Harnessing Filamentous Fungi for Enzyme Cocktail Production Through Rice Bran Bioprocessing
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolation and Growth Conditions
2.2. Phenotypic Characterization of the Fungal Strains
2.3. Molecular Identification of the Fungal Strains
2.4. Phylogenetic Analysis
2.5. Protein Production, Quantification, and SDS-PAGE Analyses
2.6. Enzymatic Activities
2.7. Proteomics Analysis
2.8. Identification of Antifungal Protein- (AFP) Coding Sequences
2.9. Statistical Analysis
3. Results
3.1. Fungal Isolation from Rice Bran and Species Identification
3.2. Protein Secretion by Different Fungal Isolates
3.3. Assessment of Rice Bran Degradation Abilities Through Enzymatic Activity Determination in Different Fungal Isolates
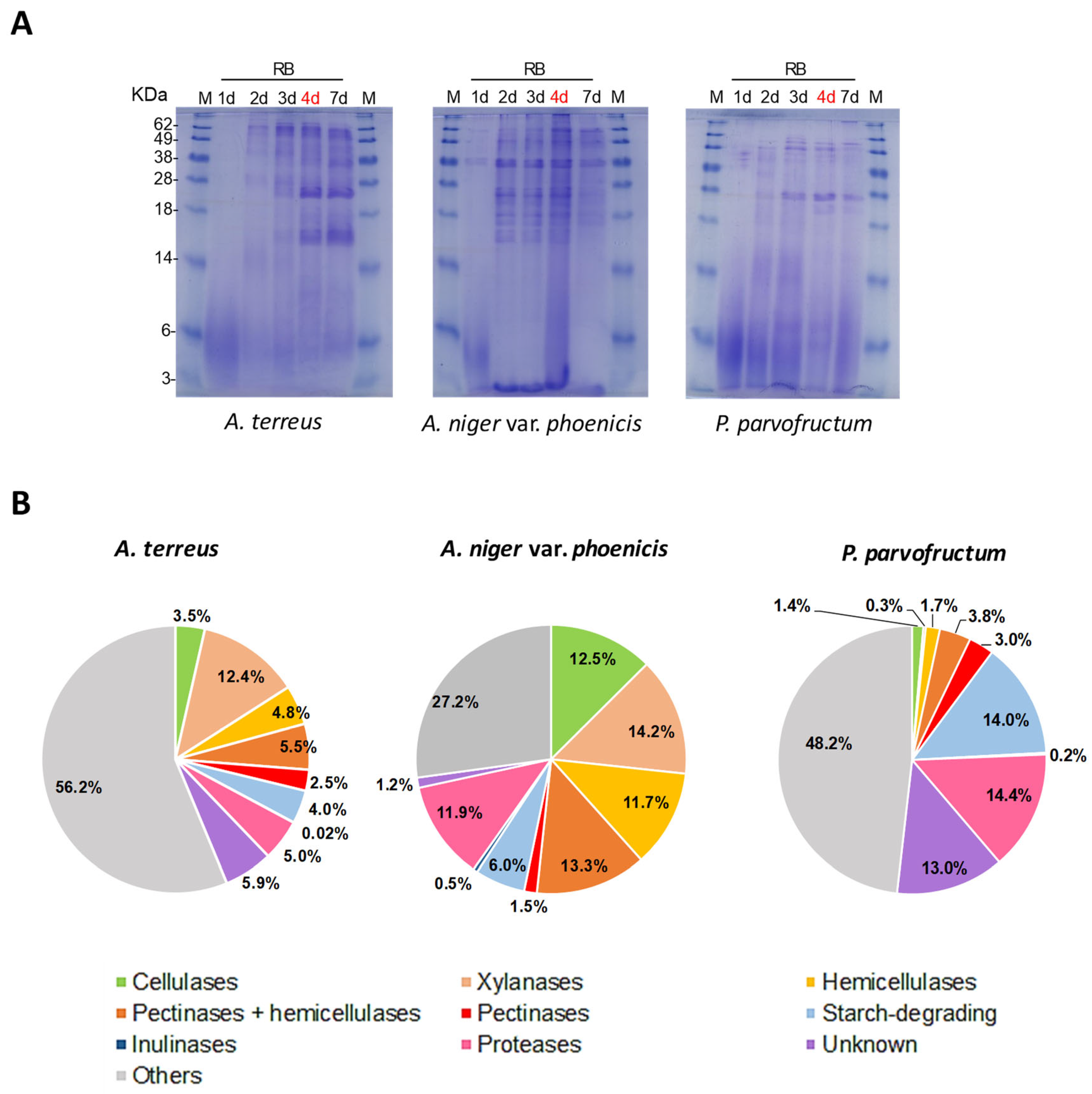
3.4. Proteomic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Otten, L.G.; Quax, W.J. Directed evolution: Selecting today’s biocatalysts. Biomol. Eng. 2005, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sarangi, P.K.; Singh, A.K.; Ganachari, S.V.; Pengadeth, D.; Mohanakrishna, G.; Aminabhavi, T.M. Biobased heterogeneous renewable catalysts: Production technologies, innovations, biodiesel applications and circular bioeconomy. Environ. Res. 2024, 261, 119745. [Google Scholar] [CrossRef]
- de Vries, R.P.; Patyshakuliyeva, A.; Garrigues, S.; Agarwal-Jans, S. The current biotechnological status and potential of plant and algal biomass degrading/modifying enzymes from ascomycete fungi. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 81–120. [Google Scholar]
- de Vries, R.P.; Makela, M.R. Genomic and postgenomic diversity of fungal plant biomass degradation approaches. Trends Microbiol. 2020, 28, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Bouws, H.; Wattenberg, A.; Zorn, H. Fungal secretomes—Nature’s toolbox for white biotechnology. Appl. Microbiol. Biotechnol. 2008, 80, 381–388. [Google Scholar] [CrossRef]
- Sohail, M.; Rakha, A.; Butt, M.S.; Iqbal, M.J.; Rashid, S. Rice bran nutraceutics: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2017, 57, 3771–3780. [Google Scholar] [CrossRef] [PubMed]
- FAO. Paddy Rice Production Worldwide in 2022, by Country (in Million Metric Tons). Statista. 2024. Available online: https://www.statista.com/statistics/255937/leading-rice-producers-worldwide/#statisticContainer (accessed on 30 May 2024).
- Sapwarobol, S.; Saphyakhajorn, W.; Astina, J. Biological functions and activities of rice bran as a functional ingredient: A review. Nutr. Metab. Insights 2021, 14, 11786388211058559. [Google Scholar] [CrossRef] [PubMed]
