ||
| P.thomii | UJNSF0008 (PQ152247) | NR 077159.1 | 99.47% | 0 | 1 | 0 | 0 | 0 | 0 | ||
| P kananaskense | UJNSF0009 (PQ152248) | NR 171583.1 | 100.00% | 0 | 2 | 0 | 0 | 0 | 0 | ||
| P. panissanguineum | UJTSF0010 (PQ152249) | NR 158825.1 | 98.14% | 0 | 0 | 0 | 1 | 0 | 0 | ||
| P. glandicola | UJNSF0011 (PQ152250) | NR 119395.1 | 99.82% | 0 | 0 | 0 | 0 | 1 | 0 | ||
| P. murcianum | UJNSF0012 (PQ152251) | NR 138358.1 | 99.15% | 0 | 0 | 0 | 0 | 0 | 4 | ||
| Eurotiomycetes | Aspergillus | A. versicolor | UJNSF0013 (PQ152252) | NR 131277.1 | 98.07% | 2 | 0 | 0 | 0 | 0 | 0 |
| A. terreus | UJNSF0014 (PQ152253) | NR 131276.1 | 99.83% | 1 | 0 | 0 | 0 | 0 | 0 | ||
| A. flavus | UJNSF0015 (PQ152254) | NR 111041.1 | 99.50% | 0 | 0 | 0 | 1 | 0 | 0 | ||
| Sordariomycetes | Fusarium | F. pseudoanthophilum | UJNSF0016 (PQ152255) | NR 163682.1 | 99.08% | 1 | 0 | 0 | 0 | 0 | 0 |
| F.salinense | UJNSF0017 (PQ152256) | NR 172264.1 | 99.46% | 0 | 1 | 0 | 0 | 0 | 0 | ||
| F. foetens | UJNSF0018 (PQ152257) | NR 159865.1 | 99.22% | 0 | 0 | 0 | 1 | 2 | 0 | ||
| F. iranicum | UJNSF0019 (PQ152258) | NR 175069.1 | 99.46% | 0 | 0 | 0 | 0 | 1 | 1 | ||
| F. asiaticum | UJNSF0020 (PQ152259) | NR 121320.1 | 99.27% | 0 | 0 | 0 | 0 | 1 | 0 | ||
| F. annulatum | UJNSF0021 (PQ152260) | NR 138275.1 | 99.64% | 0 | 0 | 0 | 0 | 1 | 0 | ||
| F. witzenhausenense | UJNSF0022 (PQ152261) | NR 172276.1 | 99.63% | 0 | 0 | 0 | 0 | 0 | 1 | ||
| F. hostae | UJNSF0023 (PQ152262) | NR 171109.1 | 99.81% | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Sordariomycetess | Fusicolla | F. aquaeductuum | UJNSF0024 (PQ152263) | NR 173406.1 | 99.11% | 0 | 1 | 0 | 1 | 0 | 0 |
| Sordariomycetess | Volutella | V. salvadorae | UJNSF0025 (PQ152264) | NR 173060.1 | 94.58% | 0 | 0 | 0 | 0 | 0 | 1 |
| Saccharomycetes | Kodamaea | K. ohmeri | UJNSF0026 (PQ152265) | NR 121464.1 | 94.88% | 0 | 0 | 2 | 2 | 2 | 1 |
| Sordariomycetes | Humicola | H. nigrescens | UJNSF0027 (PQ152266) | NR 077125.1 | 99.46% | 0 | 3 | 1 | 0 | 0 | 1 |
| Sordariomycetes | Condenascu | C. tortuosus | UJNSF0028 (PQ152267) | NR 160164.1 | 99.19% | 0 | 0 | 0 | 0 | 0 | 1 |
| Sordariomycetes | Tolypocladium | T. album | UJNSF0029 (PQ152268) | NR 155018.1 | 97.38% | 1 | 0 | 0 | 0 | 0 | 0 |
| T. inflatum | UJNSF0030 (PQ152269) | NR 171737.1 | 100.0% | 0 | 3 | 0 | 0 | 0 | 0 | ||
| T. cylindrosporum | UJNSF0031 (PQ152270) | NR 167967.1 | 99.82% | 0 | 0 | 0 | 1 | 0 | 0 | ||
| Sordariomycetes | Albophoma | A. yamanashiensis | UJNSF0032 (PQ152271) | NR 171222.1 | 99.65% | 0 | 0 | 0 | 0 | 1 | 0 |
| Sordariomycetes | Purpureocillium | P. lilacinum | UJNSF0033 (PQ152272) | NR 165946.1 | 100.00% | 0 | 0 | 0 | 1 | 0 | 0 |
