Echinocandin Adaptation in Candida albicans Is Accompanied by Altered Chromatin Accessibility at Gene Promoters and by Cell Wall Remodeling
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, M.C.; Gurr, S.J.; Cuomo, C.A.; Blehert, D.S.; Jin, H.; Stukenbrock, E.H.; Stajich, J.E.; Kahmann, R.; Boone, C.; Denning, D.W.; et al. Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. mBio 2020, 11, e00449-20. [Google Scholar] [CrossRef]
- Kainz, K.; Bauer, M.A.; Madeo, F.; Carmona-Gutierrez, D. Fungal infections in humans: The silent crisis. Microb. Cell 2020, 7, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The Still Underestimated Problem of Fungal Diseases Worldwide. Front. Microbiol. 2019, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Denning, D.W. The WHO fungal priority pathogens list as a game-changer. Nat. Rev. Microbiol. 2023, 21, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond. Chem. Rev. 2021, 121, 3390–3411. [Google Scholar] [CrossRef] [PubMed]
- Puumala, E.; Fallah, S.; Robbins, N.; Cowen, L.E. Advancements and challenges in antifungal therapeutic development. Clin. Microbiol. Rev. 2024, 37, e0014223. [Google Scholar] [CrossRef] [PubMed]
- Yiu, B.; Robbins, N.; Cowen, L.E. Interdisciplinary approaches for the discovery of novel antifungals. Trends Mol. Med. 2024, 30, 723–735. [Google Scholar] [CrossRef]
- Perlin, D.S. Echinocandin resistance, susceptibility testing and prophylaxis: Implications for patient management. Drugs 2014, 74, 1573–1585. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R., 3rd; Soriano, A.; Honore, P.M.; Bassetti, M.; Cornely, O.A.; Kollef, M.; Kullberg, B.J.; Pullman, J.; Hites, M.; Fortun, J.; et al. Efficacy and safety of rezafungin and caspofungin in candidaemia and invasive candidiasis: Pooled data from two prospective randomised controlled trials. Lancet Infect. Dis. 2024, 24, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Perlin, D.S. Echinocandin resistance: An emerging clinical problem? Curr. Opin. Infect. Dis. 2014, 27, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Woosley, L.N.; Jones, R.N.; Castanheira, M. Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: Application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J. Clin. Microbiol. 2013, 51, 2571–2581. [Google Scholar] [CrossRef] [PubMed]
