Abstract
Chemical disinfection of removable acrylic dental prostheses minimizes the risk of denture stomatitis caused by the opportunistic fungal pathogen Candida albicans. We previously reported that neutral-pH electrolysed oxidising water (EOW), a hypochlorous acid (HOCl)-based biocide, is effective at inhibiting C. albicans biofilm formation on denture resins. Knowledge about the mechanism of action of EOW on C. albicans is lacking. This study investigated the molecular mechanism of action of neutral-pH EOW against C. albicans cells that were incubated with sub-inhibitory concentrations of EOW-HOCl (treatment with 0.125× MIC90 EOW-HOCl (15 µM; T0.125) or treatment with 0.5× MIC90 EOW-HOCl (59 µM; T0.5)). RNA-sequencing (RNA-seq) was used to identify differentially expressed genes (DEGs) which were validated by qRT-PCR. Ninety-five DEGs were identified between the treated and untreated cells after a 60 min exposure. A moderate sub-inhibitory EOW-HOCl concentration (T0.125) caused significant upregulation (log2 fold change > +2) of genes responsive to oxidative stress (EBP1, GAP6, PRN1, HSP21), weak organic acid stress (PRN1), and heat-shock (HSP21). A higher sub-inhibitory concentration (T0.5) caused a significant downregulation of most DEGs (notably, −1.9 to −3 log2 fold reduction in SUT1, HNM3, STP4 expression), cessation of growth, and an upregulation of genes involved in ammonia transport, carbohydrate metabolism, and the unfolded protein and apoptotic response pathways (ATO2, IRE1). Our findings reveal HSP21 and PRN1 to be possible key players in protecting C. albicans cells against HOCl, a natural biocide of the innate immune system.
1. Introduction
Investigations of the effects of hypochlorous acid (HOCl) and hypochlorite anions (ClO−) on microbial cells have predominantly focussed on the effects of sodium hypochlorite (bleach, NaOCl) on bacteria [1,2,3,4,5,6,7,8,9]. The active chlorine species in NaOCl solutions (pH~11–13) is the weakly oxidising ClO−. However, NaOCl at neutral pH has been used to study the effects of HOCl on microbial cells. HOCl is a weak acid (pKa 7.53) that exists as a mixture of the acid and the partially dissociated proton and hypochlorite anion (HOCl/H+ plus ClO−) at near-neutral pH [10]. The antibacterial action of bleach at neutral or near-neutral pH is largely due to the ability of HOCl to aggregate bacterial proteins. Bacteria have mechanisms that respond to this protein aggregation. Activation of the redox-regulated chaperone Hsp33 has been identified as a potential defence mechanism of Escherichia coli in response to the HOCl-induced aggregation of essential proteins [8]. In addition, HOCl was reported to induce enzymes of the oxidative stress response pathway, including catalases, peroxidases, and superoxide dismutase in Gram-negative bacteria [11]. HOCl was also found to activate other protein chaperones, DNA and protein repair systems, and methionine sulfoxide reductases (MSRs), increase membrane hydrophobicity, and reduce membrane permeability and the expression of porins in Gram-negative bacteria [11]. Thus, the lethal effect of HOCl on bacterial cells is thought to be due to oxidative burst-induced stress [4]. Some Gram-negative bacteria can also adapt to sublethal HOCl concentrations by adopting a ‘viable but non-culturable’ (VBNC) state, and by biofilm formation [11].
Knowledge of the mechanism of HOCl-triggered death in yeast is limited, although oxidative damage is possibly key in Saccharomyces cerevisiae [12,13]. However, a recent study found that HOCl induces a rather different transcriptional response in C. albicans than the oxidant H2O2 [14]. Both C. albicans and S. cerevisiae have been studied extensively to understand yeast responses to various important stress conditions. Such investigations have centred on macrophage/neutrophil phagocyte-survival models to understand the pathways triggered by reactive oxygen species (ROS) and the response to H2O2-, weak-acid-, and NaOCl-induced stress [10,12,13,14,15,16,17,18,19,20,21,22,23,24]. C. albicans responds to oxidative stress by upregulating enzymes involved in the removal of H2O2 and free radicals, such as superoxide dismutase (SOD1, SOD2, SOD5), enzymes of the thioredoxin (TSA1, TRX1, TRR1) pathway, and genes encoding the glutathione/glutaredoxin (GPX1, GSH1) system [17,25]. The CAT1 (also known as CTA1 [26]) gene, which encodes the key antioxidant enzyme catalase, is another C. albicans gene that responds to osmotic, oxidative, and heavy metal stress [17]. Osmotic (NaCl), oxidative (H2O2), and heavy metal (CdSO4) stresses significantly upregulate similar core response genes, probably regulated by the fungal-specific Hog1-SAPK (stress-activated protein kinase) pathway. Other pathways, such as the Cap1 (AP-1 bZIP transcription factor) pathway, either acting alone or in parallel, contribute to the transcriptional response to oxidative stress [17,18]. The small heat shock protein Hsp21 of C. albicans has been implicated in protecting cells against neutrophil attack and oxidative stress (induced by menadione). Hsp21 is a virulence factor that enables C. albicans to adapt to elevated temperatures via the Cek1 kinase stress-response pathway [22]. Heat-shock proteins (HSPs) help organisms adapt to many stress conditions such as elevated growth temperatures, oxidative stress, or starvation [27]. There are five major HSP families, the rather highly conserved ATP-dependent Hsp100s, Hsp90s, Hsp70s, and Hsp60s families and the less conserved ATP-independent family of small HSPs ranging in size from 12 to 42 kDa [28]. HSP21 homologs are only found in some other Candida CTG clade species [22]. The relatively recently discovered pirin-like protein, Prn1 is possibly another important factor involved in protecting C. albicans cells from oxidative and weak organic acid stress [15,29]. Although S. cerevisiae has been used as a model yeast to study stress-response mechanisms, the responses of C. albicans to thermal, osmotic, and oxidative (H2O2) stress is quite unique, with no significant induction of genes common to the three main stress response pathways in S. cerevisiae. Apparently, C. albicans lacks the strong general stress response pathway exhibited by S. cerevisiae [18].
Response to HOCl in yeasts relates to its oxidative activity and the production of ROS typically found in NaOCl solutions [10,12,20,24]. The active chlorine species in NaOCl solution at pH 13–13.5, is the ClO− anion [30] (Figure 1). Thus, to study the effects of HOCl, pH adjustments are necessary to bring the NaOCl solution into the moderately acidic-to-neutral-pH range in which HOCl predominates. In moderately acidic-to-neutral-pH EOW, the active component is HOCl (Figure 1), which makes this a suitable agent to study the molecular mechanism of action of HOCl against C. albicans and related yeast species.
