Genomic and Transcriptomic Insights into Carbon-Source and Temporal Induction of a Diverse Set of Lignocellulolytic Enzymes in Irpex lacteus QJ
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Substrate
2.2. Sample Culture
2.3. Genomic Sequencing and De Novo Assembly
2.4. Genome Component Prediction and Gene Function Annotation
2.5. RNA Extraction, Library Construction, and Sequencing
2.6. Quality Control and Read Mapping
2.7. Differential Expression and Functional Enrichment Analyses
2.8. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Validation
2.9. Statistical Analysis
3. Results
3.1. General Genome Features and Phylogenetic Comparison of I. lacteus QJ
3.2. Gene Prediction and Classification
3.3. Lignocellulolytic Enzymes of I. lacteus QJ
3.4. RNA-Sequencing-Based Transcriptome Analysis of the I. lacteus QJ
3.5. KEGG Pathway and GO Enrichment Analyses of the DEGs
3.6. Expression Patterns of Lignocellulolytic Enzyme Genes Under Different Cultivation Conditions
3.7. Validation of Transcriptomic Data by qRT–PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sun, R.; Zhu, X.; Wang, C.; Yue, J.; Pan, L.; Song, C.; Zhao, Y. Effect of NH4+ and NO3− Cooperatively Regulated Carbon to Nitrogen Ratio on Organic Nitrogen Fractions during Rice Straw Composting. Bioresour. Technol. 2024, 395, 130316. [Google Scholar] [CrossRef]
- Lv, X.; Qiao, Z.; Chen, C.; Hua, J.; Zhou, C. Exploration of Multi-Source Lignocellulose-Degrading Microbial Resources and Bioaugmentation Strategies: Implications for Rumen Efficiency. Animals 2025, 15, 1920. [Google Scholar] [CrossRef]
- Sorek, N.; Yeats, T.H.; Szemenyei, H.; Youngs, H.; Somerville, C.R. The Implications of Lignocellulosic Biomass Chemical Composition for the Production of Advanced Biofuels. BioScience 2014, 64, 192–201. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, J.; Cao, S.; Yu, T.; Niu, N.; Chen, L. Development of Efficient and Simple Straw Biochar: In-Depth Analysis of Its Performance and Mechanism in Eliminating Chemical Oxygen Demand. Environ. Monit. Assess. 2025, 197, 921. [Google Scholar] [CrossRef]
- Al-Naghi, A.A.A.; Ali, T.; Inam, I.; Qureshi, M.Z.; Kahla, N.B.; Ghazouani, N.; Ahmed, H. Sustainable Concrete: Investigating the Synergistic Effects of Coconut Fiber, Wheat Straw Ash, and Silica Fume on RAC Strength and Durability. Sci. Rep. 2025, 15, 24542. [Google Scholar] [CrossRef] [PubMed]