- Ran, Q.; Li, H.; Liu, J.; Huang, M.; Zhou, Y.; Zhang, L.; Gu, L.; Jiang, Z. An effective strategy for improving the specific activity and saccharification efficiency of cellulase by pre-incubation with phenolic acids. Bioresour. Technol. 2022, 346, 126644. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Donofrio, N.; de Vries, R.P. Plant biomass degradation by fungi. Fungal Genet. Biol. 2014, 72, 2–9. [Google Scholar] [CrossRef] [PubMed]
- de Vries, R.P.; Burgers, K.; van de Vondervoort, P.; Frisvad Jens, C.; Samson Robert, A.; Visser, J. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl. Environ. Microbiol. 2004, 70, 3954–3959. [Google Scholar] [CrossRef]
- Nyongesa, B.W.; Okoth, S.; Ayugi, V. Identification key for Aspergillus species isolated from maize and soil of nandi county, Kenya. Adv. Microbiol. 2015, 05, 205–229. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Le Cam, L. Maximum likelihood: An introduction. Int. Stat. Rev. 1990, 58, 153–171. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kwon, Y.K.; Kim, J.H.; Heo, S.J.; Lee, Y.; Lee, S.J.; Shim, W.B.; Jung, W.K.; Hyun, J.H.; Kwon, K.K.; et al. Effective microwell plate-based screening method for microbes producing cellulase and xylanase and its application. J. Microbiol. Biotechnol. 2014, 24, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Kun, R.S.; Garrigues, S.; Di Falco, M.; Tsang, A.; de Vries, R.P. Blocking utilization of major plant biomass polysaccharides leads Aspergillus niger towards utilization of minor components. Microb. Biotechnol. 2021, 14, 1683–1698. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [PubMed]
- Gruben, B.S.; Zhou, M.; de Vries, R.P. GalX regulates the D-galactose oxido-reductive pathway in Aspergillus niger. FEBS Lett. 2012, 586, 3980–3985. [Google Scholar] [CrossRef] [PubMed]
- Kun, R.S.; Meng, J.; Salazar-Cerezo, S.; Mäkelä, M.R.; de Vries, R.P.; Garrigues, S. CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass. Enzyme Microb. Technol. 2020, 136, 109508. [Google Scholar] [CrossRef]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef]
- Klich, M.A.; Mullaney, E.J.; Daly, C.B.; Cary, J.W. Molecular and physiological aspects of aflatoxin and sterigmatocystin biosynthesis by Aspergillus tamarii and A. ochraceoroseus. Appl. Microbiol. Biotechnol. 2000, 53, 605–609. [Google Scholar] [CrossRef]