| Eurotiomycetes | Talaromyces | T. adpressus | UJNSF0034 (PQ152273) | NR 171595.1 | 98.63% | 0 | 0 | 0 | 0 | 1 | 0 |
| T. atricola | UJNSF0035 (PQ152274) | NR 147429.1 | 99.64% | 0 | 0 | 0 | 0 | 0 | 2 | ||
| Dothideomycetes | Cladosporium | C.velox | UJNSF0036 (PQ152275) | NR 119604.1 | 100.00% | 0 | 0 | 1 | 0 | 0 | 0 |
| C. oxysporum | UJNSF0037 (PQ152276) | NR 119855.1 | 99.62% | 0 | 0 | 0 | 1 | 1 | 0 | ||
| Dothideomycetes | Subplenodomus | S. galicola | UJNSF0038 (PQ152277) | NR 154454.1 | 95.06% | 0 | 1 | 0 | 1 | 0 | 0 |
| Sordariomycetes | Trichoderma | T. longipile | UJNSF0039 (PQ152278) | NR 134354.1 | 99.83% | 2 | 0 | 0 | 0 | 0 | 0 |
| T. vinosum | UJNSF0040 (PQ152279) | NR 144870.1 | 99.81% | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Sordariomycetes | Apodus | A. deciduus | UJNSF0041 (PQ152280) | NR 145141.1 | 87.55% | 0 | 1 | 0 | 2 | 0 | 0 |
| Sordariomycetes | Apiosordaria | A. microcarpa | UJNSF0042 (PQ152281) | NR 175133.1 | 95.32% | 0 | 0 | 1 | 0 | 0 | 0 |
| Dothideomycetes | Alternaria | A. doliconidium | UJNSF0043 (PQ152282) | NR 137143.1 | 99.62% | 0 | 0 | 0 | 1 | 0 | 0 |
| Dothideomycetes | Paraboeremia | P. camelliae | UJNSF0044 (PQ152283) | NR 158251.1 | 99.38% | 0 | 0 | 0 | 0 | 0 | 1 |
| Dothideomycetes | Coniothyrium | C. hakeae | UJNSF0045 (PQ152284) | NR 154839.1 | 93.70% | 0 | 0 | 0 | 0 | 0 | 1 |
| Dothideomycetes | Preussia | P. flanaganii | UJNSF0046 (PQ152285) | NR 077168.1 | 97.74% | 1 | 0 | 0 | 0 | 0 | 0 |
| Sordariomycetes | Pochonia | P. chlamydosporia var. chlamydosporia | UJNSF0047 (PQ152286) | NR 171784.1 | 84.15% | 0 | 0 | 0 | 1 | 0 | 0 |
| Saccharomycetes | Geotrichum | G. silvicola | UJNSF0048 (PQ152287) | NR 077071.1 | 97.01% | 0 | 0 | 0 | 0 | 0 | 1 |
| Eurotiomycetes | Exophiala | E. radicis | UJNSF0049 (PQ152288) | NR 158397.1 | 100.00% | 0 | 0 | 0 | 0 | 0 | 1 |
| Sordariomycetes | Clonostachys | C. aranearum | UJNSF0050 (PQ152289) | NR 164542.1 | 99.28% | 0 | 0 | 0 | 0 | 0 | 1 |
| Mucoromycetes | Linnemannia | L. zychae | UJNSF0051 (PQ152290) | NR 111576.1 | 97.47% | 0 | 1 | 0 | 0 | 0 | 0 |
| Mucoromycetes | Podila | P. horticola | UJNSF0052 (PQ152291) | NR 111572.1 | 99.83% | 1 | 0 | 0 | 0 | 0 | 0 |
| Mucoromycetes | Mucor | M. hiemalis f. hiemalis | UJNSF0053 (PQ152292) | NR 152948.1 | 99.20% | 0 | 2 | 0 | 1 | 0 | 0 |
| M. moelleri | UJNSF0054 (PQ152293) | NR 111659.1 | 100.00% | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Mucoromycetes | Mortierella | M. rishikesha | UJNSF0055 (PQ152294) | NR 111564.1 | 98.16% | 1 | 1 | 0 | 1 | 0 | 0 |