- Baig, K.S. Interaction of Enzymes with Lignocellulosic Materials: Causes, Mechanism and Influencing Factors. Bioresour. Bioprocess. 2020, 7, 21. [Google Scholar] [CrossRef]
- Ashokkumar, V.; Venkatkarthick, R.; Jayashree, S.; Chuetor, S.; Dharmaraj, S.; Kumar, G.; Chen, W.-H.; Ngamcharussrivichai, C. Recent Advances in Lignocellulosic Biomass for Biofuels and Value-Added Bioproducts—A Critical Review. Bioresour. Technol. 2022, 344, 126195. [Google Scholar] [CrossRef]
- Šuchová, K.; Puchart, V. Cellulolytic and Hemicellulolytic Capacity of Acetivibrio clariflavus. Appl. Microbiol. Biotechnol. 2025, 109, 105. [Google Scholar] [CrossRef] [PubMed]
- Katsimpouras, C.; Dedes, G.; Thomaidis, N.S.; Topakas, E. A Novel Fungal GH30 Xylanase with Xylobiohydrolase Auxiliary Activity. Biotechnol. Biofuels 2019, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Soleimani, A.; Lin, Y.; Alian, M.; Hosseinzadeh, H.; Suman, S.K.; Rahimi, M.; Balan, V. Enzymatic Depolymerization of Lignin and Electrocatalytic Hydrodeoxygenation for the Production of Reduced Aromatic Compounds: A Review and Perspective. Biotechnol. Adv. 2025, 83, 108647. [Google Scholar] [CrossRef]
- Wang, J.; Hong, S.; Wang, B.; Shen, X.; Wen, J.-L.; Yuan, T.-Q. Harnessing Zeolite Catalyst for the Cleavage of Targeted Chemical Bonds in Lignin. Chem. Catal. 2023, 3, 100797. [Google Scholar] [CrossRef]
- Prado, C.A.; Antunes, F.A.F.; Rocha, T.M.; Sánchez-Muñoz, S.; Barbosa, F.G.; Terán-Hilares, R.; Cruz-Santos, M.M.; Arruda, G.L.; da Silva, S.S.; Santos, J.C. A Review on Recent Developments in Hydrodynamic Cavitation and Advanced Oxidative Processes for Pretreatment of Lignocellulosic Materials. Bioresour. Technol. 2022, 345, 126458. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.S.; Williams, G.A.; Jaiswal, A.K. Emerging Technologies for the Pretreatment of Lignocellulosic Biomass. Bioresour. Technol. 2018, 262, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Navarro, D.; Chaduli, D.; Taussac, S.; Lesage-Meessen, L.; Grisel, S.; Haon, M.; Callac, P.; Courtecuisse, R.; Decock, C.; Dupont, J.; et al. Large-Scale Phenotyping of 1,000 Fungal Strains for the Degradation of Non-Natural, Industrial Compounds. Commun. Biol. 2021, 4, 871. [Google Scholar] [CrossRef] [PubMed]