- Grossman, N.T.; Chiller, T.M.; Lockhart, S.R. Epidemiology of echinocandin resistance in Candida. Curr. Fungal Infect. Rep. 2014, 8, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Bhattacharya, S.; Yadav, A.; Husain, F.; Ndiaye, A.; Kruppa, M.D.; Hayes, J.J.; Rustchenko, E. Multiple Genes of Candida albicans Influencing Echinocandin Susceptibility in Caspofungin-Adapted Mutants. Antimicrob. Agents Chemother. 2022, 66, e0097722. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Yadav, A.; Kruppa, M.D.; Rustchenko, E. Identification of 10 genes on Candida albicans chromosome 5 that control surface exposure of the immunogenic cell wall epitope beta-glucan and cell wall remodeling in caspofungin-adapted mutants. Microbiol. Spectr. 2023, 11, e0329523. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Perlin, D.S. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J. Fungi 2018, 4, 105. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Sah, S.K.; Perlin, D.S.; Rustchenko, E. Candida albicans Genes Modulating Echinocandin Susceptibility of Caspofungin-Adapted Mutants Are Constitutively Expressed in Clinical Isolates with Intermediate or Full Resistance to Echinocandins. J. Fungi 2024, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Hayes, J.J.; Rustchenko, E. The role of aneuploidy in the emergence of echinocandin resistance in human fungal pathogen Candida albicans. PLoS Pathog. 2021, 17, e1009564. [Google Scholar] [CrossRef]
- Singh-Babak, S.D.; Babak, T.; Diezmann, S.; Hill, J.A.; Xie, J.L.; Chen, Y.L.; Poutanen, S.M.; Rennie, R.P.; Heitman, J.; Cowen, L.E. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012, 8, e1002718. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, Y.; Aptekmann, A.A.; Keniya, M.V.; Gomez, R.Y.; Alayo, N.; Novi, G.; Quinteros, C.; Kaya, F.; Zimmerman, M.; Caceres, D.H.; et al. Evolutionary dynamics in gut-colonizing Candida glabrata during caspofungin therapy: Emergence of clinically important mutations in sphingolipid biosynthesis. PLoS Pathog. 2024, 20, e1012521. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, L.; Wakabayashi, H.; Myers, J.; Jiang, Y.; Cao, Y.; Jimenez-Ortigosa, C.; Perlin, D.S.; Rustchenko, E. Tolerance to Caspofungin in Candida albicans Is Associated with at Least Three Distinctive Mechanisms That Govern Expression of FKS Genes and Cell Wall Remodeling. Antimicrob. Agents Chemother. 2017, 61, e00071-17. [Google Scholar] [CrossRef]