| M. globalpina | UJNSF0056 (PQ152295) | NR 160121.1 | 94.99% | 0 | 0 | 0 | 1 | 0 | 0 | ||
| Agaricomycetes | Bjerkandera | B. ecuadorensis | UJNSF0057 (PQ152296) | NR 174062.1 | 85.63% | 0 | 0 | 0 | 1 | 0 | 0 |
| Dothideomycetes | Aaosphaeria | A. arxii | UJNSF0058 (PQ152297) | NR 169685.1 | 99.65% | 0 | 0 | 0 | 0 | 1 | 0 |
| Leotiomycetes | Scytalidium | S. circinatum | UJNSF0059 (PQ152298) | NR 160180.1 | 94.07% | 0 | 0 | 0 | 0 | 1 | 0 |
| Total | 17 | 21 | 20 | 38 | 14 | 22 | |||||
| Strain Number | Pathogenic Bacteria (Inhibition Zone Diameter/mm) | |||
|---|---|---|---|---|
| E. coli (G−) | V. parahaemolyticus (G−) | V. alginolyticus (G−) | S. aureus (G+) | |
| UJNSF0001 | - | - | 15.0 ± 0.1 | |
| UJNSF0002 | - | - | 14.5 ± 0.5 | |
| UJNSF0003 | - | 9.0 ± 0.2 | 15.0 ± 0.2 | 11.0 ± 0.4 |
| UJNSF0004 | - | - | 13.0 ± 0.2 | |
| UJNSF0006 | - | - | - | 11.0 ± 0.1 |
| UJNSF0008 | - | - | 10.0 ± 0.4 | |
| UJTSF0010 | - | - | - | 25.0 ± 0.5 |
| UJNSF0012 | - | - | - | 6.5 ± 0.5 |
| UJNSF0013 | - | - | - | 10.0 ± 0.5 |
| UJNSF0014 | 11.0 ± 0.5 | 12.0 ± 0.5 | 20.0 ± 0.4 | 16.0 ± 0.4 |
| UJNSF0017 | - | - | 16.0 ± 0.1 | 14.0 ± 0.5 |
| UJNSF0018 | - | - | - | 11.0 ± 0.5 |
| UJNSF0020 | - | - | - | 7.5 ± 0.5 |
| UJNSF0021 | - | - | - | 25.0 ± 0.2 |
| UJNSF0022 | - | - | - | 19.0 ± 0.4 |
| UJNSF0024 | - | - | - | 8.0 ± 0.2 |
| UJNSF0026 | - | 14.0 ± 0.3 | - | 27.0 ± 0.5 |
| UJNSF0034 | - | - | - | 22.5 ± 0.4 |
| UJNSF0035 | - | - | 12.0 ± 0.4 | 10.0 ± 0.4 |
| UJNSF0038 | - | - | - | 19.0 ± 0.5 |
| UJNSF0039 | - | 10.0 ± 0.2 | 22.0 ± 0.5 | 13.0 ± 0.5 |
| UJNSF0040 | - | - | - | 21.0 ± 0.4 |
| UJNSF0047 | - | - | - | 6.5 ± 0.1 |
| UJNSF0050 | - | - | - | 9.0 ± 0.5 |
| UJNSF0056 | - | - | - | 9.0 ± 0.2 |
| UJNSF0057 | - | - | - | 19.0 ± 0.5 |
| UJNSF0059 | - | - | - | 12.0 ± 0.2 |
| Strain Number | Medium | Pathogenic Bacteria (Inhibition Zone Diameter/mm) | |||
|---|---|---|---|---|---|
| E. coli (G−) | V. parahaemolyticus (G−) | V. alginolyticus (G−) | S. aureus (G+) | ||
| UJNSF0003 | 1# | - | - | - | - |
| 2# | - | - | - | - | |
| 3# | - | - | - | 10.50 ± 0.22 | |
| 4# | - | 7.83 ± 0.28 | - | 6.67 ± 0.17 | |
| UJNSF0014 | 1# | 7.67 ± 0.12 | 7.67 ± 0.33 | 10.33 ± 0.27 | 15.33 ± 0.25 |
| 2# | 9.33 ± 0.10 | 8.67 ± 0.23 | 9.67 ± 0.10 | 13.33 ± 0.24 | |
| 3# | 10.33 ± 0.35 | 9.33 ± 0.34 | 11.33 ± 0.15 | 7.33 ± 0.21 | |
| 4# | - | 7.33 ± 0.37 | - | 7.33 ± 0.26 | |
| UJNSF0017 | 1# | 7.33 ± 0.14 | - | 7.67 ± 0.37 | 12.67 ± 0.33 |
| 2# | 10.50 ± 0.33 | - | - | 15.33 ± 0.37 | |
| 3# | 9.33 ± 0.23 | - | 9.33 ± 0.33 | 13.33 ± 0.23 | |
| 4# | 7.67 ± 0.24 | - | - | 7.33 ± 0.32 | |
| UJNSF0026 | 1# | - | 7.33 ± 0.32 | - | 7.67 ± 0.17 |