- Theobald, S.; Vesth, T.C.; Geib, E.; Nybo, J.L.; Frisvad, J.C.; Larsen, T.O.; Kuo, A.; LaButti, K.; Lyhne, E.K.; Kjærbølling, I.; et al. Genomic analysis of Aspergillus section Terrei reveals a high potential in secondary metabolite production and plant biomass degradation. J. Fungi 2024, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, M.; Malusá, E.; Eichler-Löbermann, B.; Vassilev, N. Aspegillus terreus: From soil to industry and back. Microorganisms 2020, 8, 1655. [Google Scholar] [CrossRef]
- Justice, M.C.; Hsu, M.-J.; Tse, B.; Ku, T.; Balkovec, J.; Schmatz, D.; Nielsen, J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998, 273, 3148–3151. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Hajdu, D.; Bratschun-Khan, D.; Gáspári, Z.; Varbanov, M.; Philippot, S.; Fizil, Á.; Czajlik, A.; Kele, Z.; Sonderegger, C.; et al. New antimicrobial potential and structural properties of PAFB: A cationic, cysteine-rich protein from Penicillium chrysogenum Q176. Sci. Rep. 2018, 8, 1751. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, L.; Manzanares, P.; Marcos, J.F.; Martínez-Culebras, P.V. Effect of antifungal proteins (AFPs) on the viability of heat-resistant fungi (HRFs) and the preservation of fruit juices. Int. J. Food Microbiol. 2024, 425, 110886. [Google Scholar] [CrossRef]
- Holzknecht, J.; Marx, F. Navigating the fungal battlefield: Cysteine-rich antifungal proteins and peptides from Eurotiales. Front. Fungal Biol. 2024, 5, 1451455. [Google Scholar] [CrossRef]
- Kaiserer, L.; Oberparleiter, C.; Weiler-Görz, R.; Burgstaller, W.; Leiter, E.; Marx, F. Characterization of the Penicillium chrysogenum antifungal protein PAF. Arch. Microbiol. 2003, 180, 204–210. [Google Scholar] [CrossRef]
- Holzknecht, J.; Kühbacher, A.; Papp, C.; Farkas, A.; Váradi, G.; Marcos, J.F.; Manzanares, P.; Tóth, G.K.; Galgóczy, L.; Marx, F. The Penicillium chrysogenum Q176 antimicrobial protein PAFC effectively inhibits the growth of the opportunistic human pathogen Candida albicans. J. Fungi 2020, 6, 141. [Google Scholar] [CrossRef] [PubMed]
- Koutinas, A.A.; Vlysidis, A.; Pleissner, D.; Kopsahelis, N.; Lopez-Garcia, I.; Kookos, I.K.; Papanikolaou, S.; Kwan, T.H.; Ki-Lin, C.S. Valorization of industrial waste and by-product streams via fermentation for the production of chemicals and biopolymers. Chem. Soc. Rev. 2014, 43, 2587. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, J.; Chang, X.; Chen, D.; Xue, Y.; Liu, P.; Lin, H.; Han, S. A review on the pretreatment of lignocellulose for high-value chemicals. Fuel Process Technol. 2017, 160, 196–206. [Google Scholar] [CrossRef]