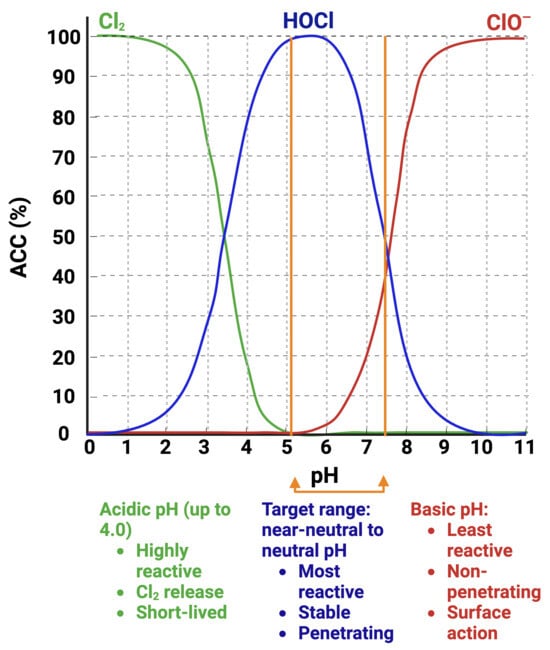
Figure 1.
Interrelationship of pH and chlorine species (available chlorine content, ACC) present in EOW. The predominant chlorine species (Cl2, HOCl, or ClO−) is pH-dependent which determines the solution stability and activity [31,32]. Created with BioRender.com.
Ready-to-use (RTU), neutral-pH EOW is an effective, surface and medical equipment disinfectant and sanitizer that lacks the toxicity associated with NaOCl (bleach) [33]. However, its mechanism of action against C. albicans is not well understood. The aim of this study was to explore the molecular mechanism of action of sub-growth-inhibitory concentrations of neutral-pH EOW-HOCl on C. albicans by analysing the transcriptional response via mRNA sequencing and RT-qPCR. We hypothesized that sub-growth-inhibitory concentrations of EOW (HOCl) would elicit a change in the expression of genes involved in the CAT1, SOD1, and TRR1 pathways, similar to the response to ROS and HOCl generated by neutrophils [20,25].
2. Materials and Methods
2.1. Determination of the Minimum Growth Inhibitory Concentration of EOW-HOCl Against C. albicans SC5314
MIC values refer to the undissociated HOCl concentration in the EOW (MIC90 EOW-HOCl). The [HOCl] of bottled EOW (Envirolyte, Auckland, New Zealand) was determined by UV–Vis spectrophotometry [34]. MIC assays were performed in 96-well microtitre plates using a method adapted from the EUCAST EDef7.1 2008 protocol [35] to determine the MIC90 of EOW-HOCl against C. albicans SC5314 at the higher cell densities required to generate sufficient RNA for RNA sequencing. C. albicans SC5314 (Molecular Biosciences Laboratory, Faculty of Dentistry, University of Otago, New Zealand) was propagated on sabouraud dextrose agar (SDA; Fort Richard, Auckland, New Zealand). Individual colonies were suspended in sterile ddH2O, and cell densities were adjusted to 107 cfu/mL (OD540 = 1.0) and 108 cfu/mL (OD540 = 10.0). Aliquots (10 µL) were added to microtitre wells containing 90 µL two-fold serial dilutions of EOW (80% to 0.3%) in sterile ddH2O, including a 90 µL no-EOW control, allowed to react for 1 and 5 min, and quenched by the addition of 100 µL 2× RPMI-1640-2%G medium (RPMI-1640; Sigma-Aldrich, Merck (Darmstadt, Germany), containing 0.165 mM MOPS buffer, Sigma-Aldrich, Merck (Darmstadt, Germany), and 2% glucose, pH 7.0). Cell suspensions were incubated statically at 37 °C for 20 h and the OD540 determined with a plate reader (Synergy 2, Bio Tek Instruments, Winooski, VT, USA). Wells with the lowest EOW concentration that had an OD540 reading at least 90% lower than the non-treated control wells indicated the MIC90 and the corresponding HOCl concentration was calculated. In other experiments, MIC assays were performed in a similar fashion with an inoculum of 108 cfu/mL, but with YNBG medium (YNB media w/o amino acids, Formedium Ltd., Hunstanton, UK, with 2% glucose). Parallel assays involved adding 10 µL of the inoculum to 90 µL of EOW dilutions pre-mixed with 100 µL 2× YNBG medium to rule out quenching effects of YNBG.
2.2. Isolation of Total RNA from C. albicans SC5314 Cells
C. albicans SC5314 was grown in YNBG (250 mL) inoculated to an OD540 of 0.05 with cells from a 16 h, 37 °C, YNBG starter culture. The culture was incubated at 37 °C with shaking until the OD540 was between 1.0 to 2.0 at which point the cultures were treated (T) with sub-growth-inhibitory concentrations of EOW-HOCl for different amounts of time; control cultures were not treated with EOW. Cells from 100 mL of the culture were harvested by filtration through a glass fibre filter No. 6 (Schleicher & Schuell BioScience GmbH, Dassel, Germany) using a vacuum manifold and washed once with 5 mL ice-cold distilled water [36]. The ‘cell cake’ was scraped off the filter with a scalpel, quickly transferred into a 1.5 mL microcentrifuge tube, snap frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted from frozen cell pellets (∼100 mg wet weight) using a hot-phenol extraction protocol [36]. Frozen cell pellets were placed in 15 mL Corex tubes containing a mixture of 1 mL acid phenol (saturated with SAB buffer; 50 mM sodium acetate, 10 mM EDTA, pH 5.0), 2 mL SAB buffer, 100 μL 10% SDS, and ∼1 g zirconia beads (0.5 mm diameter; BioSpec Products, Bartlesville, OK, USA) and heated to 65 °C in a water bath. The cells were broken by five cycles of 30 s vortexing (max speed 2500 rpm; IKA Vortex, Genius 3, Selangor Malaysia) with 1.5 min incubation at 65 °C between each cycle. Liquid phases were separated by centrifugation at 10,000× g for 10 min and traces of phenol in the ∼2 mL upper phase removed by two rounds of extraction with ~1/3 volume of chloroform. Total RNA remaining in the supernatant was precipitated with 1/10 volume of 3 M sodium acetate, pH 6.0, and 3 volumes of 100% EtOH and harvested by centrifugation at 30,600× g for 30 min at 4 °C. The RNA pellets were then washed with 1 mL 70% EtOH to remove traces of salt, air dried for 10 min, and dissolved in 200 μL RNAse-free H2O. Traces of gDNA were removed by DNase treatment (PureLink DNase kit, Invitrogen Inc., Carlsbad, CA, USA) followed by ethanol precipitation and resuspending the pellets in 0.5 mL RNAse-free H2O. RNA purity and concentrations were determined with a NanoDrop UV–Vis spectrophotometer (GE Healthcare, Amersham, UK), adjusted with RNAse-free H2O to 0.5 µg/µL, and the quantity and integrity of the total RNA extracts were confirmed by gel electrophoresis.
2.3. cDNA Template Synthesis
First strand cDNA was synthesised from 2 μg total RNA in a 20 µL reaction volume using the SuperScript IV VILO Master Mix (Invitrogen Inc., Carlsbad, CA, USA) following the manufacturer’s instructions. Control reactions without reverse transcriptase (minus RT controls; −RT) were included for each sample to confirm that the samples were not contaminated with residual gDNA.