- Zuber, J.; Sah, S.K.; Mathews, D.H.; Rustchenko, E. Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants. Microorganisms 2023, 11, 1870. [Google Scholar] [CrossRef] [PubMed]
- Torres-Garcia, S.; Yaseen, I.; Shukla, M.; Audergon, P.; White, S.A.; Pidoux, A.L.; Allshire, R.C. Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 2020, 585, 453–458. [Google Scholar] [CrossRef]
- Husain, F.; Yadav, A.; Sah, S.K.; Hayes, J.J.; Rustchenko, E. Candida albicans Strains Adapted to Caspofungin Due to Aneuploidy Become Highly Tolerant under Continued Drug Pressure. Microorganisms 2022, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zibetti, C.; Shang, P.; Sripathi, S.R.; Zhang, P.; Cano, M.; Hoang, T.; Xia, S.; Ji, H.; Merbs, S.L.; et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat. Commun. 2018, 9, 1364. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Hou, X.; Liu, X.; Wang, L.; Gao, H.; Zhao, F.; Shi, L.; Shi, L.; Yan, H.; Deng, T.; et al. The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs. J. Anim. Sci. Biotechnol. 2021, 12, 56. [Google Scholar] [CrossRef]
- Wilbanks, E.G.; Facciotti, M.T. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE 2010, 5, e11471. [Google Scholar] [CrossRef]
- Jenull, S.; Tscherner, M.; Mair, T.; Kuchler, K. ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. J. Fungi 2020, 6, 182. [Google Scholar] [CrossRef] [PubMed]
- Mille, C.; Bobrowicz, P.; Trinel, P.A.; Li, H.; Maes, E.; Guerardel, Y.; Fradin, C.; Martinez-Esparza, M.; Davidson, R.C.; Janbon, G.; et al. Identification of a new family of genes involved in beta-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans. J. Biol. Chem. 2008, 283, 9724–9736. [Google Scholar] [CrossRef] [PubMed]
- Pedreno, Y.; Maicas, S.; Arguelles, J.C.; Sentandreu, R.; Valentin, E. The ATC1 gene encodes a cell wall-linked acid trehalase required for growth on trehalose in Candida albicans. J. Biol. Chem. 2004, 279, 40852–40860. [Google Scholar] [CrossRef]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Copping, V.M.; Barelle, C.J.; Hube, B.; Gow, N.A.; Brown, A.J.; Odds, F.C. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 2005, 55, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, N.P.; Kontoyiannis, D.P.; Prince, R.A.; Lewis, R.E. Attenuation of the activity of caspofungin at high concentrations against candida albicans: Possible role of cell wall integrity and calcineurin pathways. Antimicrob. Agents Chemother. 2005, 49, 5146–5148. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.S.; Lumsdaine, S.W.; Mangrum, M.M.; Reynolds, T.B. Caspofungin-induced beta(1,3)-glucan exposure in Candida albicans is driven by increased chitin levels. mBio 2023, 14, e0007423. [Google Scholar] [CrossRef]
- Han, Q.; Wang, N.; Pan, C.; Wang, Y.; Sang, J. Elevation of cell wall chitin via Ca2+-calcineurin-mediated PKC signaling pathway maintains the viability of Candida albicans in the absence of beta-1,6-glucan synthesis. Mol. Microbiol. 2019, 112, 960–972. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Sahni, N.; Daniels, K.J.; Lu, K.L.; Huang, G.; Garnaas, A.M.; Pujol, C.; Srikantha, T.; Soll, D.R. Utilization of the mating scaffold protein in the evolution of a new signal transduction pathway for biofilm development. mBio 2011, 2, e00237-10. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, B.; Ketela, T.; Lemieux, S.; Veillette, K.; Martel, N.; Davison, J.; Sillaots, S.; Trosok, S.; Bachewich, C.; et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007, 3, e92. [Google Scholar] [CrossRef]
- Liu, T.T.; Lee, R.E.; Barker, K.S.; Lee, R.E.; Wei, L.; Homayouni, R.; Rogers, P.D. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Liston, S.D.; Lee, D.; Robbins, N.; Cowen, L.E. Functional analysis of the Candida albicans kinome reveals Hrr25 as a regulator of antifungal susceptibility. iScience 2022, 25, 104432. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wagner, A.S.; Reynolds, T.B. When Is It Appropriate to Take Off the Mask? Signaling Pathways That Regulate ss(1,3)-Glucan Exposure in Candida albicans. Front. Fungal Biol. 2022, 3, 842501. [Google Scholar] [CrossRef] [PubMed]
- Iyer, K.R.; Robbins, N.; Cowen, L.E. The role of Candida albicans stress response pathways in antifungal tolerance and resistance. iScience 2022, 25, 103953. [Google Scholar] [CrossRef]