- El-Gendi, H.; Saleh, A.K.; Badierah, R.; Redwan, E.M.; El-Maradny, Y.A.; El-Fakharany, E.M. A comprehensive insight into fungal enzymes: Structure, classification, and their role in mankind’s challenges. J. Fungi 2021, 8, 23. [Google Scholar] [CrossRef]
- Shrestha, P.; Ibáñez, A.B.; Bauer, S.; Glassman, S.I.; Szaro, T.M.; Bruns, T.D.; Taylor, J.W. Fungi isolated from Miscanthus and sugarcane: Biomass conversion, fungal enzymes, and hydrolysis of plant cell wall polymers. Biotechnol. Biofuels 2015, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Effiong, T.E.; Abdulsalami, M.S.; Egbe, N.E.; Bakare, V. Screening of fungi isolates from soil, pulp waste water and rotten wood for cellulase producing potentials. J. Appl. Sci. Environ. Manag. 2019, 23, 1051–1055. [Google Scholar] [CrossRef]
- Abdel-Sater, M.A.; El-Said, A.H.M. Xylan-decomposing fungi and xylanolytic activity in agricultural and industrial wastes. Int. Biodeter. Biodegr. 2001, 47, 15–21. [Google Scholar] [CrossRef]
- Herculano, P.N.; Lima, D.M.M.; Fernandes, M.J.S.; Neves, R.P.; Souza-Motta, C.M.; Porto, A.L.F. Isolation of cellulolytic fungi from waste of castor (Ricinus communis L.). Curr. Microbiol. 2011, 62, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Schuster, E.; Dunn-Coleman, N.; Frisvad, J.; van Dijck, P. On the safety of Aspergillus niger—A review. Appl. Microbiol. Biotechnol. 2002, 59, 426–435. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.M.; Abdel-Azeem, M.A.; Abdul-Hadi, S.Y.; Darwish, A.G. Aspergillus: Biodiversity, ecological significances, and industrial applications. In Recent Advancement in White Biotechnology Through Fungi: Volume 1: Diversity and Enzymes Perspectives; Yadav, A.N., Mishra, S., Singh, S., Gupta, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 121–179. [Google Scholar]
- Ashtekar, N.; Anand, G.; Thulasiram, H.V.; Rajeshkumar, K.C. Chapter 14—Genus Penicillium: Advances and application in the modern era. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, J., Gehlot, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 201–213. [Google Scholar]
- Zhang, X.; Yang, H.; Cui, Z. Mucor circinelloides: Efficiency of bioremediation response to heavy metal pollution. Toxicol. Res. 2017, 6, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Morin-Sardin, S.; Nodet, P.; Coton, E.; Jany, J.-L. Mucor: A janus-faced fungal genus with human health impact and industrial applications. Fungal Biol. Rev. 2017, 31, 12–32. [Google Scholar] [CrossRef]