2.4. RT-qPCR Assays
Forward and reverse primers to amplify genes of interest (Table S1) were custom-synthesised (Sigma-Aldrich, Merck, Darmstadt, Germany) and are listed in Table 1. Primers were designed to amplify similar size (~125 bp) amplicons for each gene. Amplification efficiencies for each qPCR primer pair were determined using dilutions of first strand cDNA templates starting with 10, 5, 4.2, and 3.8 ng total RNA (Figure S1). RT-qPCRs were performed in a reaction volume of 10 µL and contained 1× PowerTrack SYBR Green Master Mix (Applied Biosystems Inc., Foster City, CA, USA), 0.4 µM of each primer, and one of the cDNA dilutions. Reactions were carried out in a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems Inc., Foster City, CA, USA). Thermal cycling steps were 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s, followed by a melt (dissociation) cycle to assess the quality of the amplification products. Negative controls (no template and −RT) were included. There were two technical replicates within each RT-qPCR assay, and there were 3–6 independent repeats of each assay. The mean Cq values from all the repeat assays were used for gene expression analyses. mRNA transcript levels (2−∆Cq) were normalised to the housekeeping gene TDH3. The fold-change of mRNA levels in the treated group relative to the control was calculated with the ∆∆Cq method (2−∆∆Cq) [37]. The mean fold-change of mRNA levels (±SD) was plotted on X-Y bar graphs using GraphPad PRISM, v9.0.

Table 1.
DNA oligonucleotide primers used in the study.
2.5. RNA Sequencing
Total RNA samples (5 µg) from the C60, T60 0.125, and T60 0.5 groups (three independent replicates per group) were individually packaged as dry samples in RNA stabilization tubes (Genewiz, Azenta Life Sciences, Suzhou, China) following the manufacturer’s instructions for next generation sequencing (NGS) of mRNA using the Illumina sequencing platform (Illumina Novaseq6000, San Diego, CA, USA). RNA sequencing was conducted by Genewiz as described in Supplementary Materials and Figure S2.
Adapter sequences, PCR primers, or fragments thereof, and sequences with quality values < 20 were removed and raw sequence data (.bcl files) generated were converted into fastq files and filtered with Cutadapt (v1.9.1, phred cutoff: 20, error rate: 0.1, adapter overlap: 1 bp, minimum length: 75, proportion of N: 0.1). Sequencing reads that were <75 bases were also removed by this process to leave only high quality clean data [38,39]. The filtered transcripts were aligned against the reference genome sequence of C. albicans SC5314 (Assembly 22, ASM18296v3, available from: https://www.ncbi.nlm.nih.gov/genome/?term=ASM18296v3, accessed on 1 June 2023) using Hisat2 (v2.0.1) [40] with default parameters. Transcripts in fasta format were converted from known General Feature Format (gff) annotation files and indexed; gene and isoform expression levels from the pair-end clean data were estimated using HTSeq, (v0.6.1) [41]. Gene expression was calculated as FPKM (fragments per kilobase per million reads), based on read counts from HT-seq (v0.6.1) [42]:
Statistically significant differences of gene-specific fold change (log2 transformation) were generated with the untreated group serving as the control. Differential expression analysis was performed using the DESeq2 Bioconductor package (v1.6.3) [43] based on a model with negative binomial distribution; p (adjusted) of genes was set at <0.05 to detect differentially expressed genes (DEGs). The results from the DESeq2 analysis were further analysed to identify genes with significant differential expression according to the criteria of fold change greater than 2 (up- or down-regulated) and q value (false discovery rate, p adj < 0.05). These log2 fold-change values were used to plot column-clustered heatmaps using the WGCNA R package(v2.10.0): http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA, accessed on 15 June 2023 [44]. The correlation between fold-changes in gene expression determined by qRT-PCR and by RNAseq was determined using the Pearson’s correlation coefficient test.
GO (Gene Ontology Consortium Database; [45]) functional enrichment analysis was performed to classify the biological functions of significant DEGs (GOSeq, v1.34.1) [46]. Gene length and read count biases were included in the GO analysis using a filtering threshold for over-represented sequences by applying p < 0.05. Directed Acyclic Graph (DAG) graphical representations of the results of enrichment analysis of the differentially expressed genes were obtained using TopGO (v2.18.0). In-house scripts were used in pathway enrichment analysis based on KEGG pathway units [47] from the public pathway database, KEGG (Kyoto Encyclopedia of Genes and Genomes), using a hypergeometric test to find the pathways of the differentially expressed genes that were significantly enriched compared to the transcriptome background.
3. Results
3.1. MIC90 of EOW-HOCl at High Inoculum Densities and Culture Volumes
Microtitre MICs
Initially, C. albicans SC5314 cells were grown in 50 mL liquid medium at 37 °C with shaking at 200 rpm to ascertain whether the YNBG minimal medium lacking amino acids was suitable for growth of C. albicans (Figure 2a). These tests were necessary because typical yeast media contain sulphur containing compounds that quench the activity of EOW. YNBG medium was suitable for growing C. albicans cells, and it caused no quenching of EOW. The MIC90 of EOW-HOCl (named HOCl from now on) determined in YNBG in microtitre assays with an inoculum of 107 cells/mL was 117.5 ± 7.32 µM (n = 16 replicates). The growth cutoff in HOCl MIC assays was sharp and MIC50 values were the same as MIC90 values. To determine the effect of HOCl on the growth of larger cultures, 250 mL YNBG cultures inoculated with an overnight YNBG culture to an OD540 ≃ 0.05 were incubated at 37 °C with shaking (200 rpm) until the OD540 = ~1.0. At this point (t = 0), cultures were exposed to 0.25×-, 0.5×-, 1×- or 2×- the HOCl MIC90 and the OD540 values were measured after 60 min and 16.5 h (Figure 2b). A culture that was not treated with HOCl served as the negative control (C). OD540 measurements revealed a continuation of growth for the untreated (control) culture at 60 min from t = 0. Among the HOCl-treated (T) cultures, the T0.25× treated cells showed a mild growth inhibition after 60 min; however, cells recovered and continued to grow. Culture dilutions plated at t = 60 min after T0.25× treatment revealed survivor colony counts similar to the control culture. OD540 measurements for the T0.5×-, T1×-, and T2×- MIC90 cultures revealed no further growth after the respective treatments. Survivor colony counts of cell dilutions of the T0.5×, T1×, and T2× cultures treated for 60 min were below the limit of detection. Further tests revealed a dramatically reduced (>104 times) but still measurable survival rate (~103 cfu/mL) for the T0.5× treated culture. A 60 min exposure time of late-logarithmic cells (grey shaded area, Figure 2b) was chosen to evaluate the effects of sub-growth-inhibitory HOCl concentrations (0.125×, 0.5× MIC90) on transcription.
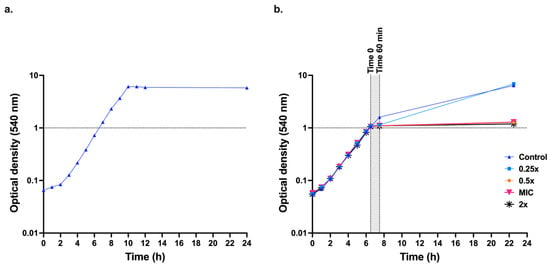
Figure 2.
Effect of EOW on C. albicans SC5314 logarithmic phase YNBG cultures. (a) Growth curve for C. albicans SC5314 cells incubated in 50 mL YNBG at 37 °C with shaking (200 rpm). (b) Growth-MIC assay; when 250 mL YNBG cultures of C. albicans SC5314 reached an OD540 ≃1.0 (t = 0), EOW was added to achieve concentrations of 0 (control), 0.25×, 0.5×, 1×, or 2× MIC90 EOW-HOCl. OD540 measurements were taken 60 min and 16.5 h after t = 0.
