From Isolation to Genomics: Characterization of Aspergillus uvarum HT4 as a Novel Producer of Extracellular Tannase
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Isolation of Tannase-Producing Fungi
2.3. Selection of Fungi with Extracellular Tannase Activity
2.3.1. Extracellular Tannase Production in Liquid Medium
2.3.2. Tannase Activity Assay
2.4. Molecular Identification of Tannase Producers
2.4.1. DNA Extraction
2.4.2. Calmodulin Gene Amplification
2.4.3. Phylogenetic Analysis of Calmodulin Sequences
2.5. Whole-Genome Sequencing, Assembly, and Annotation of the Selected Isolate
Identification and Phylogenetic Analysis of Tannase Sequences
2.6. Characterization of TE Produced by the Selected Isolate
2.6.1. Protein Determination
2.6.2. Effect of pH and Temperature on Tannase Activity
2.6.3. Verification of Tannase Activity by Zymogram
2.6.4. Protein Identification by Nano LC-MS/MS
2.6.5. Secretory Proteins in the HT4 Enzymatic Extract from HT4
3. Results
3.1. Isolation and Screening of Tannase-Producing Fungi
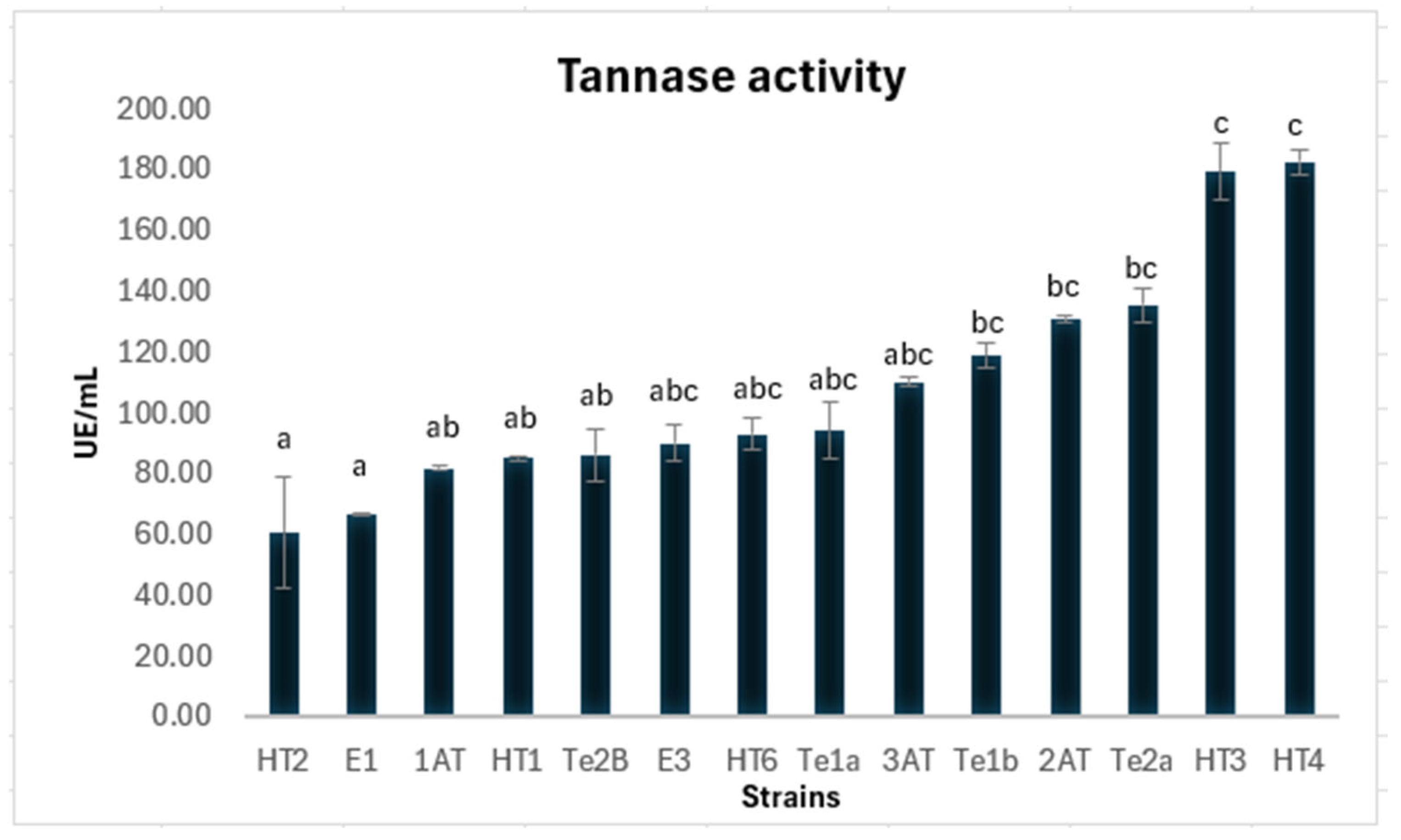
3.2. Extracellular Tannase Production in Liquid Medium
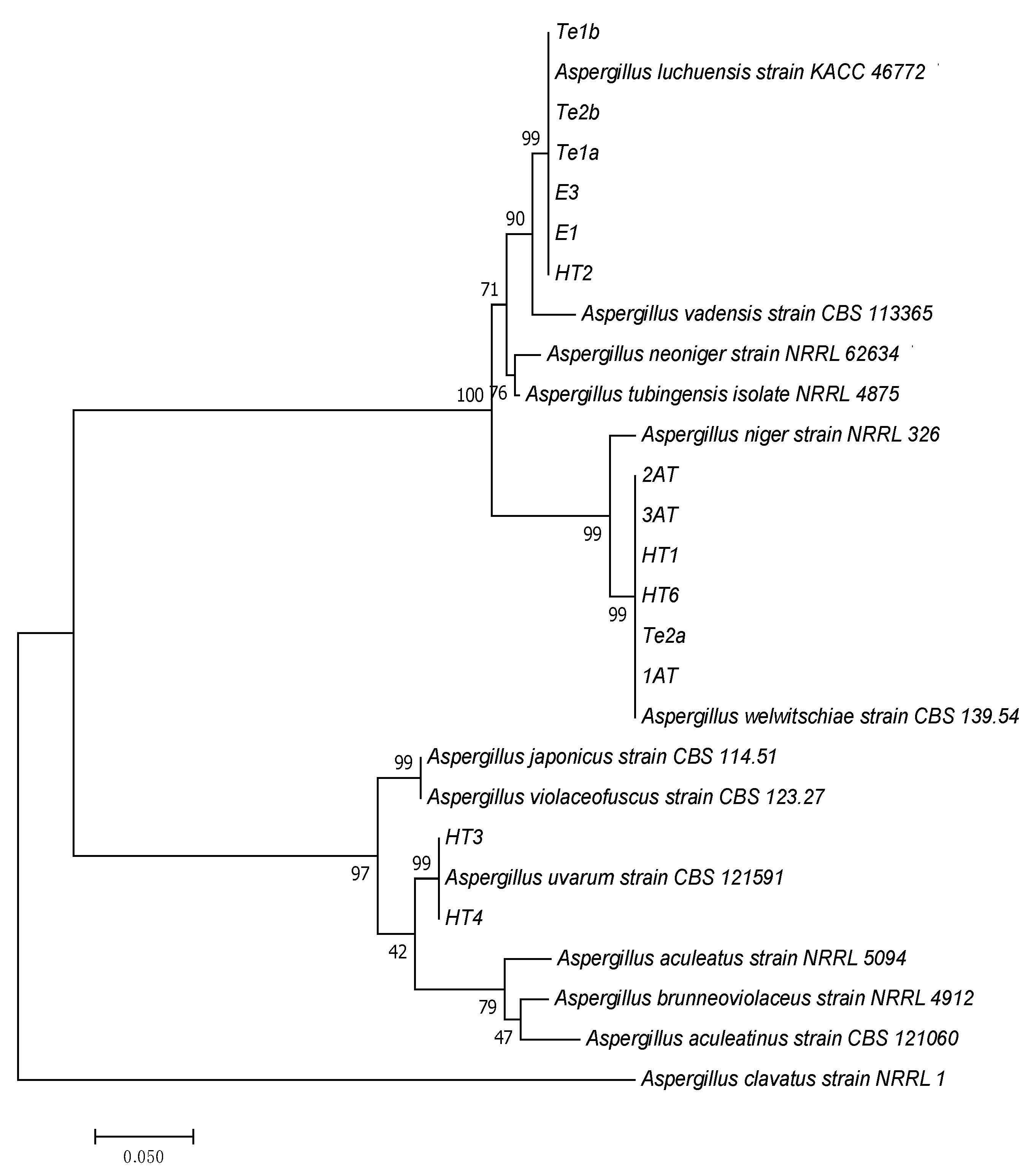
3.3. Molecular Identification of Tannase Producers
3.4. Genome Analysis of the Selected Strain
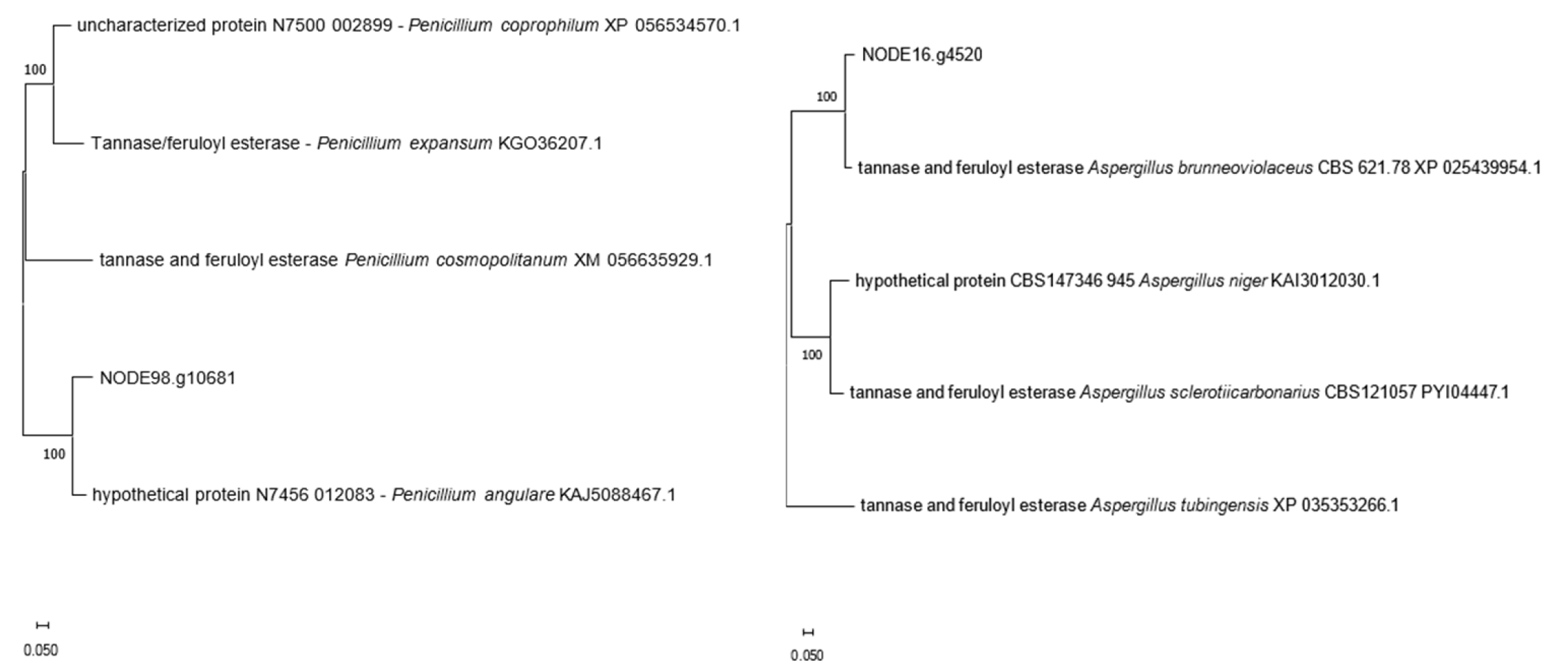
3.4.1. Tannase Sequences in HT4 Genome
3.4.2. Identification of Secondary Metabolite Gene Clusters
3.5. Characterization of TE Produced by the Selected Isolate
3.5.1. Protein Determination
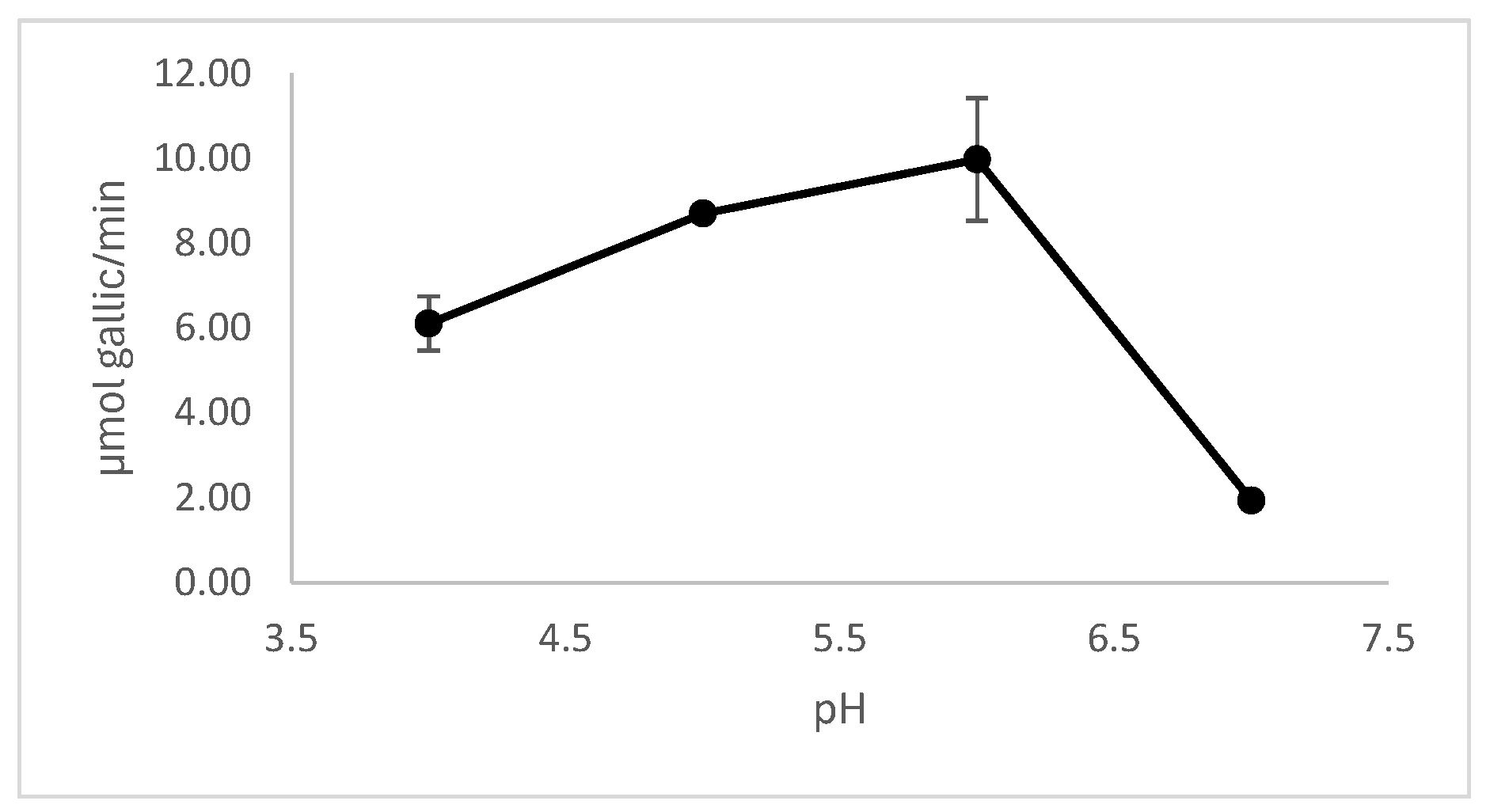
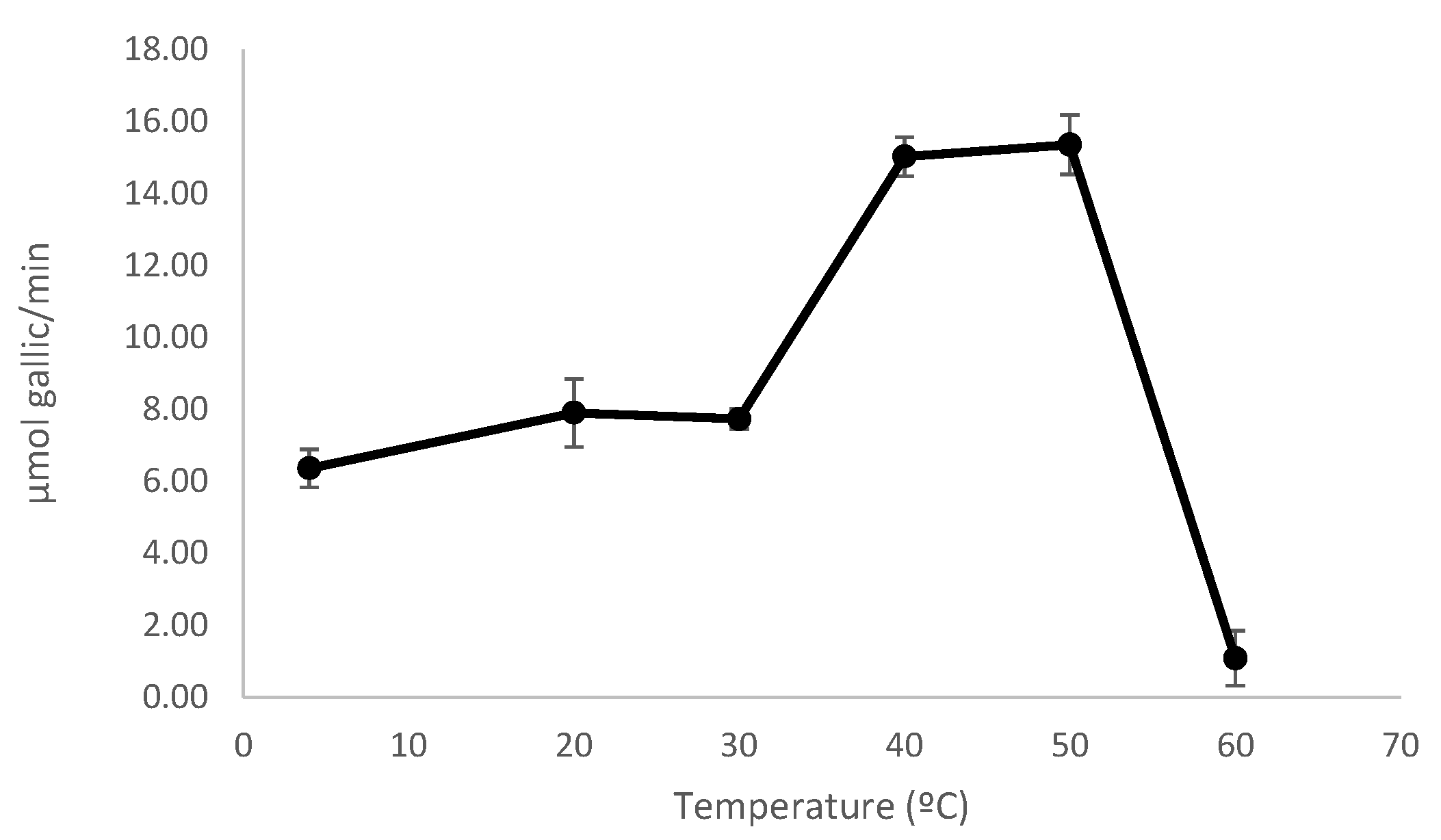
3.5.2. Effect of pH and Temperature on Enzyme Activity
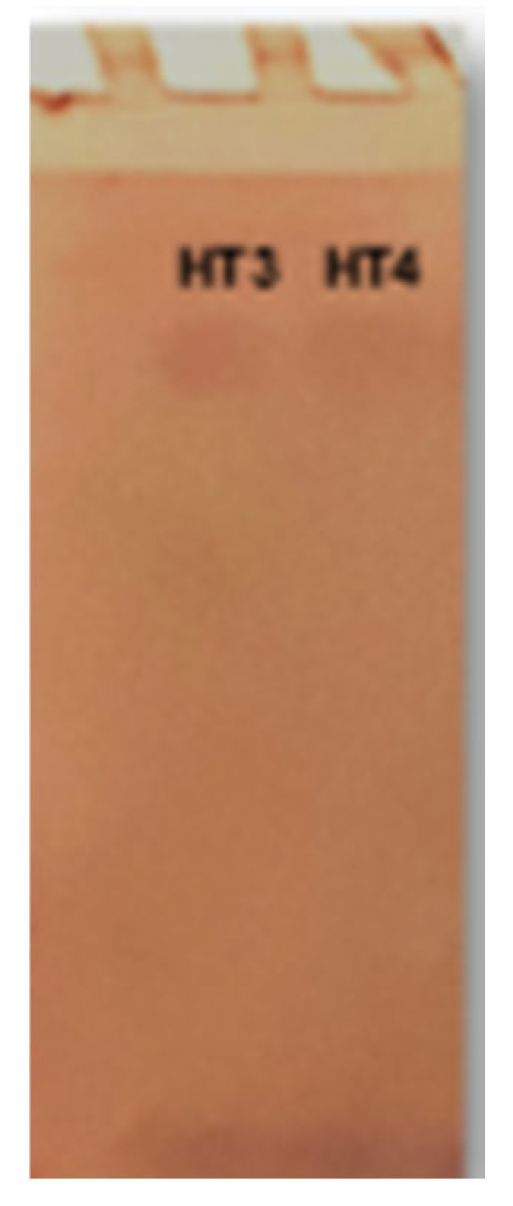
3.5.3. Tannase Verification by Zymogram
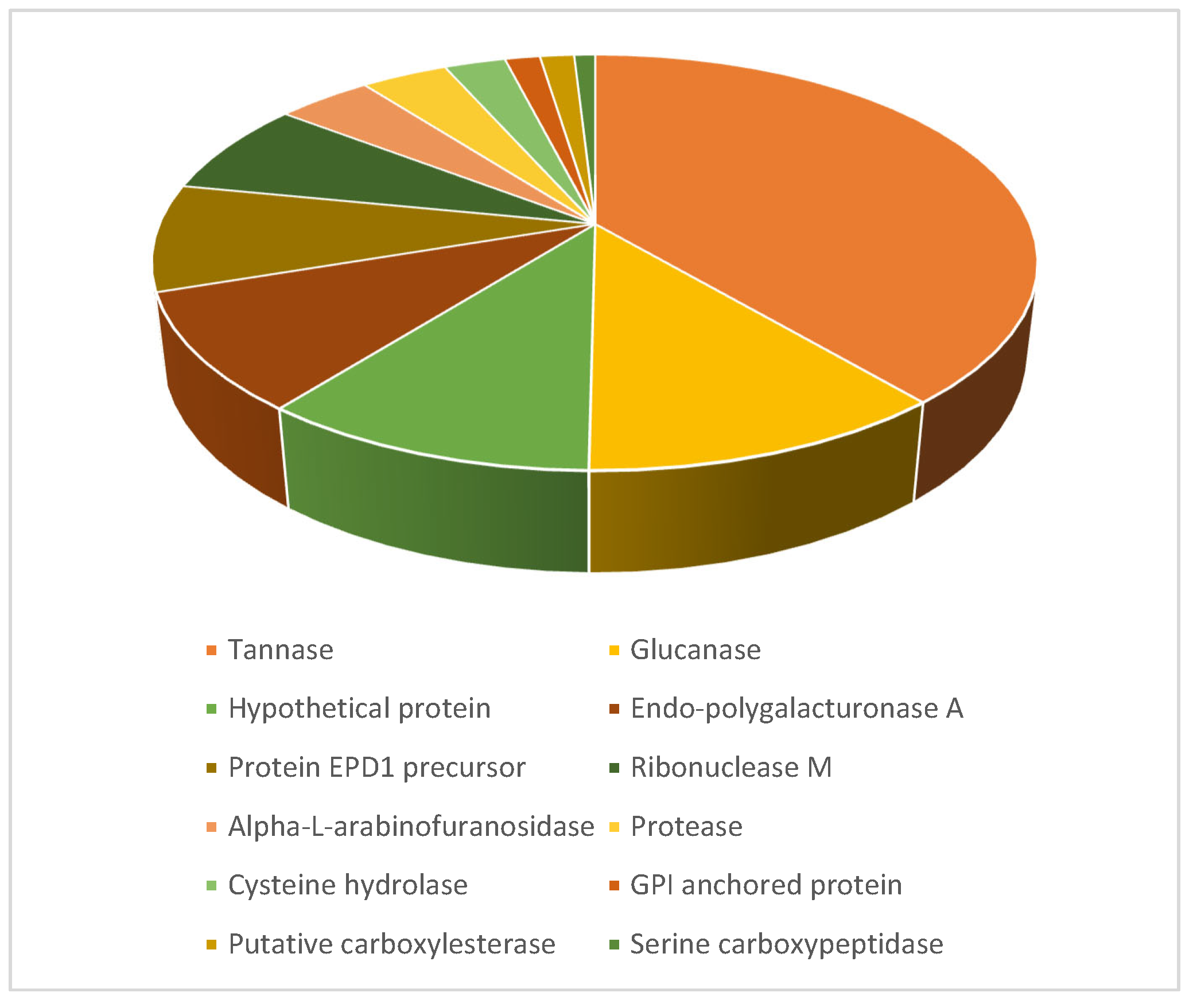