- Rodrigues Reis, C.E.; Bento, H.B.S.; Carvalho, A.K.F.; Rajendran, A.; Hu, B.; De Castro, H.F. Critical applications of Mucor circinelloides within a biorefinery context. Crit. Rev. Biotechnol. 2019, 39, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Lee Soo, C.; Billmyre, R.B.; Li, A.; Carson, S.; Sykes Sean, M.; Huh Eun, Y.; Mieczkowski, P.; Ko Dennis, C.; Cuomo Christina, A.; Heitman, J. Analysis of a food-borne fungal pathogen outbreak: Virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 2014, 5, 10-1128. [Google Scholar] [CrossRef]
- Sørensen, A.; Lübeck, M.; Lübeck, P.S.; Ahring, B.K. Fungal beta-glucosidases: A bottleneck in industrial use of lignocellulosic materials. Biomolecules 2013, 3, 612–631. [Google Scholar] [CrossRef]
- Knob, A.; Terrasan, C.R.F.; Carmona, E.C. β-Xylosidases from filamentous fungi: An overview. World J. Microbiol. Biotechnol. 2010, 26, 389–407. [Google Scholar] [CrossRef]
- Poria, V.; Saini, J.K.; Singh, S.; Nain, L.; Kuhad, R.C. Arabinofuranosidases: Characteristics, microbial production, and potential in waste valorization and industrial applications. Bioresour. Technol. 2020, 304, 123019. [Google Scholar] [CrossRef]
- Singh, R.V.; Sambyal, K. β-galactosidase as an industrial enzyme: Production and potential. Chem. Pap. 2023, 77, 11–31. [Google Scholar] [CrossRef]
- Bhatia, S.; Singh, A.; Batra, N.; Singh, J. Microbial production and biotechnological applications of α-galactosidase. Int. J. Biol. Macromol. 2020, 150, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, S.; Shang, W.; Yan, Z.; Wu, X.; Li, Y.; Chen, G.; Liu, X.; Wang, L. Synergistic mechanism of GH11 xylanases with different action modes from Aspergillus niger An76. Biotechnol. Biofuels 2021, 14, 118. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Kun Roland, S.; Peng, M.; Gruben Birgit, S.; Benoit Gelber, I.; Mäkelä, M.; de Vries, R.P. The cultivation method affects the transcriptomic response of Aspergillus niger to growth on sugar beet pulp. Microbiol. Spectr. 2021, 9, e01064-21. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 2016, 15, 106. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, N.; Singh, A.; Adsul, M.; Dixit, P.; Sandhu, S.K.; Mathur, A.; Puri, S.K.; Singhania, R.R. Penicillium: The next emerging champion for cellulase production. Bioresour. Technol. Rep. 2018, 2, 131–140. [Google Scholar] [CrossRef]