- Navarro-Garcia, F.; Alonso-Monge, R.; Rico, H.; Pla, J.; Sentandreu, R.; Nombela, C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology (Reading) 1998, 144 Pt 2, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, Y.; Zhang, B.; Zheng, W.; Xing, L.; Li, M. Alkaline stress triggers an immediate calcium fluctuation in Candida albicans mediated by Rim101p and Crz1p transcription factors. FEMS Yeast Res. 2011, 11, 430–439. [Google Scholar] [CrossRef]
- Thewes, S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell 2014, 13, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Qasim, M.N.; Valle Arevalo, A.; Nobile, C.J.; Hernday, A.D. The Roles of Chromatin Accessibility in Regulating the Candida albicans White-Opaque Phenotypic Switch. J. Fungi 2021, 7, 37. [Google Scholar] [CrossRef]
- Zhou, X.; O’Shea, E.K. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell 2011, 42, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Ikeh, M.A.; Kastora, S.L.; Day, A.M.; Herrero-de-Dios, C.M.; Tarrant, E.; Waldron, K.J.; Banks, A.P.; Bain, J.M.; Lydall, D.; Veal, E.A.; et al. Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol. Biol. Cell 2016, 27, 2784–2801. [Google Scholar] [CrossRef] [PubMed]
- Douglas, C.M. Fungal beta(1,3)-D-glucan synthesis. Med. Mycol. 2001, 39 (Suppl. S1), 55–66. [Google Scholar] [CrossRef]
- Mio, T.; Adachi-Shimizu, M.; Tachibana, Y.; Tabuchi, H.; Inoue, S.B.; Yabe, T.; Yamada-Okabe, T.; Arisawa, M.; Watanabe, T.; Yamada-Okabe, H. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol. 1997, 179, 4096–4105. [Google Scholar] [CrossRef]
- Lussier, M.; Sdicu, A.M.; Shahinian, S.; Bussey, H. The Candida albicans KRE9 gene is required for cell wall beta-1, 6-glucan synthesis and is essential for growth on glucose. Proc. Natl. Acad. Sci. USA 1998, 95, 9825–9830. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Gow, N.A. Mannosylation in Candida albicans: Role in cell wall function and immune recognition. Mol. Microbiol. 2013, 90, 1147–1161. [Google Scholar] [CrossRef]
- Kruppa, M.; Jabra-Rizk, M.A.; Meiller, T.F.; Calderone, R. The histidine kinases of Candida albicans: Regulation of cell wall mannan biosynthesis. FEMS Yeast Res. 2004, 4, 409–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Munro, C.A.; de Bruijn, I.; Lenardon, M.D.; McKinnon, A.; Gow, N.A.R. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008, 4, e1000040. [Google Scholar] [CrossRef]