| 2# | - | - | - | ||
| 3# | 7.83 ± 0.28 | - | 9.33 ±0.42 | ||
| 4# | - | 8.33 ± 0.27 | - | 8.67 ± 0.23 | |
| UJNSF0035 | 1# | - | 8.33 ± 0.33 | - | 9.67 ± 0.17 |
| 2# | - | 9.67 ± 0.25 | 7.33 ± 0.24 | 11.67 ± 0.27 | |
| 3# | - | 8.67 ± 0.17 | 8.00 ± 0.10 | 8.33 ± 0.12 | |
| 4# | - | 7.67 ± 0.22 | 7.67 ± 0.12 | 9.67 ± 0.13 | |
| UJNSF0039 | 1# | - | 9.33 ± 0.27 | 11.33 ± 0.27 | 7.67 ± 0.14 |
| 2# | 7.50 ± 0.23 | 7.33 ± 0.11 | - | 15.00 ± 0.22 | |
| 3# | 7.33 ± 0.21 | 7.67 ± 0.23 | 9.33 ± 0.23 | 10.67 ± 0.24 | |
| 4# | - | 9.33 ± 0.13 | - | 7.67 ± 0.16 | |
| Strain Number | Medium | E. coli (G−) | V. parahaemolyticus (G−) | V. alginolyticus (G−) | S. aureus (G+) |
|---|---|---|---|---|---|
| UJNSF0003 | 3# | - | - | - | 90.35% |
| UJNSF0014 | 1# | - | - | 80.30% | 97.18% |
| 2# | - | - | 83.18% | 89.57% | |
| 3# | 60.18% | 35.43% | 96.14% | - | |
| UJNSF0017 | 1# | - | - | 84.31% | - |
| 2# | 69.75% | - | - | 97.49% | |
| 3# | - | - | 58.87% | 96.76% | |
| 4# | - | - | - | - | |
| UJNSF0026 | 3# | - | - | - | 96.76% |
| 4# | - | 38.30% | - | 87.23% | |
| UJNSF0035 | 1# | - | - | - | 96.84% |
| 2# | - | 71.56% | 88.11% | 95.87% | |
| 3# | - | - | - | - | |
| 4# | - | - | 53.38% | - | |
| UJNSF0039 | 1# | - | 35.29% | 92.98% | - |
| 2# | - | - | - | 99.85% | |
| 3# | - | - | 73.79% | 93.32% | |
| 4# | - | 37.73% | - | - | |
| enrofloxacin | 100% | 74.03% | 100% | 97.83% | |
| norfloxacin | 89.12% | 100% | 98.29% | 99.86% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.-S.; Liu, H.; Li, X.-F.; Wang, C.-Y.; Zhang, X.; Wang, J.-J.; Song, F.; Bao, J.; Zhang, H. Exploring the Biodiversity and Antibacterial Potential of the Culturable Soil Fungi in Nyingchi, Tibet. J. Fungi 2025, 11, 276. https://doi.org/10.3390/jof11040276
Huang S-S, Liu H, Li X-F, Wang C-Y, Zhang X, Wang J-J, Song F, Bao J, Zhang H. Exploring the Biodiversity and Antibacterial Potential of the Culturable Soil Fungi in Nyingchi, Tibet. Journal of Fungi. 2025; 11(4):276. https://doi.org/10.3390/jof11040276
Chicago/Turabian StyleHuang, Shan-Shan, Haishan Liu, Xia-Fei Li, Chun-Ying Wang, Xiujun Zhang, Juan-Juan Wang, Fuhang Song, Jie Bao, and Hua Zhang. 2025. "Exploring the Biodiversity and Antibacterial Potential of the Culturable Soil Fungi in Nyingchi, Tibet" Journal of Fungi 11, no. 4: 276. https://doi.org/10.3390/jof11040276
APA StyleHuang, S.-S., Liu, H., Li, X.-F., Wang, C.-Y., Zhang, X., Wang, J.-J., Song, F., Bao, J., & Zhang, H. (2025). Exploring the Biodiversity and Antibacterial Potential of the Culturable Soil Fungi in Nyingchi, Tibet. Journal of Fungi, 11(4), 276. https://doi.org/10.3390/jof11040276