- Vieira, R.I.M.; Peixoto, A.d.S.; Monclaro, A.V.; Ricart, C.A.O.; Filho, E.X.F.; Miller, R.N.G.; Gomes, T.G. Fungal Coculture: Unlocking the Potential for Efficient Bioconversion of Lignocellulosic Biomass. J. Fungi 2025, 11, 458. [Google Scholar] [CrossRef]
- Suryadi, H.; Judono, J.J.; Putri, M.R.; Eclessia, A.D.; Ulhaq, J.M.; Agustina, D.N.; Sumiati, T. Biodelignification of Lignocellulose Using Ligninolytic Enzymes from White-Rot Fungi. Heliyon 2022, 8, e08865. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Yu, H.; Zhang, J.; Li, F.; Li, C.; Zhang, X.; Ma, F. The Potential of Cottonseed Hull as Biorefinery Substrate after Biopretreatment by Pleurotus ostreatus and the Mechanism Analysis Based on Comparative Proteomics. Ind. Crops Prod. 2019, 130, 151–161. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Asfour, H.Z.; Alhakamy, N.A.; Al-Rabia, M.W.; Abdallah, H.M.; ALsiyud, D.F.; AlZahrany, T.A.; Mohamed, G.A. Irpex lacteus: A Multifaceted Fungal Powerhouse—Exploring Its Industrial Applications, Secondary Metabolites, and Biological Activities. Arab. J. Sci. Eng. 2025, 50, 14371–14414. [Google Scholar] [CrossRef]
- Niu, D.; Zuo, S.; Jiang, D.; Tian, P.; Zheng, M.; Xu, C. Treatment Using White Rot Fungi Changed the Chemical Composition of Wheat Straw and Enhanced Digestion by Rumen Microbiota in Vitro. Anim. Feed Sci. Technol. 2018, 237, 46–54. [Google Scholar] [CrossRef]
- Kato, H.; Miura, D.; Kato, M.; Shimizu, M. Metabolic Mechanism of Lignin-Derived Aromatics in White-Rot Fungi. Appl. Microbiol. Biotechnol. 2024, 108, 532. [Google Scholar] [CrossRef]
- Wang, Z.-D.; Wang, B.-T.; Jin, L.; Ruan, H.-H.; Jin, F.-J. Implications of Carbon Catabolite Repression for Aspergillus-Based Cell Factories: A Review. Biotechnol. J. 2024, 19, 2300551. [Google Scholar] [CrossRef]
- Kerkaert, J.D.; Huberman, L.B. Regulation of Nutrient Utilization in Filamentous Fungi. Appl. Microbiol. Biotechnol. 2023, 107, 5873–5898. [Google Scholar] [CrossRef] [PubMed]
- Franzino, T.; Boubakri, H.; Cernava, T.; Abrouk, D.; Achouak, W.; Reverchon, S.; Nasser, W.; Haichar, F.e.Z. Implications of Carbon Catabolite Repression for Plant–Microbe Interactions. Plant Commun. 2022, 3, 100272. [Google Scholar] [CrossRef] [PubMed]