3.6. Protein Identification by Nano LC-MS/MS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TE | Tannase extract |
| BGCs | Biosynthetic gene clusters |
References
- Tang, Z.; Ma, Y.; Li, Y.; Xu, Y. Recent advances of tannase: Production, characterization, purification, and application in the tea industry. Foods 2024, 14, 79. [Google Scholar] [CrossRef]
- Pan, L.; Feng, S.; Li, W.; Zhu, W. Sorghum tannin extract impedes in vitro digestibility and fermentability of nutrients in the simulated porcine gastrointestinal tract. J. Anim. Sci. 2023, 101, skad126. [Google Scholar] [CrossRef] [PubMed]
- Magaya, R.P.; Mutibvu, T.; Ncube, S.; Nyahangare, E.T.; Mapfumo, P.; Mtambanengwe, F.; Nyakudya, E.; Nhamo, A. Effects of Phytase and Tannase Enzyme Supplementation on In Vitro Nutrient Digestibility and Tannin Degradation of Sorghum-Based Broiler Diets. Int. J. Life Sci. Agric. Res. 2024, 3, 346–354. [Google Scholar] [CrossRef]
- Chao, B.; Liu, R.; Zhang, X.; Zhang, X.; Tan, T. Tannin extraction pretreatment and very high gravity fermentation of acorn starch for bioethanol production. Bioresour. Technol. 2017, 241, 900–907. [Google Scholar] [CrossRef]
- Lekshmi, R.; Arif Nisha, S.; Thirumalai Vasan, P.; Kaleeswaran, B. A comprehensive review on tannase: Microbes associated production of tannase exploiting tannin rich agro-industrial wastes with special reference to its potential environmental and industrial applications. Environ. Res. 2021, 201, 111625. [Google Scholar] [CrossRef]
- da Silva, G.R.; dos Santos, E.S.; de Souza, A.P.; de Oliveira, R.A.; de Macedo, G.R. Production of lignocellulolytic enzymatic complex using pretreated carnauba straw as carbon source and application on sugarcane bagasse hydrolysis. Biomass Convers. Biorefin. 2020, 12, 85–95. [Google Scholar] [CrossRef]