- da Silva, R.R. Bacterial and fungal proteolytic enzymes: Production, catalysis and potential applications. Appl. Biochem. Biotechnol. 2017, 183, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.G.; Khan, K.; Aishwarya, S.; Padyana, S.; Huchegowda, R.; Reddy, K.R.; Pais, R.; Alrafas, H.; Dsouza, R.; Madhavi, J.; et al. Fungal amylases and their industrial applications. In Industrially Important Fungi for Sustainable Development: Volume 2: Bioprospecting for Biomolecules; Abdel-Azeem, A.M., Yadav, A.N., Yadav, N., Sharma, M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 407–434. [Google Scholar]
- Garrigues, S.; Gandía, M.; Castillo, L.; Coca, M.; Marx, F.; Marcos, J.F.; Manzanares, P. Three antifungal proteins from Penicillium expansum: Different patterns of production and antifungal activity. Front. Microbiol. 2018, 9, 2370. [Google Scholar] [CrossRef]
- Gandía, M.; Kakar, A.; Giner-Llorca, M.; Holzknecht, J.; Martínez-Culebras, P.; Galgóczy, L.; Marx, F.; Marcos, J.F.; Manzanares, P. Potential of antifungal proteins (AFPs) to control Penicillium postharvest fruit decay. J. Fungi 2021, 7, 449. [Google Scholar] [CrossRef] [PubMed]
- Abad, A.V.; Manzanares, P.; Marcos, J.F.; Martínez-Culebras, P.V. The Penicillium digitatum antifungal protein PdAfpB shows high activity against mycobiota involved in sliced bread spoilage. Food Microbiol. 2023, 109, 104142. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Gandía, M.; Marcos, J.F. Occurrence and function of fungal antifungal proteins: A case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 2016, 100, 2243–2256. [Google Scholar] [CrossRef]
- Huber, A.; Lerchster, H.; Marx, F. Nutrient excess triggers the expression of the Penicillium chrysogenum antifungal protein PAFB. Microorganisms 2019, 7, 654. [Google Scholar] [CrossRef]
- Okabe, M.; Lies, D.; Kanamasa, S.; Park, E.Y. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl. Microbiol. Biotechnol. 2009, 84, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Boruta, T.; Bizukojc, M. Production of lovastatin and itaconic acid by Aspergillus terreus: A comparative perspective. World J. Microbiol. Biotechnol. 2017, 33, 34. [Google Scholar] [CrossRef]
- Ling, K.H. Territrems, tremorgenic mycotoxins isolated from Aspergillus terreus. J. Toxicol. Toxin Rev. 1994, 13, 243–252. [Google Scholar] [CrossRef]
- Blumenthal, C.Z. Production of toxic metabolites in Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei: Justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi. Regul. Toxicol. Pharmacol. 2004, 39, 214–228. [Google Scholar] [CrossRef]
- Thakur, R.; Shishodia, S.K.; Sharma, A.; Chauhan, A.; Kaur, S.; Shankar, J. Accelerating the understanding of Aspergillus terreus: Epidemiology, physiology, immunology and advances. Curr. Res. Microb. Sci. 2024, 6, 100220. [Google Scholar] [CrossRef] [PubMed]