- Metreveli, E.; Khardziani, T.; Elisashvili, V. The Carbon Source Controls the Secretion and Yield of Polysaccharide-Hydrolyzing Enzymes of Basidiomycetes. Biomolecules 2021, 11, 1341. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yang, K.; Wang, X.; Tu, T.; Wang, Y.; Zhang, J.; Su, X.; Yao, B.; Huang, H.; Luo, H. Insights into the H2O2-Driven Lytic Polysaccharide Monooxygenase Activity on Efficient Cellulose Degradation in the White Rot Fungus Irpex lacteus. J. Agric. Food Chem. 2023, 71, 8104–8111. [Google Scholar] [CrossRef] [PubMed]
- Civzele, A.; Stipniece-Jekimova, A.A.; Mezule, L. Fungal Ligninolytic Enzymes and Their Application in Biomass Lignin Pretreatment. J. Fungi 2023, 9, 780. [Google Scholar] [CrossRef]
- Cui, M.J.; Sun, Y.X.; Hu, X.M. Regulation of cellulase biosynthesis by transcription factor TeClr1 in Talaromyces sp. J. Northeast Agric. Univ. 2025, 56, 47–58. [Google Scholar] [CrossRef]
- Zhong, Z.; Norvienyeku, J.; Chen, M.; Bao, J.; Lin, L.; Chen, L.; Lin, Y.; Wu, X.; Cai, Z.; Zhang, Q.; et al. Directional Selection from Host Plants Is a Major Force Driving Host Specificity in Magnaporthe Species. Sci. Rep. 2016, 6, 25591. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R Package for Identifying Differentially Expressed Genes from RNA-Seq Data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Yao, M.; Li, W.; Duan, Z.; Zhang, Y.; Jia, R. Genome Sequence of the White-Rot Fungus Irpex lacteus F17, a Type Strain of Lignin Degrader Fungus. Stand. Genomic Sci. 2017, 12, 55. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, K.; Gong, H.; Yang, K.; Wang, X.; Zhou, G.; Cui, W.; Chen, Y.; Yang, Y. Whole Genome Sequencing and the Lignocellulose Degradation Potential of Bacillus subtilis RLI2019 Isolated from the Intestine of Termites. Biotechnol. Biofuels Bioprod. 2023, 16, 130. [Google Scholar] [CrossRef] [PubMed]
- Hage, H.; Miyauchi, S.; Virágh, M.; Drula, E.; Min, B.; Chaduli, D.; Navarro, D.; Favel, A.; Norest, M.; Lesage-Meessen, L.; et al. Gene Family Expansions and Transcriptome Signatures Uncover Fungal Adaptations to Wood Decay. Environ. Microbiol. 2021, 23, 5716–5732. [Google Scholar] [CrossRef]
- Ohm, R.A.; Riley, R.; Salamov, A.; Min, B.; Choi, I.-G.; Grigoriev, I.V. Genomics of Wood-Degrading Fungi. Fungal Genet. Biol. 2014, 72, 82–90. [Google Scholar] [CrossRef]