3.2. Harvest of Cells After EOW Exposure and RNA Extraction and Purification
Cultures of C. albicans SC5314 (YNBG, 250 mL) were incubated at 37 °C with shaking until the OD540 = 1.0. Cultures were then either untreated (control) or treated for 60 min with 0.125×, or 0.5× MIC90 HOCl. Samples (100 mL) of triplicate cultures were harvested at t = 0 and t = 60 min after treatment and total RNA was extracted and purified as described in the methods section. All DNAse-treated RNA samples showed A260 nm/A280 nm absorption ratios of between 1.8 and 2.2, and the A260 nm/A230 nm ratios were 2 to 2.3 which indicated acceptable levels of sample purity for downstream qPCR and RNA-seq analysis.
Gel electrophoresis confirmed RNA integrity and the detection of equal amounts of RNA confirmed the purity of the RNA samples. The triplicate control, T0.125, and T0.5 RNA samples harvested at t = 60 min were analysed first by RT-qPCR and then sent for mRNA sequencing.
3.3. RT-qPCR Assays
The –RT and cDNA samples were subjected to RT-qPCR. Both housekeeping genes, ACT1 and TDH3, were stably expressed. The expression levels (2−∆∆Cq) of the genes of interest were normalized to TDH3 because it was expressed at higher levels than ACT1 (lower Cq value).
There was no noticeable change to the mean Cq values for any of the candidate genes in response to 60 min exposure of cells to sub-growth inhibitory concentrations (0.125× or 0.5× MIC90) of HOCl (Table 2, Figure S3). The expression levels of ACT1, CAT1, SOD1, and TRR1 (relative to TDH3) varied (Figure S3a,b), but treatment with either 0.125× or 0.5× MIC90 HOCl, did not significantly alter the expression of any of the genes relative to the control (Figure S3c,d, Table 3).

Table 2.
mRNA expression levels of C. albicans SC5314 cells exposed to sub-growth-inhibitory EOW-HOCl concentrations for 60 min.

Table 3.
TDH3 normalised mean ∆∆ Cq values and log2 fold changes in mRNA expression levels relative to the TDH3 normalised mean ∆∆ Cq values of the untreated control (C).
Thus, none of the candidate genes nor, as expected, any of the two housekeeping genes were up- or down-regulated in response to HOCl. In order to determine which genes did actually respond to HOCl treatment, we undertook a transcriptome analysis.
3.4. Transcriptome Response of C. albicans SC5314 to HOCl
3.4.1. Filtered (Clean) Data and Assembly of Transcripts
Analysis of the mRNA sequencing data (Table S2) revealed mean total reads of 16,357,328, 18,578,325, and 16,453,547, for the control, T0.125, and the T0.5 groups, respectively. Of the total number of bases, 98% had quality scores higher than 20 (Q20); 94% (control), and 93% (T0.125, T0.5) had quality scores higher than 30 (Q30) (Q-Phred), indicating high sample purity. Of the total reads, 95%, 94%, and 93% from the control, T0.125, and T0.5 groups, respectively, aligned successfully to the reference genome.
3.4.2. Differential Expression of C. albicans Genes Following Exposure to HOCl
A total of 95 genes were significantly differentially expressed (designated ‘differentially expressed genes’; DEGs) between the EOW treatment groups and the untreated control group (Figure 3). A heatmap dendrogram of the DEGs (Figure 3a) and a Principal Component Analysis (PCA) chart (Figure S4) show the relationship (clustering) between the triplicate repeats of EOW treated samples (T0.125-1, -2, -3; T0.5-1, -2, -3) and the control samples (C-1, C-2, C-3). One sample each from the treated (T0.5-1) and the untreated (C-1) groups was an outlier. The outliers could possibly be due to multiple freeze–thaw cycles of these total RNA samples. However, in the overall analyses, all triplicates were included in the log2 fold change calculations and in the downstream qPCR validation assays.
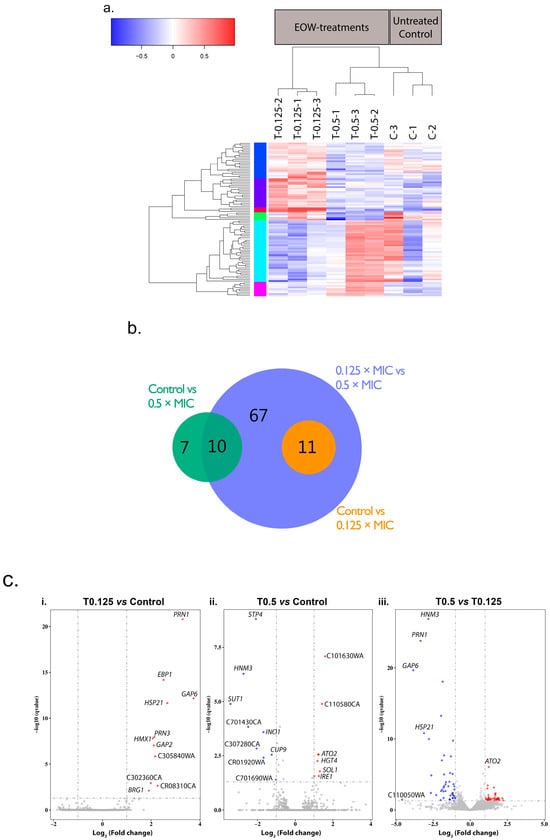
Figure 3.
Comparison of gene expression in samples within and between treatment groups. (a) Heatmap dendrogram for the normalized mRNA expression levels (log2 of FPKMs) of the DEGs. Gene expression is relative to their combined average expression levels for each of the three biological replicates grown in the absence (60 min; C-1, -2, -3) or presence (60 min; T0.125-1, -2, -3; T0.5-1, -2, -3) of sub-growth inhibitory concentrations of HOCl. Colours of the vertical bar next to the y-axis of the dendrogram indicate different groups of transcripts with similar patterns of down- (blue) and/or up-regulation (red) between the various treatment groups. (b) Venn diagram showing the number of DEGs between the indicated pairs of treatment groups. (c) Genes differentially expressed in response to sub-growth-inhibitory HOCl. Volcano plots of the DEGs between the three pairs of treatment comparisons (i–iii) displaying the –log10 q value (y-axis) vs. log2 fold change (x-axis). The cut-off values (dotted grey lines) used to define significantly DEGs were q < 0.05 and a log2 fold change > |1|, i.e., larger than 2-fold (up- or down-regulated). Red data points—significantly upregulated genes; blue data points—significantly down regulated genes; grey data points—no significant change in expression.
A Venn diagram (Figure 3b) and volcano plots (Figure 3c) showed that in the three comparisons of gene expression, 61 genes were upregulated and 55 were downregulated. When cells were treated with the lower HOCl concentration (T0.125), 11 genes were upregulated compared to the control. With the higher HOCl concentration (T0.5), eight genes were upregulated, and nine genes were downregulated. Forty-two and 46 genes were upregulated and downregulated, respectively, when expression in T0.5 cells was compared to expression in T0.125 cells. For all three comparisons there was a total of 116 DEGs; however there were 21 DEGs that were common between comparison groups (Figure 3b), hence there were 95 unique DEGs. GO annotations of the 56 most prominent GO terms enriched in the DEGs revealed that 33 genes are involved in molecular function, 17 are involved in cellular components, and 34 genes are involved in biological processes (Table S3).