- Cavalcanti, R.M.F.; Jorge, J.A.; Guimarães, L.H.S. Characterization of Aspergillus fumigatus CAS-21 tannase with potential for propyl gallate synthesis and treatment of tannery effluent from leather industry. 3 Biotech 2018, 8, 270. [Google Scholar] [CrossRef]
- Govindarajan, R.K.; Revathi, S.; Rameshkumar, N.; Krishnan, M.; Kayalvizhi, N. Microbial tannases: Biosynthesis, purification, and industrial applications. Int. J. Biol. Macromol. 2025, 311 Pt 1, 143376. [Google Scholar] [CrossRef]
- Saad, M.M.; Ibrahim, N.; Abdel-Ghany, T.M.; Hassan, S.; El-Gendy, A.O.; Mikhail, M.S. Optimization of Tannase Production by Aspergillus glaucus in Solid-State Fermentation Using Agricultural Waste Substrates. Bioresour. Bioprocess. 2023, 10, 45. [Google Scholar] [CrossRef]
- Núñez Sellés, A.J.; Vélez Castro, H.T.; Agüero-Agüero, J.; González-González, J.; Naddeo, F.; De Simone, F.; Rastrelli, L. Isolation and quantitative analysis of phenolic antioxidants, free sugars, and polyols from mango (Mangifera indica L.) stem bark aqueous decoction used in Cuba as a nutritional supplement. J. Agric. Food Chem. 2002, 50, 762–766. [Google Scholar] [CrossRef]
- Bradoo, S.; Gupta, R.; Saxena, R.K. Screening of extracellular tannase-producing fungi: Development of a rapid and simple plate assay. J. Gen. Appl. Microbiol. 1996, 42, 325–329. [Google Scholar] [CrossRef]
- Sharma, S.; Bhat, T.K.; Dawra, R.K. A spectrophotometric method for assay of tannase using rhodanine. Anal. Biochem. 2000, 279, 85–89. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; González, L.; Tablada, M.; Robledo, C.W. InfoStat (Versión 2015). Grupo InfoStat, Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina. 2015. Available online: http://www.infostat.com.ar (accessed on 30 March 2025).