- Ren, Z.; Chhetri, A.; Guan, Z.; Suo, Y.; Yokoyama, K.; Lee, S.Y. Structural basis for inhibition and regulation of a chitin synthase from Candida albicans. Nat. Struct. Mol. Biol. 2022, 29, 653–664. [Google Scholar] [CrossRef]
- Mio, T.; Yabe, T.; Sudoh, M.; Satoh, Y.; Nakajima, T.; Arisawa, M.; Yamada-Okabe, H. Role of three chitin synthase genes in the growth of Candida albicans. J. Bacteriol. 1996, 178, 2416–2419. [Google Scholar] [CrossRef] [PubMed]
- Iracane, E.; Vega-Estevez, S.; Buscaino, A. On and Off: Epigenetic Regulation of C. albicans Morphological Switches. Pathogens 2021, 10, 1463. [Google Scholar] [CrossRef] [PubMed]
- Jenull, S.; Mair, T.; Tscherner, M.; Penninger, P.; Zwolanek, F.; Silao, F.S.; de San Vicente, K.M.; Riedelberger, M.; Bandari, N.C.; Shivarathri, R.; et al. The histone chaperone HIR maintains chromatin states to control nitrogen assimilation and fungal virulence. Cell Rep. 2021, 36, 109406. [Google Scholar] [CrossRef]
- Lopes da Rosa, J.; Kaufman, P.D. Chromatin-mediated Candida albicans virulence. Biochim. Biophys. Acta 2012, 1819, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Ma, Z.; Tang, Z.; Yu, L.; Liu, S.; Huang, T.; Wang, P.; Wu, T.; Song, Z.; Zhang, H.; et al. Integrative ATAC-seq and RNA-seq Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 742672. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Huang, W.; Liu, G.; Zhai, B.; Yan, H.; Zhang, Y. Integration of ATAC-Seq and RNA-Seq reveals FOSL2 drives human liver progenitor-like cell aging by regulating inflammatory factors. BMC Genom. 2023, 24, 260. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.J. Action at a distance: Epigenetic silencing of large chromosomal regions in carcinogenesis. Hum. Mol. Genet. 2007, 16, R88–R95. [Google Scholar] [CrossRef] [PubMed]
- Rustchenko-Bulgac, E.P. Variations of Candida albicans electrophoretic karyotypes. J. Bacteriol. 1991, 173, 6586–6596. [Google Scholar] [CrossRef] [PubMed]
- Rustchenko, E. Chromosome instability in Candida albicans. FEMS Yeast Res. 2007, 7, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. Gencode 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Stark, R.; Brown, G. DiffBind: Differential Binding Analysis of ChIP-Seq Peak Data, 3.2. 2011. Available online: http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf (accessed on 18 April 2024).
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