- Lin, L. Bottom-up synthetic ecology study of microbial consortia to enhance lignocellulose bioconversion. Biotechnol. Biofuels Bioprod. 2022, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Puentes-Téllez, P.E.; Falcao Salles, J. Construction of effective minimal active microbial consortia for lignocellulose degradation. Microb. Ecol. 2018, 76, 419–429. [Google Scholar] [CrossRef]
- Hernanz-Koers, M.; Gandía, M.; Garrigues, S.; Manzanares, P.; Yenush, L.; Orzaez, D.; Marcos, J.F. FungalBraid: A GoldenBraid-based modular cloning platform for the assembly and exchange of DNA elements tailored to fungal synthetic biology. Fungal Genet. Biol. 2018, 116, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Mozsik, L.; Pohl, C.; Meyer, V.; Bovenberg, R.A.L.; Nygard, Y.; Driessen, A.J.M. Modular synthetic biology toolkit for filamentous fungi. ACS Synth. Biol. 2021, 10, 2850–2861. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Giménez, E.; Gandía, M.; Sáez, Z.; Manzanares, P.; Yenush, L.; Orzáez, D.; Marcos, J.F.; Garrigues, S. FungalBraid 2.0: Expanding the synthetic biology toolbox for the biotechnological exploitation of filamentous fungi. Front. Bioeng. Biotechnol. 2023, 11, 1222812. [Google Scholar] [CrossRef]
- Liao, B.; Chen, X.; Zhou, X.; Zhou, Y.; Shi, Y.; Ye, X.; Liao, M.; Zhou, Z.; Cheng, L.; Ren, B. Applications of CRISPR/Cas gene-editing technology in yeast and fungi. Arch. Microbiol. 2021, 204, 79. [Google Scholar] [CrossRef]
- Kun, R.S.; Gomes, A.C.S.; Hildén, K.S.; Salazar Cerezo, S.; Mäkelä, M.R.; de Vries, R.P. Developments and opportunities in fungal strain engineering for the production of novel enzymes and enzyme cocktails for plant biomass degradation. Biotechnol. Adv. 2019, 37, 107361. [Google Scholar] [CrossRef]
- Gao, L.; Xia, C.; Xu, J.; Li, Z.; Yu, L.; Liu, G.; Song, X.; Qu, Y. Constitutive expression of chimeric transcription factors enables cellulase synthesis under non-inducing conditions in Penicillium oxalicum. Biotechnol. J. 2017, 12, 1700119. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Liu, K.; Ma, W.; Jiang, Y.; Hou, S.; Tan, Y.; Yuan, Q.; Niu, K.; Fang, X. Redesigning transcription factor Cre1 for alleviating carbon catabolite repression in Trichoderma reesei. Synth. Syst. Biotechnol. 2020, 5, 230–235. [Google Scholar] [CrossRef]
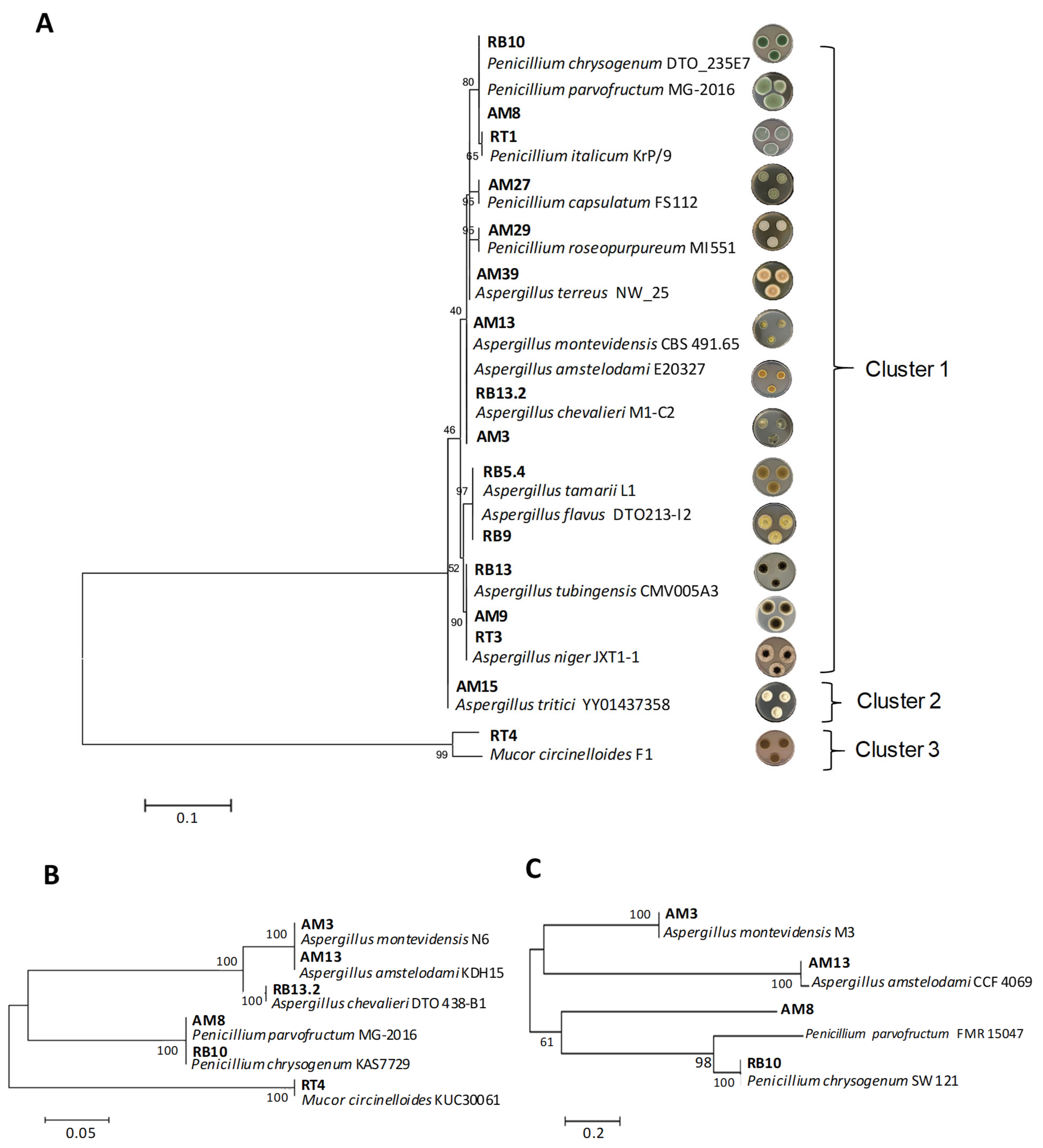
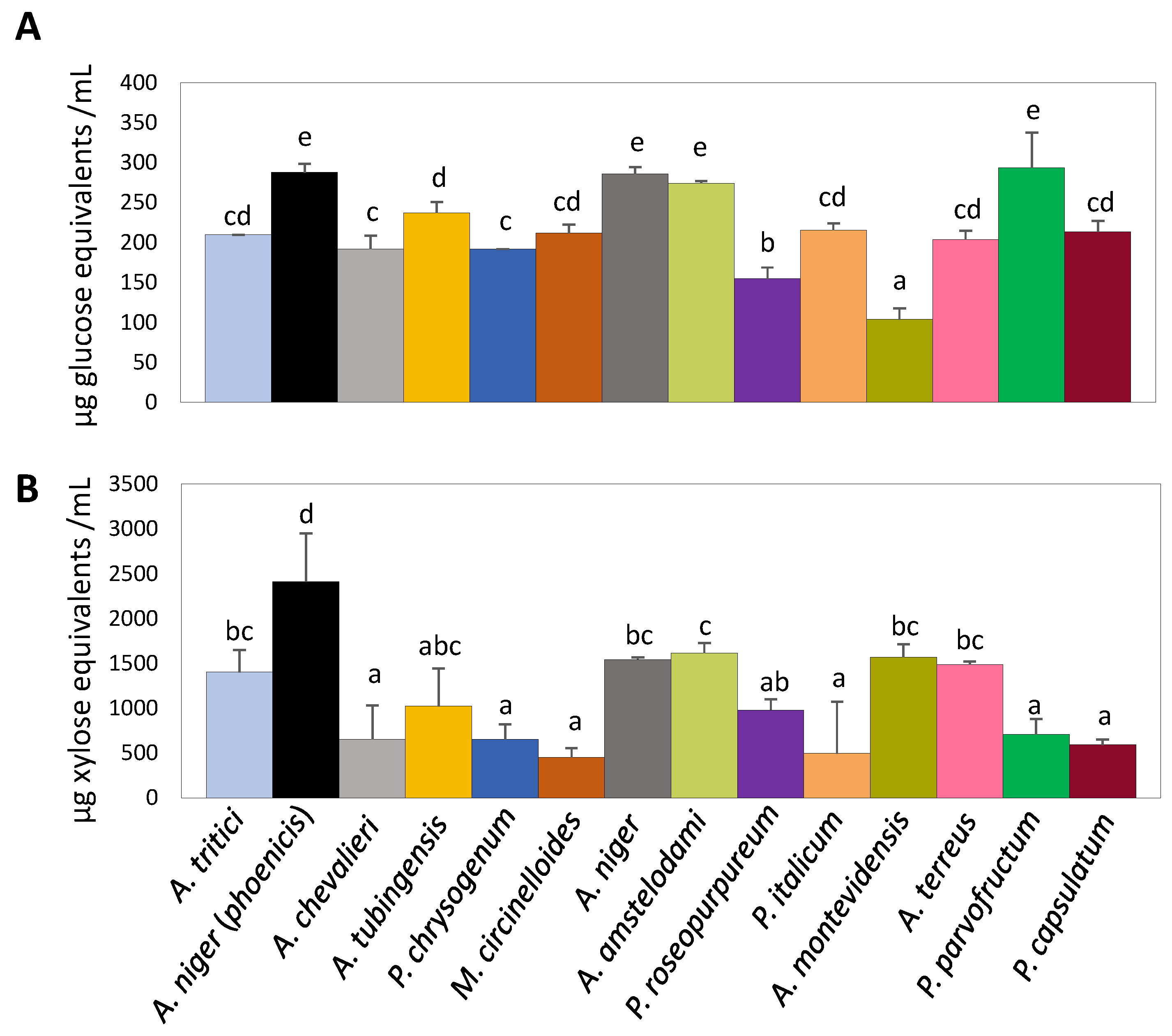

| Code | Species | GenBank Accession Number | ||
|---|---|---|---|---|