3.4.3. Transcriptional Response of C. albicans SC5314 Cells to Sub-Growth-Inhibitory Concentrations of HOCl
The transcriptional response of C. albicans SC5314 cells to EOW elicited two distinct concentration-based responses. With the 0.125× MIC90 HOCl treatment, 11 genes were upregulated with a log2 fold change in expression between 1.9 to 3.7 relative to the control (C) samples. A comparison of the mean FPKM values with those of the control and the T0.5 treatment groups demonstrated that these genes were specifically modulated in response to low concentrations of HOCl (0.125× MIC90), because the control and 0.5× MIC90-treated samples showed comparable expression levels (Table 4).

Table 4.
DEGs of C. albicans SC5314 in response to either lower (T0.125; group 1) or higher (T0.5; group 2) concentrations of HOCl.
The biggest response of cells to 0.125× MIC90 was the upregulation of GAP6, PRN1, and HSP21 (Figure 3c, Table 4; refer to Table S1 for gene characteristics), followed by two uncharacterized genes (CAALFM_CR08310CA and CAALFM_C305840WA), EBP1, GAP2, PRN3, HMX1, CAALFM_C302360CA, and BRG1 Figure 3c; Table S1). Of note in the T0.125 comparison to the control cells (Table 4) is that no genes were significantly downregulated (Figure 3c); this is possibly because most of the downregulated genes were of very low abundance.
With the 0.5× MIC90 HOCl treatment (Group 2, Table 4), eight of the significantly upregulated genes (ATO2, HGT4, SOL1, IRE1, and the uncharacterized genes CAALFM_C101630WA, CAALFM_C110580CA, CAALFM_C305250CA, and CAALFM_CR06020WA) were at most 1.8 (log2) upregulated, suggesting that the cells had little chance to adapt to the higher (59 µM) HOCl concentration. The most significant upregulation was that of two uncharacterized genes (CAALFM_C101630WA and CAALFM_C110580CA), followed by ATO2 (Figure 3c; Table S1). ATO2 stands out as the gene with the most significantly upregulated transcript compared to the control and T0.125 cells (Table 4, Figure 3c). Of the nine significantly downregulated genes, three were genes of unknown function (CAALFM_C701690WA, CAALFM_CR01920WA, and CAALFM_C701430CA). SUT1 was the most downregulated gene followed by HNM3, STP4, CAALFM_CR01920WA, INO1, and CUP9 (Table 4).
When comparing the two treatments (Table 4), it is evident that the lower concentration and the higher concentration of HOCl elicited two completely different types of responses. GAP6, PRN1, and HSP21 were the key genes upregulated in response to 0.125× MIC90 HOCl, based on their degree of upregulation and also their levels of expression (FPKM values). Some of the more significantly downregulated genes in response to the higher concentration of HOCl were involved in mitochondrial function and RNA or DNA processing. In summary, eleven genes were upregulated in response to 0.125× MIC90 HOCl treatment; the most notably differentially expressed genes were GAP6, PRN1, and HSP21. There were two uncharacterized genes (CAALFM_C101630WA, CAALFM_C110580CA) and ATO2 that were specifically upregulated in response to 0.5× MIC90 HOCl. The most significant response to 0.5× MIC90 HOCl, however, was the downregulation of a number of genes, most notably HNM3, INO1, SUT1, and three uncharacterized genes (CAALFM_CR01920WA, CAALFM_C307280CA, CAALFM_C701430CA), which had relatively high expression levels in the control and T0.125 cells.
3.5. Confirmation of Gene Expression by RT-qPCR
To validate the RNA-seq results, RT-qPCR assays were performed for five key DEGs (ATO2, EBP1, GAP6, HSP21, and PRN1) (Figure 4). The RT-qPCR results for the log2 fold change in expression of the DEGs showed a positive correlation with the RNA-seq results (Figure 4a, R2 = 0.71 for the T0.125 treatment, and Figure 4b, R2 = 0.90 for the T0.5 treatment). There was upregulation of all the DEGs that were selected for RT-qPCR, relative to the control with the 0.125× MIC90 HOCl treatment (Figure 4c); in contrast the 0.5× MIC90 HOCl treatment caused negligible changes in expression (Figure 4d). In general, there was greater upregulation by the T0.125 treatment (Figure 4e) than by the T0.5 treatment relative to the control; however, there was greater upregulation of ATO2 in the T0.5 cells than in the T0.125 cells (Figure 4f).
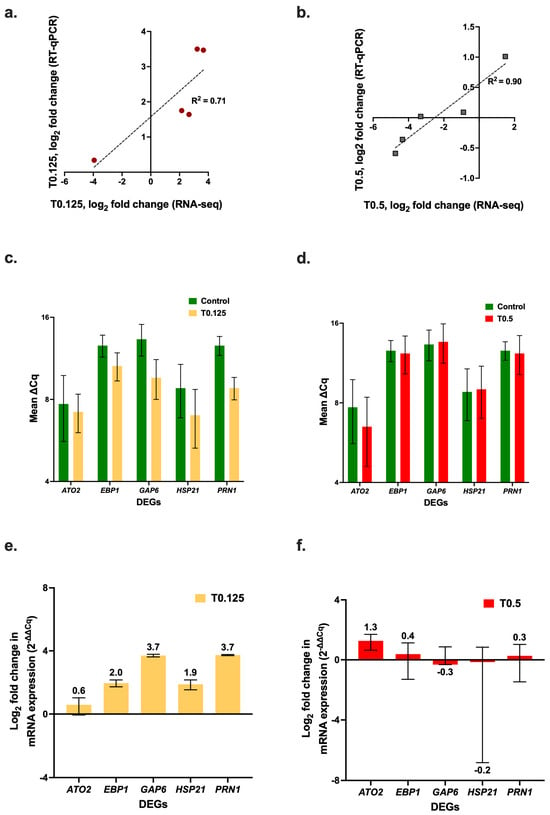
Figure 4.
Quantification of C. albicans SC5314 gene expression levels in cells treated with sub-growth-inhibitory concentrations of HOCl. Correlation between the RT-qPCR and the RNA-seq log2-fold change values for the (a) 0.125× and (b) 0.5× MIC90 HOCl treatment samples. RT-qPCR quantification of C. albicans SC5314 ATO1, EBP1, GAP6, HSP21, and PRN1 mRNA expression levels (∆Cq) normalized with TDH3 for the (c) 0.125× MIC90 and (d) 0.5× MIC90 HOCl treatment samples. Log2 fold change in expression (2−∆∆Cq) of logarithmic phase cells in response to 60 min exposure to (e) 0.125× MIC90 and (f) 0.5× MIC90 HOCl.