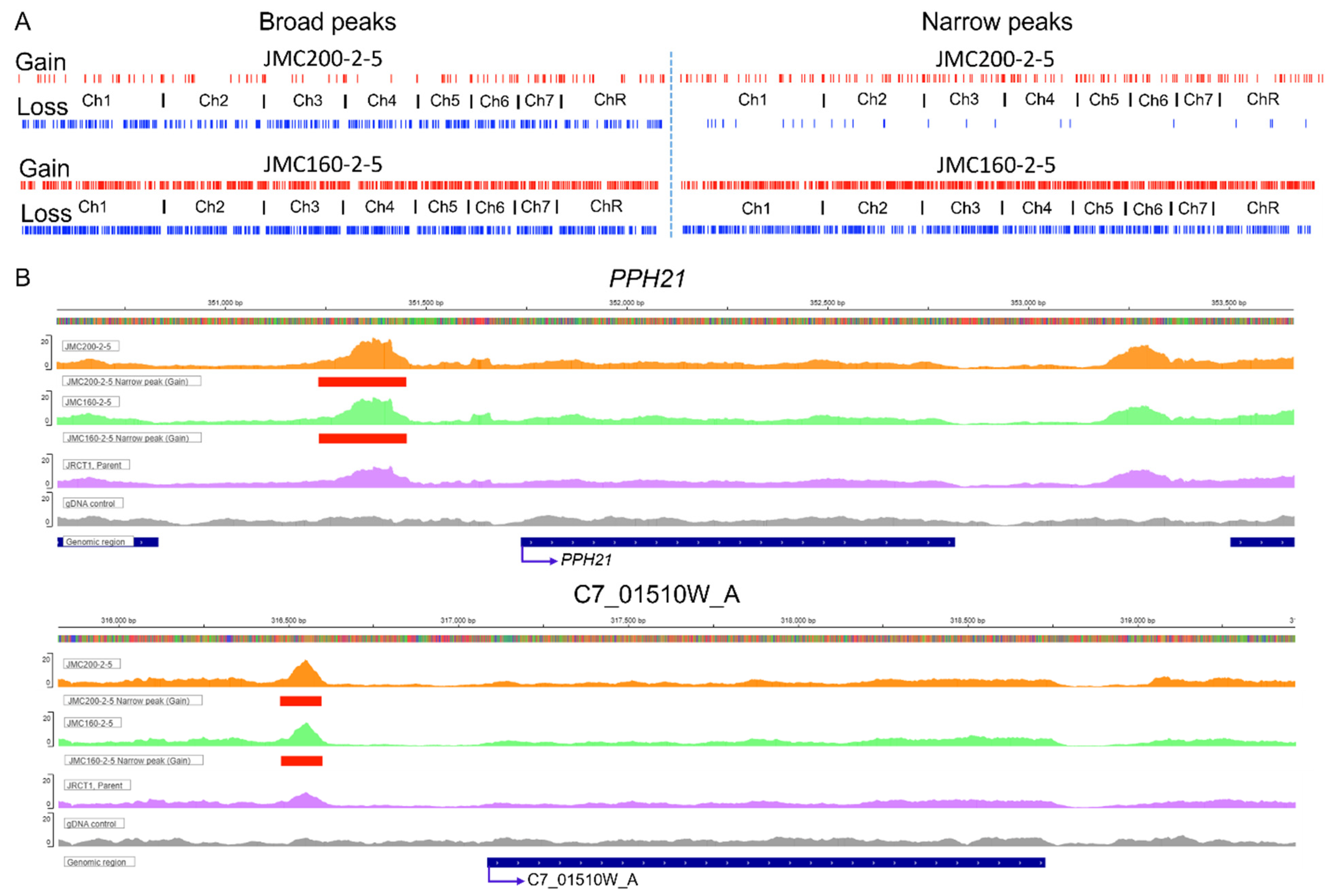
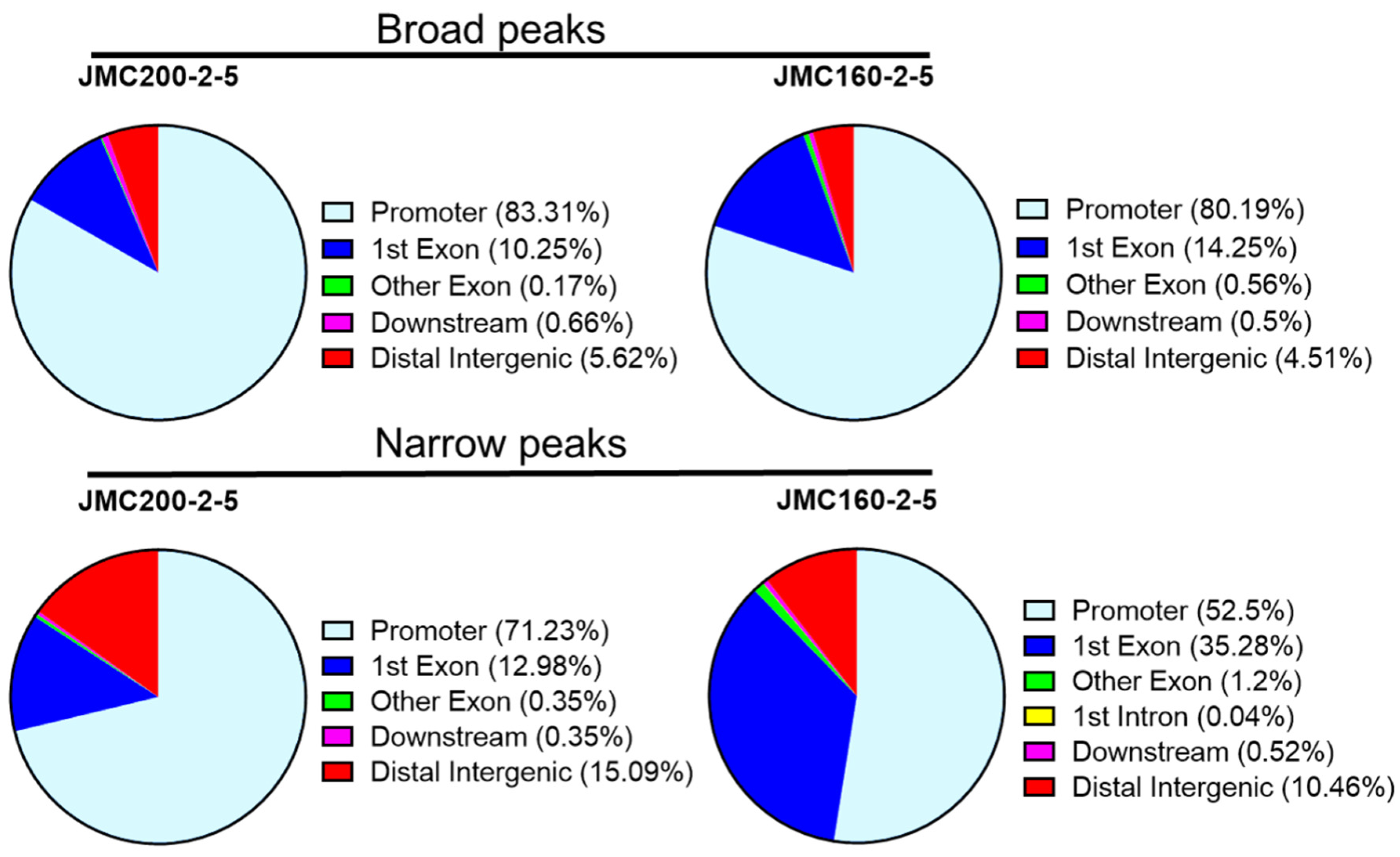
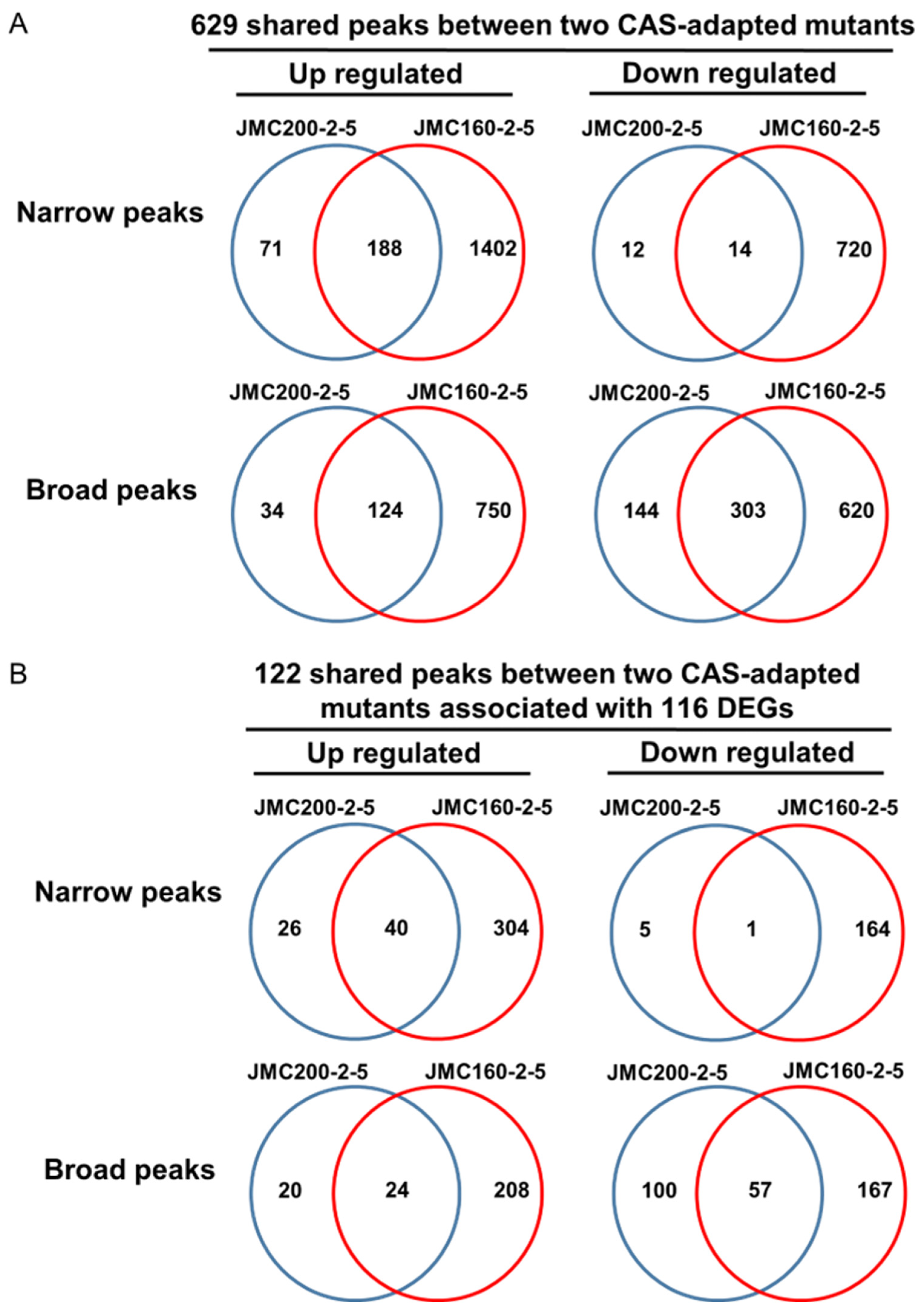
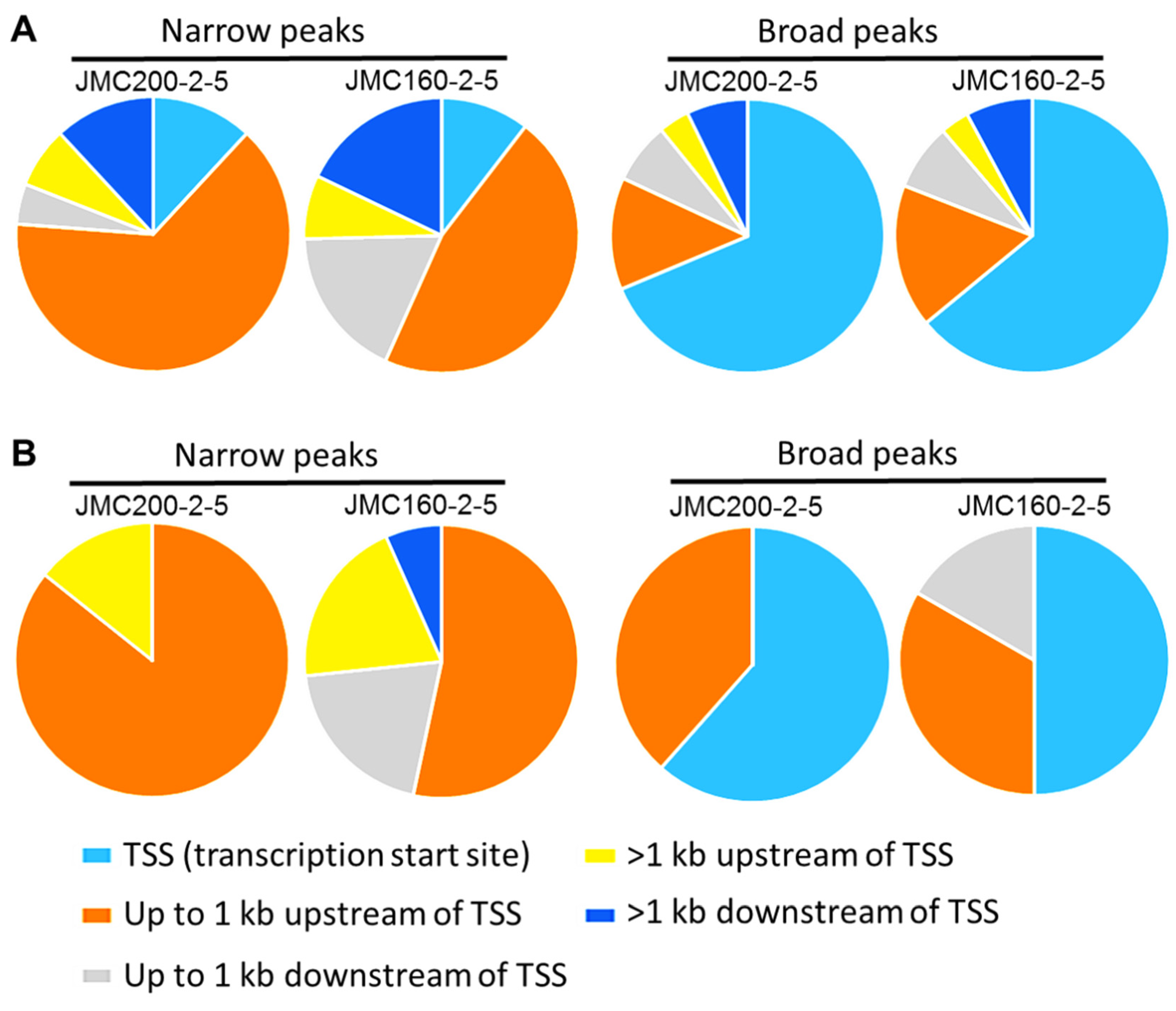

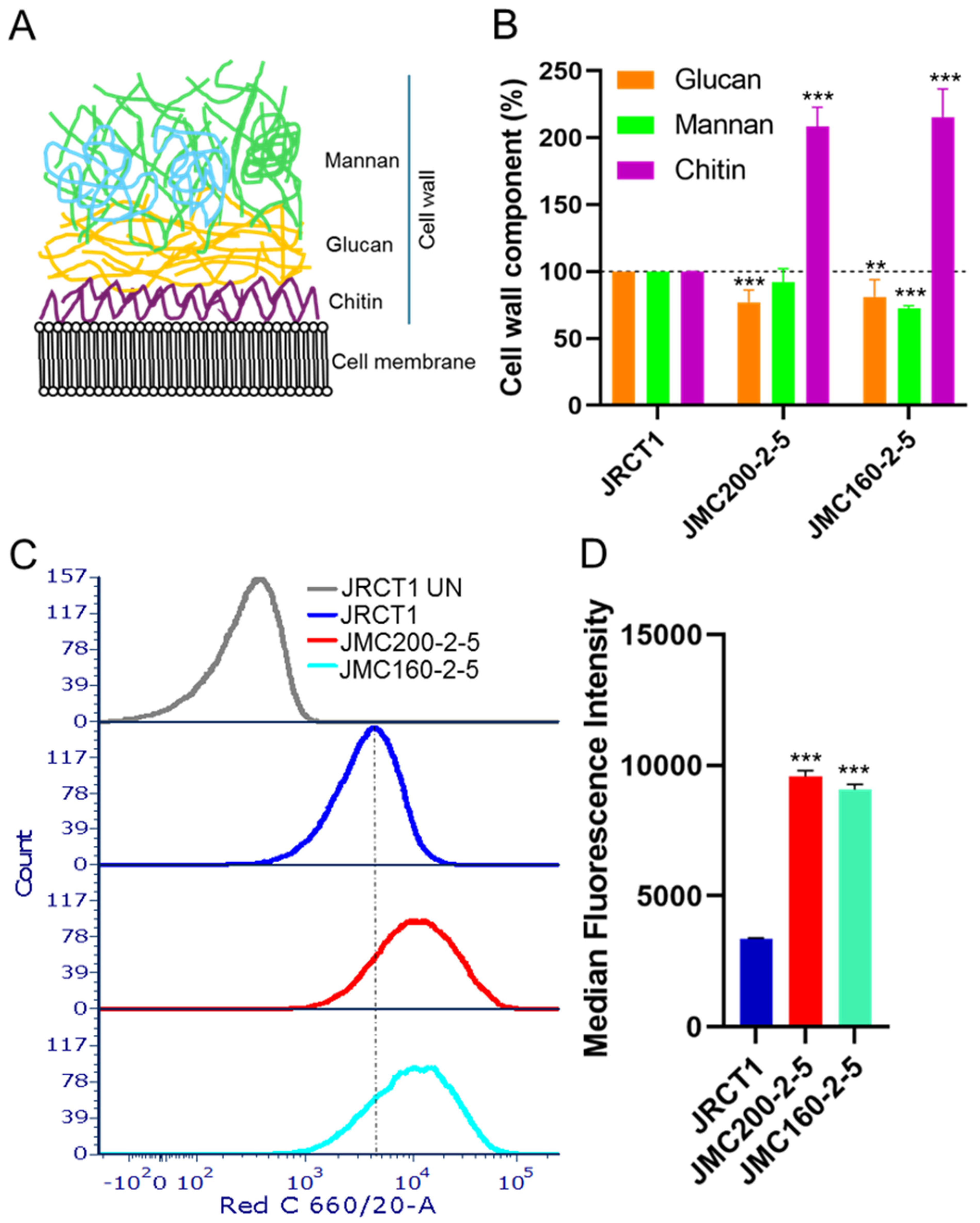
| Ch | Peaks in JMC200-2-5 | Peaks in JMC160-2-5 | ||||||
|---|---|---|---|---|---|---|---|---|
| Broad | Narrow | Broad | Narrow | |||||
| Gain | Loss | Gain | Loss | Gain | Loss | Gain | Loss | |
| 1 | 34 | 115 | 48 | 10 | 179 | 233 | 300 | 191 |
| 2 | 22 | 47 | 32 | 5 | 140 | 133 | 243 | 98 |
| 3 | 15 | 59 | 39 | 4 | 106 | 129 | 205 | 102 |
| 4 | 6 | 47 | 26 | 2 | 89 | 97 | 173 | 78 |
| 5 | 12 | 53 | 26 | 0 | 71 | 79 | 144 | 66 |
| 6 | 19 | 35 | 22 | 1 | 68 | 63 | 131 | 67 |
| 7 | 21 | 29 | 26 | 0 | 61 | 57 | 125 | 37 |
| R | 29 | 62 | 40 | 4 | 160 | 132 | 269 | 95 |
| Total in column | 158 | 447 | 259 | 26 | 874 | 923 | 1590 | 734 |
| Total | 605 | 285 | 1797 | 2324 | ||||
| Mutant | Total DEGs | Total ATAC-Seq Peaks | % Peaks and Associated DEGs Changing in the Same Direction |
|---|---|---|---|
| JMC200-2-5 | 2664 | 890 | 24.26 |
| JMC160-2-5 | 3135 | 4121 | 23.41 |
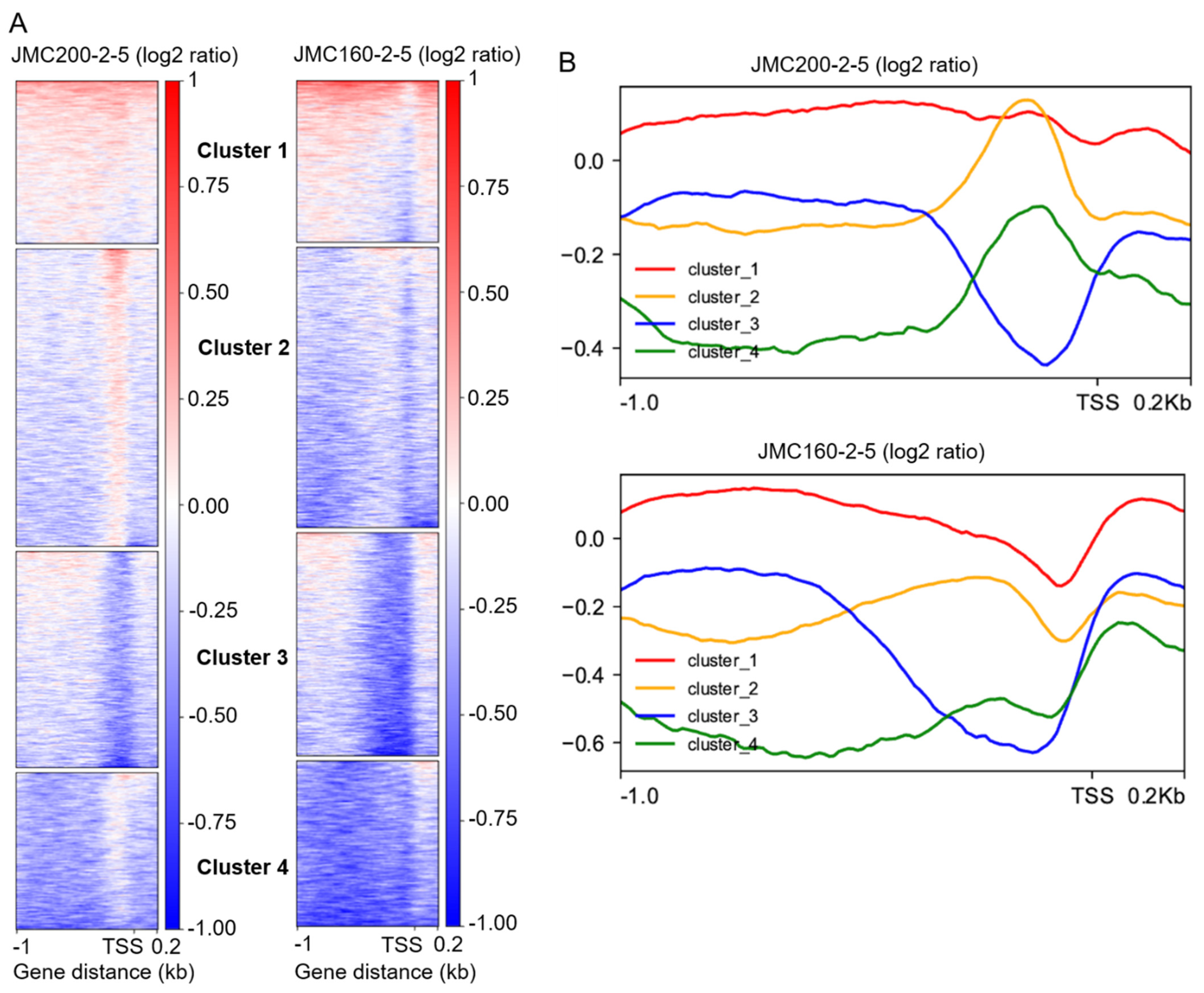
| Cluster | JMC200-2-5 | JMC160-2-5 | DEGs in Common | |
|---|---|---|---|---|
| Total | With Function/No Function | |||
| 1 | 33 | 27 | 25 | 11/14 |
| 2 | 22 | 35 | 13 | 9/4 |
| 3 | 32 | 21 | 14 | 10/4 |
| 4 | 22 | 33 | 19 | 13/6 |
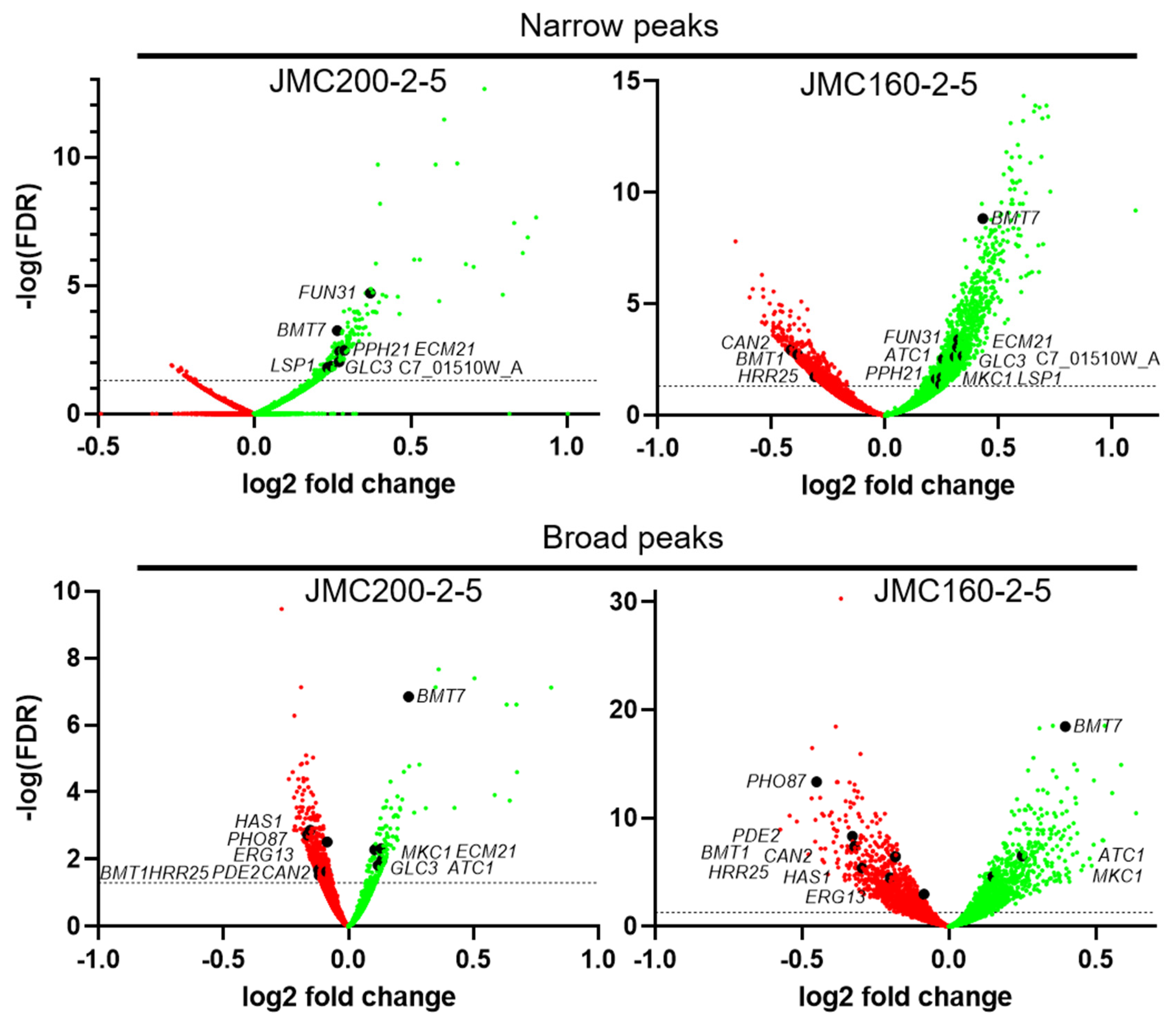
| Standard Name | Assembly 19/21 Identifier | Systematic Name | RNA-Seq Ratio | ATAC-Seq Peak Ratio | ||||
|---|---|---|---|---|---|---|---|---|
| JMC200-2-5 | JMC160-2-5 | JMC200-2-5 | JMC160-2-5 | |||||
| Narrow | Broad | Narrow | Broad | |||||
| Caspofungin responsive genes | ||||||||
| NA * | orf19.6578 | C7_01510W_A | 2.52 | 3.12 | 1.21 | NA | 1.27 | NA |
| ECM21 | orf19.4887 | C1_10180C_A | 2.03 | 1.63 | 1.21 | 1.1 | 1.25 | NA |
| LSP1 | orf19.3149 | C2_06730W_A | 1.28 | 1.25 | 1.18 | NA | 1.17 | NA |
| PPH21 | orf19.1683 | C3_01600W_A | 1.66 | 2.06 | 1.22 | NA | 1.19 | NA |
| MKC1 ** | orf19.7523 | CR_00120C_A | 1.46 | 1.72 | NA | 1.08 | 1.18 | 1.11 |
| HAS1 | orf19.3962 | C5_04750C_A | 0.56 | 0.35 | NA | 0.9 | NA | 0.87 |
| CAN2 † | orf19.111 | C6_01060C_A | 0.36 | 0.36 | NA | 0.94 | 0.75 0.83 | 0.88 |
| PHO87 | orf19.2454 | C1_05940W_A | 0.76 | 0.49 | NA | 0.89 | NA | 0.73 |