| ITS | ß-Tubulin | Calmodulin | ||
| AM3 | Aspergillus montevidensis | PQ483919 | PQ520635 | PQ609700 |
| RB9 | Aspergillus flavus | PQ483907 | - | - |
| RB5.4 | Aspergillus tamarii | PQ483908 | - | - |
| AM9 | Aspergillus niger | PQ483905 | - | - |
| AM13 | Aspergillus amstelodami | PQ483911 | PQ520634 | PQ609696 |
| AM15 | Aspergillus tritici | PQ483909 | - | - |
| AM39 | Aspergillus terreus | PQ483910 | - | - |
| RT3 | Aspergillus niger var. phoenicis | PQ483906 | - | - |
| RB13.2 | Aspergillus chevalieri | PQ483912 | PQ520633 | - |
| RB13 | Aspergillus tubingensis | PQ483904 | - | - |
| AM29 | Penicillium roseopurpureum | PQ483916 | - | - |
| AM8 | Penicillium parvofructum | PQ483914 | PQ520632 | PQ609699 |
| AM27 | Penicillium capsulatum | PQ483917 | - | - |
| RB10 | Penicillium chrysogenum | PQ483915 | PQ520631 | PQ609698 |
| RT1 | Penicillium italicum | PQ483913 | - | - |
| RT4 | Mucor circinelloides | PQ483918 | PQ609697 | - |
| Fungi | [Protein] (mg/mL) |
|---|---|
| A. montevidensis AM3 | 0.93 ± 0.01 b |
| A. niger AM9 | 1.27 ± 0.09 bcde |
| A. amstelodami AM13 | 1.38 ± 0.04 cde |
| A. tritici AM15 | 1.30 ± 0.03 bcde |
| A. terreus AM39 | 1.64 ± 0.08 e |
| A. niger var. phoenicis RT3 | 2.17 ± 0.01 f |
| A. chevalieri RB13.2 | 1.48 ± 0.11 de |
| A. tubingensis RB13 | 1.27 ± 0.08 bcde |
| P. roseopurpureum AM29 | 1.57 ± 0.73 de |
| P. parvofructum AM8 | 1.64 ± 0.03 e |
| P. capsulatum AM27 | 0.99 ± 0.06 bc |
| P. chrysogenum RB10 | 1.29 ± 0.01 bcde |
| P. italicum RT1 | 1.14 ± 0.09 bcd |
| M. circinelloides RT4 | 0.43 ± 0.01 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yélamos, A.M.; Marcos, J.F.; Manzanares, P.; Garrigues, S. Harnessing Filamentous Fungi for Enzyme Cocktail Production Through Rice Bran Bioprocessing. J. Fungi 2025, 11, 106. https://doi.org/10.3390/jof11020106
Yélamos AM, Marcos JF, Manzanares P, Garrigues S. Harnessing Filamentous Fungi for Enzyme Cocktail Production Through Rice Bran Bioprocessing. Journal of Fungi. 2025; 11(2):106. https://doi.org/10.3390/jof11020106
Chicago/Turabian StyleYélamos, Ana M., Jose F. Marcos, Paloma Manzanares, and Sandra Garrigues. 2025. "Harnessing Filamentous Fungi for Enzyme Cocktail Production Through Rice Bran Bioprocessing" Journal of Fungi 11, no. 2: 106. https://doi.org/10.3390/jof11020106
APA StyleYélamos, A. M., Marcos, J. F., Manzanares, P., & Garrigues, S. (2025). Harnessing Filamentous Fungi for Enzyme Cocktail Production Through Rice Bran Bioprocessing. Journal of Fungi, 11(2), 106. https://doi.org/10.3390/jof11020106