4. Discussion
The hypothesis underpinning this study, based on previous research [25], was that treatment of C. albicans cells with sub-growth-inhibitory concentrations of EOW (HOCl) would elicit a significant change in expression levels of genes (CAT1, SOD1, TRR1) involved in the oxidative stress response pathway. However, there was no significant change in the expression of any of these genes in response to HOCl exposure, and hence, the research hypothesis was rejected. A recent study examining the effect of HOCl on C. albicans also found a minimal change in SOD1 expression but reported increased expression of CAT1 and TRR1 as well as increased expression of MXR1 and SRX1. The reason for the differences in gene expression in that study may be the different concentration of HOCl used (5 µM) and the different exposure time (15 min) [14]. In addition, the study did not state the source of HOCl or how the concentration was measured. In the present study, the responses of C. albicans cells to the lower (15 µM) and the higher (59 µM) concentrations of HOCl were distinctly different. Eleven genes of C. albicans cells exposed to 15 µM HOCl for 60 min were between 4-fold and 14-fold upregulated: PRN1 and PRN3 (proteins with similarity to pirins), HSP21, GAP6, and GAP2 (two broad-specificity amino acid permeases), EBP1 (an NADPH oxidoreductase), HMX1 (a heme oxygenase), and two uncharacterized proteins (CAALFM_CR08310CA, CAALFM_C305840WA).
Two relatively recent reports [15,29] revealed an important function for Prn1 in the oxidative stress response of C. albicans to H2O2. An increase in Prn1 expression along with increases in the expression of Hsp21, Ebp1, and many other proteins involved in the oxidative stress response, protein folding, and proteasome-dependent catabolism were reported. In the more recent of the two studies [29], deletion of PRN1 caused cells to accumulate increased levels of ROS when exposed to high concentrations of H2O2; a higher proportion of ∆PRN1 knock-out cells showed signs of apoptosis and they also exhibited increased proteasomal activity when compared to wild-type control cells treated the same way. In C. albicans, the small heat shock protein Hsp21 enables cells to adapt to environmental stress (heat- and menadione-induced) by fine-tuning the homeostasis of intracellular stress protectants such as glycerol and trehalose via the activation of the Cek1 kinase pathway. Hsp21 was also found to be an important virulence factor of C. albicans [22]. Hsp21 is required to resist killing by human neutrophils; it is involved in regulating glycerol, glycogen, and trehalose homeostasis in response to elevated temperatures [22], and it also contributes to the response to ethanol-induced stress [48]. GAP2 and GAP6 are amino acid permeases and the uncharacterized gene, CAALFM_CR08310CA with a 6-fold upregulation, was assigned to processes involving alkanesulfonate catabolism with possible oxidoreductase and/or sulfonate dioxygenase activity. The upregulation of these genes may protect cells from the HOCl stress by removing potentially toxic proteins with HOCl modified sulphur containing amino acids and replenishing the cells with a fresh supply of amino acids from the surrounding environment.
A 60 min exposure of C. albicans cells to 59 µM HOCl mostly caused the downregulation of DEGs. Among the 17 DEGs, there was a significant downregulation of SUT1 (a Zn2Cys6 transcription factor involved in sterol uptake), HNM3 (a putative transporter), STP4 (a C2H2 transcription factor), and INO1 (inositol-1-phosphate synthase). There were only a few genes upregulated (<1.5 log2-fold) in response to 59 µM HOCl: ATO2 (an ammonia exporter), HGT4 (a high-affinity glucose transporter [26]), and IRE1 (a protein kinase involved in the regulation of the unfolded protein response (UPR) [26]), indicating that cells struggled to cope with the stress at the higher HOCl concentration. This study used neutral-pH EOW with a pH value of 6.8 and was diluted 75-fold to achieve the higher (59 µM) HOCl concentration, thus it is unlikely that the vehicle for delivering the HOCl will have had a significant effect on gene expression.
Cells exposed to 15 µM HOCl showed a slight growth lag after the 60 min time-point, but recovered from the EOW-induced effects, as reflected in optical density measurements and plate counts that were similar to those of the no-treatment control (107 cfu/mL). In contrast, exposure to 59 µM HOCl prevented further cell growth. Aliquots of these cultures from assay repeats revealed varied results, unlike those for the control and 0.125× MIC90 HOCl-treated cultures. The results ranged from significantly reduced cell counts (~103 to 104 cfu/mL) to no detectable live cell counts. Furthermore, the colony morphologies of cells grown in the presence of 0.5× MIC90 HOCl were also quite different from cells grown at the lower HOCl concentration; this raises the possibility that these cells were shutting down key cellular processes and were transitioning to a viable but nonculturable (VBNC) state, but this hypothesis requires verification. Literature on the effects of biocides on the VBNC state of fungi are limited. The VBNC state occurs in the phenol-degrading strain Candida sp. LN1 (a strain closely related to C. albicans SC5314) as an adaption to redox stress [49]. Furthermore, the initial RT-qPCR experiments showed an ~2-fold downregulation of the CAT1, SOD1, and TRR1 genes in response to 0.5× MIC90 HOCl (Figure S3d). This was also echoed in the RNA-seq transcriptomic response. It was previously reported that genes encoding proteins involved in the oxidative stress response (including superoxidase dismutase and thioredoxin reductase) were significantly downregulated when Brettanomyces bruxellensis yeast cells treated with 0.8 mg/L SO2 entered a VBNC state [50]. Similar adaptive responses may occur in C. albicans SC5314 cells that were exposed to 59 µM (0.5× MIC90) HOCl redox stress, but again this hypothesis requires verification. In another study [13], a 10 to 15 min exposure to HOCl concentrations, similar to those used in this research, significantly affected S. cerevisiae cell counts. A lack of instantaneous killing of S. cerevisiae cells was reported. Instead, the slow progressive loss of K+ from cells with increasing HOCl concentration indicated that the plasma membrane remained intact. It was proposed that the range of effects on metabolism meant that subsequent plating on rich medium may or may not give rise to cell replication and colony formation, hence, causing variable cell counts [13]. This may explain the variable cell counts observed for C. albicans cells exposed to 59 µM HOCl.
The UPR is an adaptive mechanism that is activated in response to the presence of unfolded or misfolded proteins in the ER [51]. The reliance of C. albicans on the ER-resident Ire1 protein for sensing ER stress and activating the UPR in response to antimicrobial (tunicamycin)-induced ER stress has been reported [51]. C. albicans cells respond to ER stress primarily by activating the conserved Ire1-Hac1-dependent UPR pathway and then by timely attenuation of the pathway once homeostasis is achieved by activation of the Ire-mediated HOG1-MAPK pathway [51,52]. Cell death in S. cerevisiae after exposure to 300 µM HOCl is mainly due to apoptosis that results in elevated levels of reactive oxygen species (ROS) and the formation of HOCl-modified proteins; HOCl lethality is mediated by the protease Kex1p [12]. The significant enrichment of the IRE-mediated UPR and the intrinsic apoptotic signalling pathway in response to ER stress that was triggered by exposure of C. albicans cells to 0.5× MIC90 of HOCl (59 µM) in the present study echoes these findings.
Previous studies suggested that C. albicans possesses additional mechanism(s) (absent in S. cerevisiae) by which H2O2 exposure triggers a superoxide stress response [53]. Such alternative mechanisms could possibly influence the adaptive response of C. albicans to HOCl. Taken together, our findings suggest that the stress response to sub-inhibitory concentrations of EOW triggers a unique stress response pathway that is quite distinct from the oxidative stress response pathway induced by H2O2 or menadione.