| ERG13 | orf19.7312 | CR_09160C_A | 0.39 | 0.37 | NA | 0.94 | NA | 0.94 |
| Cell wall synthesis genes | ||||||||
| GLC3 | orf19.5622 | C6_03340C_A | 1.93 | 2.20 | 1.19 | 1.1 | 1.24 | NA |
| BMT7 | orf19.342 | C3_03450C_A | 2.18 | 1.91 | 1.2 | 1.18 | 1.35 | 1.32 |
| FUN31 | orf19.7451 | C3_06620W_A | 1.89 | 1.54 | 1.29 | NA | 1.25 | NA |
| MKC1 ** | orf19.7523 | CR_00120C_A | 1.46 | 1.72 | NA | 1.08 | 1.18 | 1.11 |
| ATC1 | orf19.6214 | C1_06940C_A | 1.57 | 1.92 | NA | 1.09 | 1.19 | 1.19 |
| PDE2 ‡ | orf19.2972 | C1_02840W_A | 0.64 | 0.72 | NA | 0.92 | NA | 0.8 1.17 |
| HRR25 | orf19.3476 | C6_02340W_A | 0.78 | 0.67 | NA | 0.92 | 0.81 | 0.81 |
| BMT1 † | orf19.6782 | C3_07180C_A | 0.40 | 0.38 | NA | 0.92 | 0.77 0.83 | 0.8 |
| Mutant | Number of Peaks | Number of Genes | Ratio (Peaks/Genes) |
|---|---|---|---|
| Caspofungin-responsive and cell wall synthesis genes | |||
| JMC200-2-5 | 19 | 16 | 1.2 |
| JMC160-2-5 | 25 | 16 | 1.6 |
| Entire genome in C. albicans caspofungin-adapted mutants | |||
| JMC200-2-5 | 890 | 6182 | 0.1 |
| JMC160-2-5 | 4121 | 6182 | 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sah, S.K.; Yadav, A.; Stahl, T.; Hayes, J.J.; Bulger, M.; Rustchenko, E. Echinocandin Adaptation in Candida albicans Is Accompanied by Altered Chromatin Accessibility at Gene Promoters and by Cell Wall Remodeling. J. Fungi 2025, 11, 110. https://doi.org/10.3390/jof11020110
Sah SK, Yadav A, Stahl T, Hayes JJ, Bulger M, Rustchenko E. Echinocandin Adaptation in Candida albicans Is Accompanied by Altered Chromatin Accessibility at Gene Promoters and by Cell Wall Remodeling. Journal of Fungi. 2025; 11(2):110. https://doi.org/10.3390/jof11020110
Chicago/Turabian StyleSah, Sudisht K., Anshuman Yadav, Tyler Stahl, Jeffrey J. Hayes, Michael Bulger, and Elena Rustchenko. 2025. "Echinocandin Adaptation in Candida albicans Is Accompanied by Altered Chromatin Accessibility at Gene Promoters and by Cell Wall Remodeling" Journal of Fungi 11, no. 2: 110. https://doi.org/10.3390/jof11020110
APA StyleSah, S. K., Yadav, A., Stahl, T., Hayes, J. J., Bulger, M., & Rustchenko, E. (2025). Echinocandin Adaptation in Candida albicans Is Accompanied by Altered Chromatin Accessibility at Gene Promoters and by Cell Wall Remodeling. Journal of Fungi, 11(2), 110. https://doi.org/10.3390/jof11020110