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1989. [Google Scholar]
- Garmendia, G.; Vero, S. Occurrence and biodiversity of Aspergillus section Nigri on ‘Tannat’grapes in Uruguay. Int. J. Food Microbiol. 2016, 216, 31–39. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Smith, P.K.; Khron, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Neuhoff, V.; Stamm, R.; Pardowitz, I.; Arold, N.; Ehrhardt, W.; Taube, D. Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 1990, 11, 101–117. [Google Scholar] [CrossRef]
- Rossello, J.; Lima, A.; Gil, M.; Rodríguez Duarte, J.; Correa, A.; Carvalho, P.C.; Kierbel, A.; Durán, R. The EAL-domain protein FcsR regulates flagella, chemotaxis and type III secretion system in Pseudomonas aeruginosa by a phosphodiesterase independent mechanism. Sci. Rep. 2017, 7, 10281. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.D.; Lima, D.B.; Fischer, J.S.; Clasen, M.A.; Kurt, L.U.; Camillo-Andrade, A.C.; Carvalho, P.C. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nat. Protoc. 2022, 17, 1553–1578. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Visagie, C.M.; Yilmaz, N.; Kocsubé, S.; Frisvad, J.C.; Hubka, V.; Samson, R.A.; Houbraken, J. A Review of Recently Introduced Aspergillus, Penicillium, Talaromyces and Other Eurotiales Species. Stud. Mycol. 2024, 107, 1–66. [Google Scholar] [CrossRef]
- Godio, R.P.; Martín, J.F. Modified oxidosqualene cyclases in the formation of bioactive secondary metabolites: Biosynthesis of the antitumor clavaric acid. Fungal Genet. Biol. 2009, 46, 232–242. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Liang, Z.Y.; Shen, N.X.; Liu, W.L.; Zhou, X.J.; Fu, X.M.; Chen, M.; Wang, C.Y. New naphtho-γ-pyrones isolated from marine-derived fungus Penicillium sp. HK1-22 and their antimicrobial activities. Mar. Drugs 2019, 17, 322. [Google Scholar] [CrossRef]
- Wang, X.; Gong, X.; Li, P.; Lai, D.; Zhou, L. Structural diversity and biological activities of cyclic depsipeptides from fungi. Molecules 2018, 23, 169. [Google Scholar] [CrossRef]
- Matsuda, K. Okaramines and other plant fungal products as new insecticide leads. Curr. Opin. Insect Sci. 2018, 30, 67–72. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Thrane, U.; Samson, R.A.; Pitt, J.I. Important mycotoxins and the fungi which produce them. In Advances in Food Mycology; Pitt, J., Samson, R.A., Thrane, U., Eds.; Springer: New York, NY, USA, 2006; pp. 3–31. [Google Scholar]
- Egea, M.B.; Dantas, L.A.; Sousa, T.L.D.; Lima, A.G.; Lemes, A.C. The potential, strategies, and challenges of Monascus pigment for food application. Front. Sustain. Food Syst. 2023, 7, 1141644. [Google Scholar] [CrossRef]
- Biswas, I.; Mitra, D.; Mohapatra, P.K.D. Structural and catalytic advancement of fungal tannase: A proteomic contribution in industrial applicability. Bioresour. Technol. Rep. 2022, 19, 101103. [Google Scholar] [CrossRef]
- Aracri, F.M.; Cavalcanti, R.M.F.; Guimarães, L.H.S. Extracellular tannase from Aspergillus ochraceus: Influence of the culture conditions on biofilm formation, enzyme production, and application. J. Microbiol. Biotechnol. 2019, 29, 1749–1759. [Google Scholar] [CrossRef]
- Cruz, R.; de Lima, J.S.; Fonseca, J.C.; dos Santos Fernandes, M.J.; Lima, D.M.M.; Duda, G.P.; Moreira, K.A.; de Souza Motta, C.M. Diversity of filamentous fungi of area from Brazilian Caatinga and high-level tannase production using mango (Mangifera indica L.) and surinam cherry (Eugenia uniflora L.) leaves under SSF. Adv. Microbiol. 2013, 3, 52–60. [Google Scholar] [CrossRef]
- Thakur, N.; Nath, A.K.; Sharma, A. Optimization of production conditions, isolation, purification, and characterization of tannase from filamentous fungi. Folia Microbiol. 2024, 69, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Lal, D.; Gardner, J.J. Production, characterization and purification of tannase from Aspergillus niger. Eur. J. Exp. Biol. 2012, 2, 1430–1438. [Google Scholar]
- Sharma, K.P.; John, P.J. Purification and characterization of tannase and tannase gene from Enterobacter sp. Process Biochem. 2011, 46, 240–244. [Google Scholar] [CrossRef]
- Yao, J.; Guo, G.S.; Ren, G.H.; Liu, Y.H. Production, characterization and applications of tannase. J. Mol. Catal. B Enzym. 2014, 101, 137–147. [Google Scholar] [CrossRef]
- Batra, A.; Saxena, R.K. Potential tannase producers from the genera Aspergillus and Penicillium. Process Biochem. 2005, 40, 1553–1557. [Google Scholar] [CrossRef]