Limitations of stress-response studies include limited numbers of time points, variations of pH and growth media, and the use of varying concentrations of drugs between various research teams, all of which are likely to affect the results. Most C. albicans stress genes in response to H2O2 are induced within 10 to 30 min exposure times [17,18]. The choice of a 60 min time point is a possible limitation of the present study. Future time course studies should investigate a range of HOCl concentrations at additional time points including a five min exposure which is used in industry-standard disinfectant efficacy assays. Comparisons of global cellular responses among studies are limited by a lack of standard conditions for stress induction and transcription profile analysis; hence details of the activating process and measurement of the response varies [17]. The protocol used in the present study was limited by the quenching effects of RPMI-1640 on the EOW-HOCl [10], which is why we used YNBG without any amino acids to study the effects of HOCl on C. albicans cells.
The growing threat of opportunistic C. albicans infections in vulnerable individuals, including those residing in aged-care facilities, highlights the potential danger of C. albicans adaptive resistance to antimicrobial agents and to biocides. This study identified the genes that are possibly responsible for the stress response of C. albicans to sub-inhibitory HOCl concentrations. The role of these genes in biocide tolerance or resistance should be confirmed by gene deletions and by measuring their effects on the susceptibility of C. albicans cells to EOW and other stress inducers, including growth at elevated temperature (heat stress) or exposure to high salt (osmotic stress) or H2O2 (oxidative stress).
5. Conclusions
The present study identified Hsp21 and Prn1 as potentially important factors in protecting C. albicans cells against HOCl. A moderate sub-growth-inhibitory concentration (15 µM) of HOCl significantly upregulated GAP6, HSP21, and PRN1. A higher sub-growth-inhibitory concentration (59 µM) of HOCl induced mostly significant downregulation of C. albicans genes, reduced cell growth, and the upregulation of a few genes involved in key metabolic processes, including carbohydrate metabolism, ammonium transport, and the unfolded protein response pathway. The lethal action of HOCl on C. albicans cells may be due to ER protein unfolding events and apoptosis. Neutrophils produce high levels of myeloperoxidase, which converts H2O2 generated during the oxidative burst into the much more reactive HOCl. This study led to the hypothesis that Hsp21 and Prn1 are also key players in protecting C. albicans cells from neutrophil attack by the innate host immune system, but this requires validation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof11110761/s1.
Author Contributions
Conceptualization, C.S.K., R.D.C. and E.L.; methodology, C.S.K., R.D.C., G.R.T., T.J.M. and E.L.; software, C.S.K., T.J.M. and E.L.; validation, C.S.K., R.D.C., G.R.T., T.J.M. and E.L.; formal analysis, C.S.K.; investigation, C.S.K.; resources, C.S.K. and R.D.C.; data curation, C.S.K. and E.L.; writing—original draft preparation, C.S.K.; writing—review and editing, R.D.C., G.R.T. and E.L.; visualization, R.D.C. and E.L.; supervision, R.D.C. and G.R.T.; project administration, R.D.C.; funding acquisition, C.S.K., R.D.C. and G.R.T. All authors have read and agreed to the published version of the manuscript.
Funding
Chitra S. Krishnan was recipient of a University of Otago doctoral scholarship and a University of Otago postgraduate (Doctoral) publishing bursary. This research was supported by grants-in-aid from the Sir John Walsh Research Institute, University of Otago, the Maurice and Phyllis Paykel Trust (grant 191124), New Zealand, and the New Zealand Dental Research Foundation (grant RF8.11 2019).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| cDNA | complementaryDNA |
| CGD | Candida Genome Database |
| Cq | Quantification Cycle (threshold cycle) |
| DAG | Directed Acrylic Graph |
| DEGs | Differentially Expressed Genes |
| DEU | Differential Exon Usage |
| DNA | Deoxyribonucleic Acid |
| EOW | Electrolysed Oxidising Water |
| ER | Endoplasmic Reticulum |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| FPKM | Fragments Per Kilobase per Million reads |
| GO | Gene Ontology Consortium Database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MIC | Minimum Inhibitory Concentration |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| mRNA | messengerRNA |
| NGS | Next Generation Sequencing |
| OD | Optical Density |
| PCA | Principal Component Analysis |
| RNA-seq | Ribonucleic Acid-sequencing |
| ROS | Reactive Oxygen Species |
| RPKM | Reads Per Kilobase of transcript per Million reads |
| RPMI | Rosewell Parks Memorial Institute |
| RT | Reverse Transcriptase |
| RT-qPCR | Reverse Transcription-Quantitative Polymerase Chain Reaction |
| RTU | Ready-to-Use |
| SDA | Sabouraud’s Dextrose Agar |
| UPR | Unfolded Protein Response |
| UV–Vis | UltaViolet–Visible |
| VBNC | Viable But Non-Culturable |
| YNBG | Yeast Nitrogen Base Glucose |
References
- Dukan, S.; Touati, D. Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 1996, 178, 6145–6150. [Google Scholar] [CrossRef] [PubMed]
- Dukan, S.; Belkin, S.; Touati, D. Reactive oxygen species are partially involved in the bacteriocidal action of hypochlorous acid. Arch. Biochem. Biophys. 1999, 367, 311–316. [Google Scholar] [CrossRef]
- Dunn, L.L.; Smith, D.M.; Critzer, F.J. Transcriptomic behavior of Salmonella enterica Newport in response to oxidative sanitizers. J. Food Prot. 2020, 83, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Loi, V.V.; Busche, T.; Tedin, K.; Bernhardt, J.; Wollenhaupt, J.; Huyen, N.T.T.; Weise, C.; Kalinowski, J.; Wahl, M.C.; Fulde, M.; et al. Redox-sensing under hypochlorite stress and infection conditions by the Rrf2-family repressor Hypr in Staphylococcus aureus. Antioxid. Redox Signal. 2018, 29, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Merchel Piovesan Pereira, B.; Wang, X.; Tagkopoulos, I. Short- and long-term transcriptomic responses of Escherichia coli to biocides: A systems analysis. Appl. Environ. Microbiol. 2020, 86, e00708-20. [Google Scholar] [CrossRef]
- Wang, S.; Deng, K.; Zaremba, S.; Deng, X.; Lin, C.; Wang, Q.; Tortorello, M.L.; Zhang, W. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 2009, 75, 6110–6123. [Google Scholar] [CrossRef]
- Wang, X.; Wu, H.; Niu, T.; Bi, J.; Hou, H.; Hao, H.; Zhang, G. Downregulated expression of virulence factors induced by benzyl isothiocyanate in Staphylococcus aureus: A transcriptomic analysis. Int. J. Mol. Sci. 2019, 20, 5441. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Ilbert, M.; Graf, P.C.; Ozcelik, D.; Jakob, U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 2008, 135, 691–701. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Lu, J.; Yu, Z.; Song, H.; Bond, P.L.; Guo, J. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress. ISME J. 2021, 15, 2969–2985. [Google Scholar] [CrossRef]