- Chen, L.; Gong, Y.; Cai, Y.; Liu, W.; Zhou, Y.; Xiao, Y.; Xu, Z.; Liu, Y.; Lei, X.; Wang, G.; et al. Genome Sequence of the Edible Cultivated Mushroom Lentinula edodes (Shiitake) Reveals Insights into Lignocellulose Degradation. PLoS ONE 2016, 11, e0160336. [Google Scholar] [CrossRef] [PubMed]
- Kuuskeri, J.; Häkkinen, M.; Laine, P.; Smolander, O.-P.; Tamene, F.; Miettinen, S.; Nousiainen, P.; Kemell, M.; Auvinen, P.; Lundell, T. Time-Scale Dynamics of Proteome and Transcriptome of the White-Rot Fungus Phlebia Radiata: Growth on Spruce Wood and Decay Effect on Lignocellulose. Biotechnol. Biofuels 2016, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.V.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and Transcriptional Analysis of the Predicted Secretome in the Lignocellulose-Degrading Basidiomycete Fungus Pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef]
- Ruiz-Dueñas, F.J.; Barrasa, J.M.; Sánchez-García, M.; Camarero, S.; Miyauchi, S.; Serrano, A.; Linde, D.; Babiker, R.; Drula, E.; Ayuso-Fernández, I.; et al. Genomic Analysis Enlightens Agaricales Lifestyle Evolution and Increasing Peroxidase Diversity. Mol. Biol. Evol. 2021, 38, 1428–1446. [Google Scholar] [CrossRef]
- Qin, X.; Su, X.; Luo, H.; Ma, R.; Yao, B.; Ma, F. Deciphering Lignocellulose Deconstruction by the White Rot Fungus Irpex lacteus Based on Genomic and Transcriptomic Analyses. Biotechnol. Biofuels 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Salvachúa, D.; Martínez, A.T.; Tien, M.; López-Lucendo, M.F.; García, F.; de los Ríos, V.; Martínez, M.J.; Prieto, A. Differential Proteomic Analysis of the Secretome of Irpex lacteus and Other White-Rot Fungi during Wheat Straw Pretreatment. Biotechnol. Biofuels 2013, 6, 115. [Google Scholar] [CrossRef]
- Kijpornyongpan, T.; Schwartz, A.; Yaguchi, A.; Salvachúa, D. Systems Biology-Guided Understanding of White-Rot Fungi for Biotechnological Applications: A Review. iScience 2022, 25, 104640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Duan, Y.; Zhang, M.; Liang, C.; Li, W.; Liu, C.; Ye, Y. Genome Sequencing and Metabolic Potential Analysis of Irpex lacteus. J. Fungi 2024, 10, 846. [Google Scholar] [CrossRef] [PubMed]