- Wagner, D.K.; Collins-Lech, C.; Sohnle, P.G. Inhibition of neutrophil killing of Candida albicans pseudohyphae by substances which quench hypochlorous acid and chloramines. Infect. Immun. 1986, 51, 731–735. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving reactive chlorine stress: Responses of Gram-negative bacteria to hypochlorous acid. Microorganisms 2020, 8, 1220. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Alavian-Ghavanini, A.; Habernig, L.; Bauer, M.A.; Hammer, A.; Rossmann, C.; Zimmermann, A.S.; Ruckenstuhl, C.; Buttner, S.; Eisenberg, T.; et al. The cell death protease Kex1p is essential for hypochlorite-induced apoptosis in yeast. Cell Cycle 2013, 12, 1704–1712. [Google Scholar] [CrossRef][Green Version]
- King, D.A.; Hannum, D.M.; Qi, J.S.; Hurst, J.K. HOCl-mediated cell death and metabolic dysfunction in the yeast Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2004, 423, 170–181. [Google Scholar] [CrossRef]
- Douglas, L.M.; Min, K.; Konopka, J.B. Candida albicans resistance to hypochlorous acid. mBio 2023, 14, e02671-23. [Google Scholar] [CrossRef]
- Amador-García, A.; Zapico, I.; Borrajo, A.; Malmström, J.; Monteoliva, L.; Gil, C. Extending the proteomic characterization of Candida albicans exposed to stress and apoptotic inducers through data-independent acquisition mass spectrometry. mSystems 2021, 6, e00946-21. [Google Scholar] [CrossRef]
- Cottier, F.; Tan, A.S.; Chen, J.; Lum, J.; Zolezzi, F.; Poidinger, M.; Pavelka, N. The transcriptional stress response of Candida albicans to weak organic acids. G3 (Bethesda) 2015, 5, 497–505. [Google Scholar] [CrossRef]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.P.; Quin, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 2006, 17, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Nantel, A.; Whiteway, M. Stress-induced gene expression in Candida albicans: Absence of a general stress response. Mol. Biol. Cell 2003, 14, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, C.; Collette, J.R.; Brothers, K.M.; Shepardson, K.M.; Cramer, R.A.; Wheeler, R.T.; Lorenz, M.C. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot. Cell 2013, 12, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kwolek-Mirek, M.; Bartosz, G.; Spickett, C.M. Sensitivity of antioxidant-deficient yeast to hypochlorite and chlorite. Yeast 2011, 28, 595–609. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Jacobsen, I.D.; Miramon, P.; Slesiona, S.; Bohovych, I.M.; Brown, A.J.; Hube, B. Small but crucial: The novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS ONE 2012, 7, e38584. [Google Scholar] [CrossRef] [PubMed]
- Ramsdale, M.; Selway, L.; Stead, D.; Walker, J.; Yin, Z.; Nicholls, S.M.; Crowe, J.; Sheils, E.M.; Brown, A.J.P. MNL1 regulates weak acid induced stress responses of the fungal pathogen Candida albicans. Mol. Biol. Cell 2008, 19, 4393–4403. [Google Scholar] [CrossRef]
- Ratti, B.A.; Godoy, J.S.; de Souza Bonfim Mendonca, P.; Bidoia, D.L.; Nakamura, T.U.; Nakamura, C.V.; Lopes Consolaro, M.E.; Estivalet Svidzinski, T.I.; de Oliveira Silva, S. Microbicidal activity of neutrophils is inhibited by isolates from recurrent vaginal candidiasis (RVVC) caused by Candida albicans through fungal thioredoxin reductase. Cell. Immunol. 2015, 293, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Dantas Ada, S.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef]
- Candida Genome Database (CGD); Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. Available online: http://www.candidagenome.org/ (accessed on 1 June 2023).
- Lindquist, S. The heat-shock response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Arribas, V.; Monteoliva, L.; Hernaez, M.L.; Gil, C.; Molero, G. Unravelling the role of Candida albicans Prn1 in the oxidative stress response through a proteomics approach. Antioxidants 2024, 13, 527. [Google Scholar] [CrossRef]
- Nakagawara, S.; Goto, T.; Nara, M.; Ozawa, Y.; Hotta, K.; Arata, Y. Spectroscopic characterization and the pH dependence of bactericidal activity of the aqueous chlorine solution. Anal. Sci. Int. J. Jpn. Soc. Anal. Chem. 1998, 14, 691–698. [Google Scholar] [CrossRef]
- Fukuzaki, S. Mechanisms of actions of sodium hypochlorite in cleaning and disinfection processes. Biocontrol Sci. 2006, 11, 147–157. [Google Scholar] [CrossRef]
- Dewi, F.R.; Stanley, R.; Powell, S.M.; Burke, C.M. Application of electrolysed oxidising water as a sanitiser to extend the shelf-life of seafood products: A review. J. Food Sci. Technol. 2017, 54, 1321–1332. [Google Scholar] [CrossRef]
- Krishnan, C.S.; Tompkins, G.R.; Lyons, K.M.; Cannon, R.D. Electrolysed oxidising water as a multi-purpose biocide in dental healthcare-A scoping review. Gerodontology 2023, 40, 422–462. [Google Scholar] [CrossRef]
- Krishnan, C.S.; Lyons, K.M.; Tompkins, G.R.; Cannon, R.D. Storage-related stability and antimicrobial efficacy of bottled, neutral-pH Electrolysed Oxidising Water. J. Dent. 2023, 137, 104656. [Google Scholar] [CrossRef]
- Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing. EUCAST definitive document EDef 7.1: Method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef]
- James, J.E.; Lamping, E.; Santhanam, J.; Milne, T.J.; Abd Razak, M.F.; Zakaria, L.; Cannon, R.D. A 23 bp cyp51A promoter deletion associated with voriconazole resistance in clinical and environmental isolates of Neocosmospora keratoplastica. Front. Microbiol. 2020, 11, 272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Andrew, S. FastQC: A Quality Control Tool for High Throughput Sequence Data, 0.11.4; Babraham Institute: Cambridge, UK, 2015; Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 28 April 2023).
- Martin, M. Cutadapt: Removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Hsp21 potentiates antifungal drug tolerance in Candida albicans. PLoS ONE 2013, 8, e60417. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Xu, L.; Zhang, R.; Zhou, Y.; Xiao, Y.; Su, X.; Shen, C.; Sun, F.; Hashmi, M.Z.; Lin, H.; et al. Viable but nonculturable state of yeast Candida sp. strain Ln1 induced by high phenol concentrations. Appl. Environ. Microbiol. 2021, 87, e0111021. [Google Scholar] [CrossRef]
- Capozzi, V.; Di Toro, M.R.; Grieco, F.; Michelotti, V.; Salma, M.; Lamontanara, A.; Russo, P.; Orru, L.; Alexandre, H.; Spano, G. Viable But Not Culturable (VBNC) state of Brettanomyces bruxellensis in wine: New insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbiol. 2016, 59, 196–204. [Google Scholar] [CrossRef]
- Sircaik, S.; Roman, E.; Bapat, P.; Lee, K.K.; Andes, D.R.; Gow, N.A.R.; Nobile, C.J.; Pla, J.; Panwar, S.L. The protein kinase Ire1 impacts pathogenicity of Candida albicans by regulating homeostatic adaptation to endoplasmic reticulum stress. Cell. Microbiol. 2021, 23, e13307. [Google Scholar] [CrossRef]
- Husain, F.; Pathak, P.; Roman, E.; Pla, J.; Panwar, S.L. Adaptation to endoplasmic reticulum stress in Candida albicans relies on the activity of the Hog1 mitogen-activated protein kinase. Front. Microbiol. 2021, 12, 794855. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, D.J.; Stephen, D.W.; Terriere, E.C. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 1996, 138, 83–88. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).