- Akcay, C.; Arslan, R.; Ceylan, F. Valorization of Various Lignocellulosic Wastes to Ganoderma lucidum (Curtis) P. Karst (Reishi Mushroom) Cultivation and Their FT-IR Assessments. PLoS ONE 2025, 20, e0328732. [Google Scholar] [CrossRef]
- Zhang, J.; Meng Markillie, L.; Mitchell, H.D.; Gaffrey, M.J.; Orr, G.; Schilling, J.S. Distinctive Carbon Repression Effects in the Carbohydrate-Selective Wood Decay Fungus Rhodonia placenta. Fungal Genet. Biol. 2022, 159, 103673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Liu, W.; Song, L.; Guo, Z.; Bian, Z.; Han, Y.; Cai, H.; Yang, P.; Meng, K. The Potential of Trichoderma asperellum for Degrading Wheat Straw and Its Key Genes in Lignocellulose Degradation. Front. Microbiol. 2025, 16, 1550495. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2018, 19, 48. [Google Scholar] [CrossRef]
- Alfaro, M.; Majcherczyk, A.; Kües, U.; Ramírez, L.; Pisabarro, A.G. Glucose Counteracts Wood-Dependent Induction of Lignocellulolytic Enzyme Secretion in Monokaryon and Dikaryon Submerged Cultures of the White-Rot Basidiomycete Pleurotus ostreatus. Sci. Rep. 2020, 10, 12421. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Ma, X.; Xie, Y.; Waheed, A.; Liu, G. Heterologously Expressed Cellobiose Dehydrogenase Acts as Efficient Electron-Donor of Lytic Polysaccharide Monooxygenase for Cellulose Degradation in Trichoderma reesei. Int. J. Mol. Sci. 2023, 24, 17202. [Google Scholar] [CrossRef]
- Wang, Z.; Fu, X.; Diao, W.; Wu, Y.; Rovira, C.; Wang, B. Theoretical Study of the in Situ Formation of H2O2 by Lytic Polysaccharide Monooxygenases: The Reaction Mechanism Depends on the Type of Reductant. Chem. Sci. 2025, 16, 3173–3186. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, T.; Wu, Z.; Zeng, G.; Huang, D.; Shen, Y.; He, X.; Lai, M.; He, Y. Study on Biodegradation Process of Lignin by FTIR and DSC. Environ. Sci. Pollut. Res. 2014, 21, 14004–14013. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Feng, Y.; Wu, Z.; Saeed, M.; Hu, C.; Zhu, D.; Chen, H. Advances in Cellobiose Dehydrogenases: Molecular Modifications and High-Throughput Screening for Biotechnological Applications. J. Agric. Food Chem. 2025, 73, 15467–15479. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, Y.; Yu, J.; Wang, L. Recent Advances in the Efficient Degradation of Lignocellulosic Metabolic Networks by Lytic Polysaccharide Monooxygenase. Acta Biochim. Biophys. Sin. 2023, 55, 529–539. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 2025, 53, D30–D44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Jin, E.; Wang, A.; Chen, T.; Zhang, X.; Zhu, J.; Dong, L.; Sun, Y.; Yu, C.; et al. The GSA Family in 2025: A Broadened Sharing Platform for Multi-Omics and Multimodal Data. Genom. Proteom. Bioinform. 2025, 23, qzaf072. [Google Scholar] [CrossRef] [PubMed]
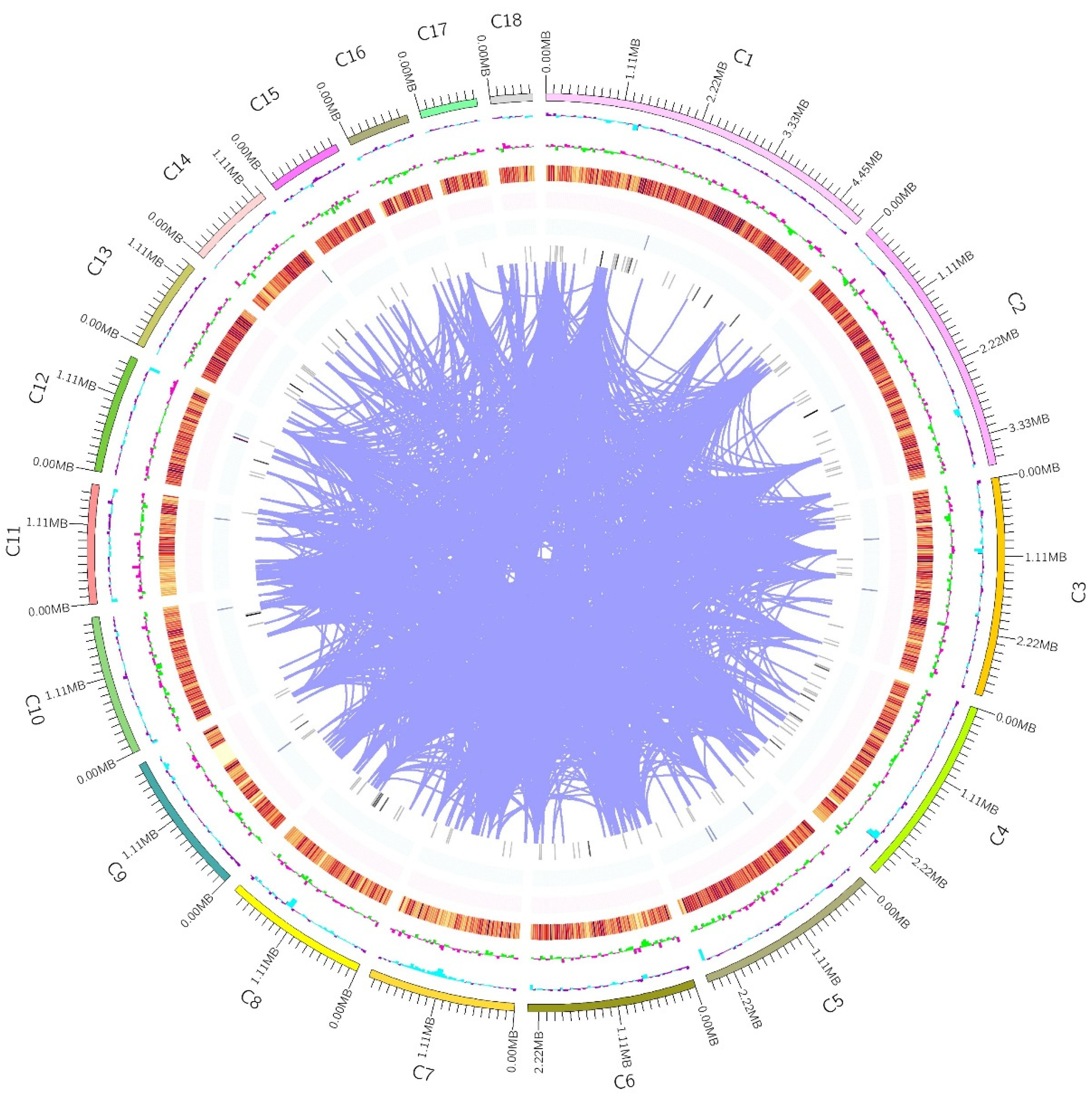
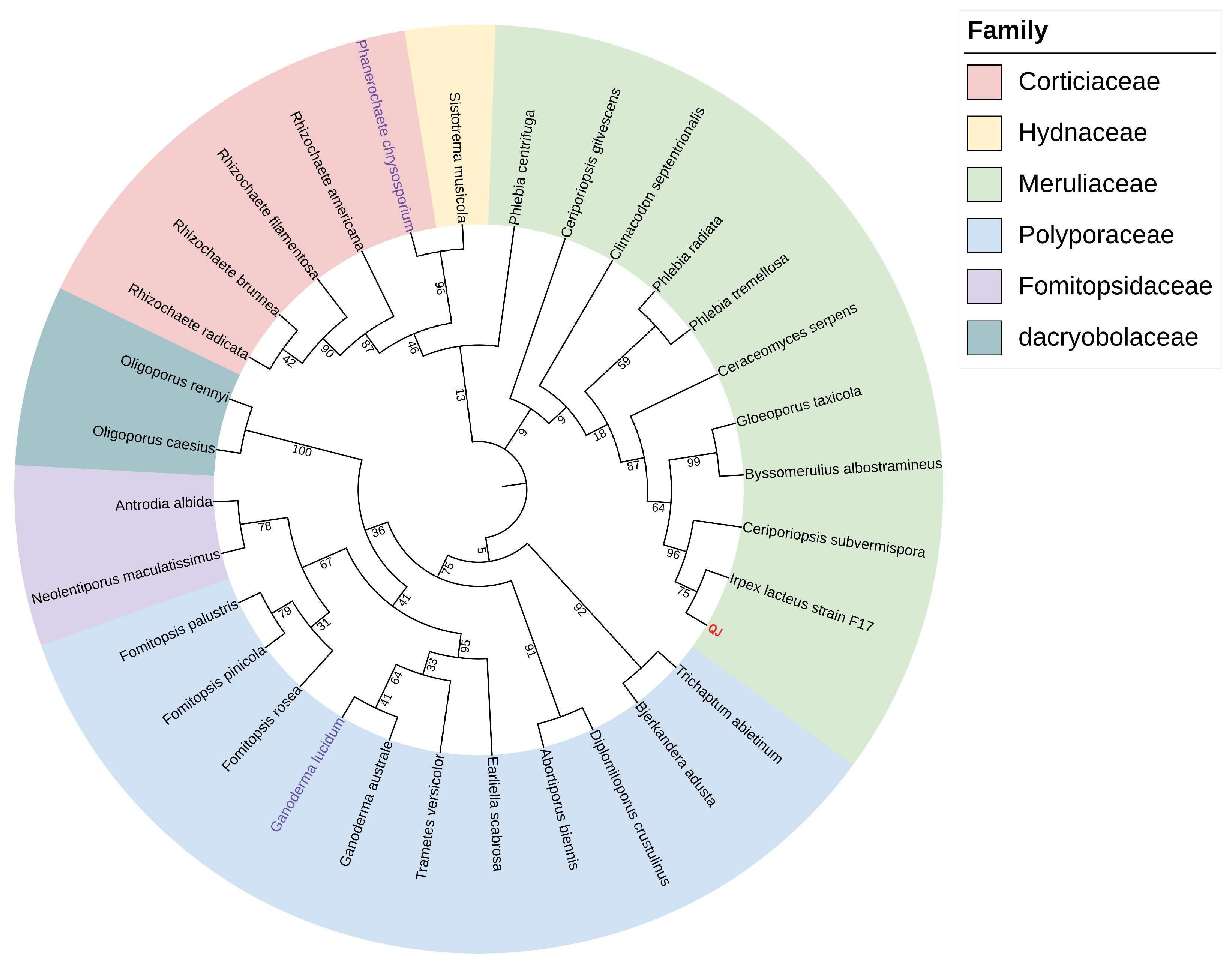
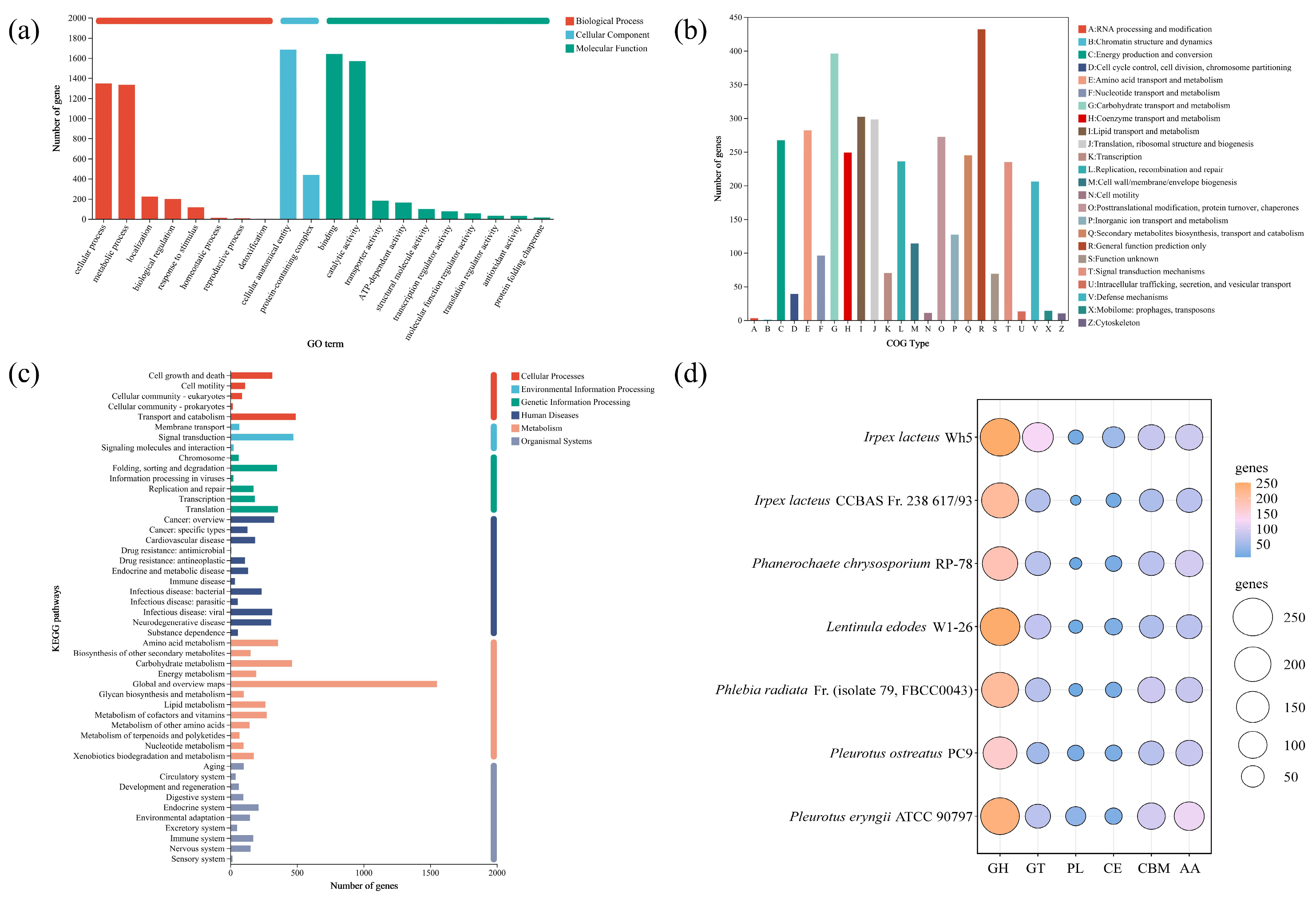
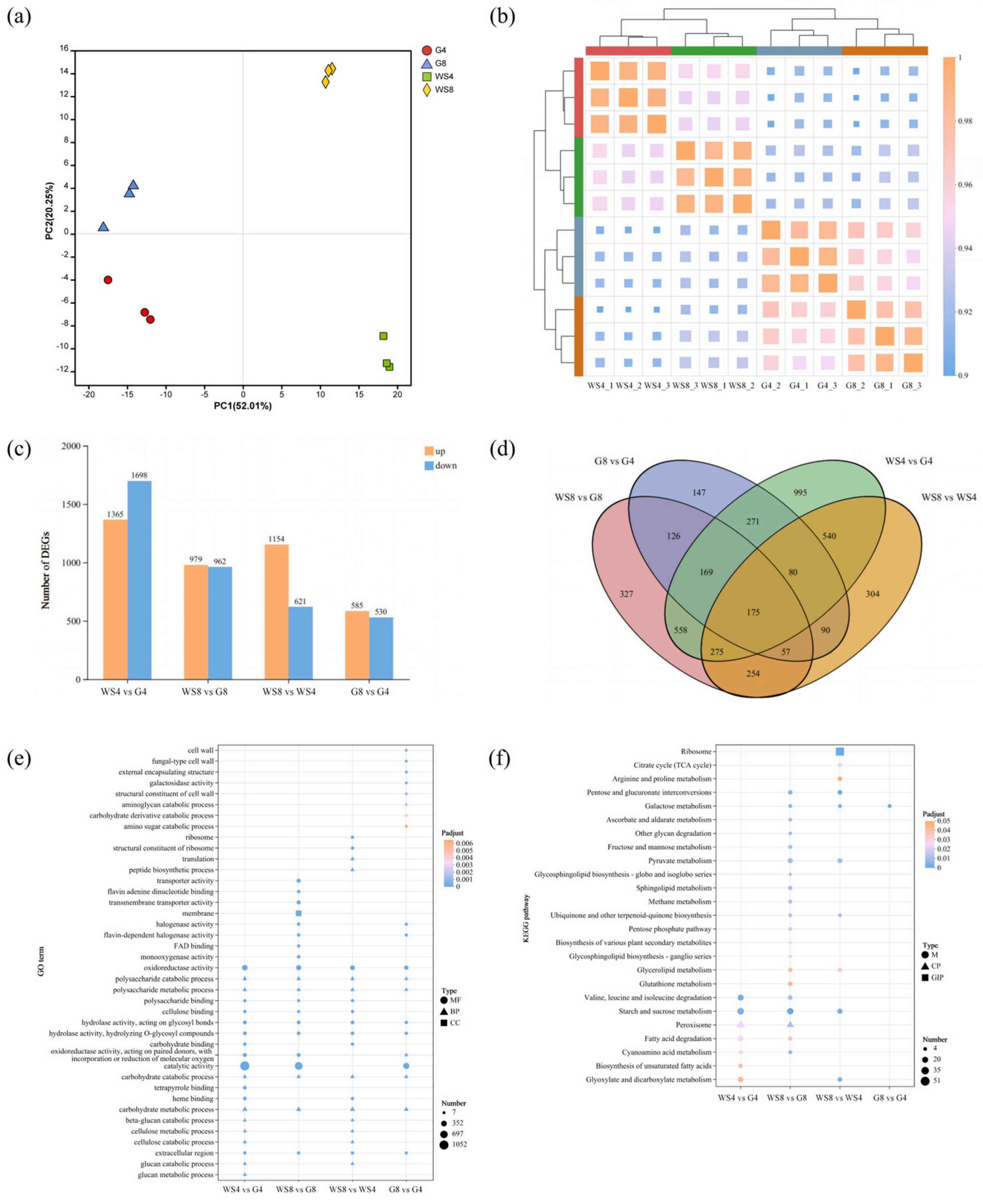



| Sample Name | Irpex lacteus QJ | Irpex lacteus F17 |
|---|---|---|
| Total Scaffold No. | 45 | - |
| Total Bases in Scaffold (bp) | 39,301,785 | 44,362,654 |
| Scaffold N50 (bp) | 2,087,568 | - |
| Scaffold N90 (bp) | 885,300 | - |
| G + C (%) | 50.67 | 49.64 |
| Gene No. | 11,843 | 10,661 |
| Gene Total Length (bp) | 26,809,457 | 15,030,327 |
| Gene/Genome (%) | 68.21 | 33.88 |
| BUSCO Complete (%) | 98.3 | - |
| BUSCO Complete Duplicated (%) | 0.3 | - |
| BUSCO Fragmented (%) | 0.5 | - |
| BUSCO Missing (%) | 1.2 | - |
| Category | Enzyme Class | Gene No. | CAZyme Family | Gene_ID |
|---|---|---|---|---|
| Cellulase | endoglucanase | 6 | GH5 | gene01385 |
| gene05383 | ||||
| gene05537 | ||||
| gene05609 | ||||
| gene11225 | ||||
| GH44 | gene04254 | |||
| exoglucanase | 4 | GH6 | gene02619 | |
| GH7 | gene07665 | |||
| gene09638 | ||||
| gene10561 | ||||
| β-glucosidase | 3 | GH1 | gene05482 | |
| gene08831 | ||||
| GH3 | gene06602 | |||
| Hemicellulase | endo-1,4-β-xylanase | 6 | GH10 | gene03404 |
| gene04780 | ||||
| gene04781 | ||||
| gene04782 | ||||
| gene11452 | ||||
| GH43 | gene06889 | |||
| β-xylosidase | 2 | GH5 | gene06616 | |
| gene06617 | ||||
| endo-1,4-β-mannosidase | 4 | GH5 | gene00338 | |
| gene01023 | ||||
| gene01542 | ||||
| gene01562 | ||||
| β-mannosidase | 2 | GH2 | gene08172 | |
| gene09683 | ||||
| endo-1,5-α-L-arabinosidase | 3 | GH43 | gene01342 | |
| gene01346 | ||||
| gene01347 | ||||
| α-L-arabinofuranosidase | 1 | GH51 | gene05705 | |
| xylosidase/arabinosidase | 1 | GH43 | gene01805 | |
| exo-β-1,3-galactanase | 1 | GH43 | gene02371 | |
| α-glucosidase | 2 | GH31 | gene00099 | |
| gene07570 | ||||
| α-xylosidase | 1 | GH31 | gene01971 | |
| acetylxylan esterase | 1 | CE1 | gene03019 | |
| α-L-fucosidase | 1 | GH95 | gene04349 | |
| LPMOs | lytic cellulose monooxygenase | 16 | AA9 | gene00110 |
| gene03689 | ||||
| gene04035 | ||||
| gene05103 | ||||
| gene05104 | ||||
| gene05105 | ||||
| gene05819 | ||||
| gene05820 | ||||
| gene06333 | ||||
| gene06336 | ||||
| gene07979 | ||||
| gene10470 | ||||
| gene11145 | ||||
| gene11482 | ||||
| gene03397 | ||||
| gene11679 | ||||
| ligninolytic enzyme | laccase | 1 | AA1 | gene06909 |
| manganese peroxidase | 9 | AA2 | gene00042 | |
| gene07987 | ||||
| gene08017 | ||||
| gene08018 | ||||
| gene09109 | ||||
| gene09110 | ||||
| gene09113 | ||||
| gene10372 | ||||
| gene10896 | ||||
| glyoxal oxidase | 3 | AA5 | gene00014 | |
| gene00015 | ||||
| gene01604 | ||||
| dye-decolorizing peroxidase | 5 | gene05240 | ||
| gene05241 | ||||
| gene06228 | ||||
| gene06238 | ||||
| gene06243 | ||||
| CYP450 monooxygenase | 3 | gene10474 | ||
| gene10531 | ||||
| gene10840 | ||||
| cellobiose dehydrogenase | 1 | AA8 | gene03913 | |
| pyranose 2-oxidase | 1 | AA3 | gene04426 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, L.; Liu, B.; Zhu, Q.; Meng, K.; Cai, H.; Han, Y.; Liu, W.; Yang, P. Genomic and Transcriptomic Insights into Carbon-Source and Temporal Induction of a Diverse Set of Lignocellulolytic Enzymes in Irpex lacteus QJ. J. Fungi 2025, 11, 882. https://doi.org/10.3390/jof11120882
Song L, Liu B, Zhu Q, Meng K, Cai H, Han Y, Liu W, Yang P. Genomic and Transcriptomic Insights into Carbon-Source and Temporal Induction of a Diverse Set of Lignocellulolytic Enzymes in Irpex lacteus QJ. Journal of Fungi. 2025; 11(12):882. https://doi.org/10.3390/jof11120882
Chicago/Turabian StyleSong, Liye, Baorui Liu, Qijun Zhu, Kun Meng, Hongying Cai, Yunsheng Han, Weiwei Liu, and Peilong Yang. 2025. "Genomic and Transcriptomic Insights into Carbon-Source and Temporal Induction of a Diverse Set of Lignocellulolytic Enzymes in Irpex lacteus QJ" Journal of Fungi 11, no. 12: 882. https://doi.org/10.3390/jof11120882
APA StyleSong, L., Liu, B., Zhu, Q., Meng, K., Cai, H., Han, Y., Liu, W., & Yang, P. (2025). Genomic and Transcriptomic Insights into Carbon-Source and Temporal Induction of a Diverse Set of Lignocellulolytic Enzymes in Irpex lacteus QJ. Journal of Fungi, 11(12), 882. https://doi.org/10.3390/jof11120882