- Osipov, D.O.; Matys, V.Y.; Nemashkalov, V.A.; Rozhkova, A.M.; Shashkov, I.A.; Satrutdinov, A.D.; Sinitsyn, A.P. Cloning, Isolation, and Properties of a New Recombinant Tannase from the Aspergillus niger Fungus. Appl. Biochem. Microbiol. 2022, 58, 958–965. [Google Scholar] [CrossRef]
- Susca, A.; Moretti, A.; Stea, G.; Villani, A.; Haidukowski, M.; Logrieco, A.; Moretti, A. Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae isolates from multiple crop and geographic origins. Front. Microbiol. 2016, 7, 1412. [Google Scholar] [CrossRef]
- Busk, P.K.; Lange, M.; Pilgaard, B.; Lange, L. Several genes encoding enzymes with the same activity are necessary for aerobic fungal degradation of cellulose in nature. PLoS ONE 2014, 9, e114138. [Google Scholar] [CrossRef]
- Alfaro, M.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative analysis of secretomes in basidiomycete fungi. J. Proteom. 2014, 102, 28–43. [Google Scholar] [CrossRef]
| Identifier | Length (AA) | Identity (%) | Homolog Source |
|---|---|---|---|
| NODE_1.g376 | 512 | 95.20 | Aspergillus uvarum CBS 121591 XP_025490290.1 |
| NODE_3.g1172 | 537 | 100 | Aspergillus uvarum CBS 121591 XP_025488689.1 |
| NODE_9.g2866 | 543 | 97.18 | Aspergillus uvarum CBS 121591 XP_025492021.1 |
| NODE_11.g3384 | 1477 | 99.60 | Aspergillus uvarum CBS 121591 XP_025487606.1 |
| NODE_11.g3515 | 548 | 100 | Aspergillus uvarum CBS 121591 XP_025485684.1 |
| NODE_16.g4469 | 588 | 99.83 | Aspergillus uvarum CBS 121591 XP_025491148 |
| NODE_16.g4509 | 572 | 99.48 | Aspergillus uvarum CBS 121591 XP_025491187 |
| NODE_16.g4520 | 585 | 92.31 | Aspergillus brunneoviolaceus CBS 621.78 XP_025439954 |
| NODE_18.g4952 | 739 | 99.30 | Aspergillus uvarum CBS121591 XP_025493482 |
| NODE_55.g9244 | 578 | 97.75 | Aspergillus uvarum CBS 121591 XP_025487768.1 |
| NODE_57.g9409 | 545 | 100 | Aspergillus uvarum CBS 121591 XP_025487823 |
| NODE_60.g9637 | 584 | 100 | Aspergillus uvarum CBS 121591 XP_025496649.1 |
| NODE_70.g10053 | 522 | 100 | Aspergillus uvarum CBS 121591 XP_025489644.1 |
| NODE_71.g10109 | 532 | 96.75 | Aspergillus uvarum CBS 121591 XP_025495855.1 |
| NODE_98.g10681 | 505 | 88.20 | Penicillium angulare N7456_012083 KAJ5088467 |
| Region | Type | Metabolite | Reported Bioactivity | Potential Application |
|---|---|---|---|---|
| 3.1 | Terpene | Clavaric acid | Farnesyltransferase inhibition [29] | Anticancer therapeutics |
| 3.2 | T1PKS | Naphtho-γ-pyrone | Antimicrobial [30] | Food preservatives, antibiotics |
| 6.1 | NRPS | Cyclic depsipeptides | Insecticidal, antimicrobial, anticancer [31] | Biopesticides, drug discovery |
| 10.1 | Indole | Okaramine D | Insecticidal [32] | Agricultural biocontrol |
| 21.4 | Indole, NRPS, NRPS-like (híbrido) | histidyltryptophanyldiketopiperazine/roquefortine C/roquefortine D/meleagrine/glandicoline A/B/ | Neurotoxicity (roquefortines), antimicrobial, cytotoxic, acetylcholinesterase inhibition (meleagrine, glandicolines) [33] | Natural product scaffolds for drug discovery; neuropharmacology; antimicrobial agents (limited due to toxicity) |
| 59.1 | T1PKS | Monascorubrin | Pigmentation, antioxidant [34] | Natural food colorants |
| Identifier | Unique Peptides | Spectrum Count | % Coverage | Protein Score | Function |
|---|---|---|---|---|---|
| NODE_18.g4952 | 17 | 93 | 21.4 | 46.99 | tannase |
| NODE_60.g9637 | 13 | 41 | 15.4 | 35.50 | tannase |
| NODE_43.g8261 | 11 | 30 | 16.2 | 25.57 | alpha/beta-hydrolase |
| NODE_58.g9479 | 8 | 14 | 24.2 | 23.12 | alpha-L-arabinofuranosidase |
| NODE_44.g8317 | 1 | 12 | 3.9 | 3.94 | translation elongation factor EF-Tu |
| NODE_25.g6245 | 4 | 11 | 6.6 | 11.21 | glucanase |
| NODE_23.g5957 | 4 | 9 | 6.5 | 6.95 | uncharacterized protein |
| NODE_86.g10522 | 2 | 3 | 5.7 | 4.28 | alpha/beta-hydrolase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arbildi, E.; Ovsejevi, K.; Roldán, D.; Durán, R.; Portela, M.; Garmendia, G.; Vero, S. From Isolation to Genomics: Characterization of Aspergillus uvarum HT4 as a Novel Producer of Extracellular Tannase. J. Fungi 2025, 11, 722. https://doi.org/10.3390/jof11100722
Arbildi E, Ovsejevi K, Roldán D, Durán R, Portela M, Garmendia G, Vero S. From Isolation to Genomics: Characterization of Aspergillus uvarum HT4 as a Novel Producer of Extracellular Tannase. Journal of Fungi. 2025; 11(10):722. https://doi.org/10.3390/jof11100722
Chicago/Turabian StyleArbildi, Erika, Karen Ovsejevi, Diego Roldán, Rosario Durán, Magdalena Portela, Gabriela Garmendia, and Silvana Vero. 2025. "From Isolation to Genomics: Characterization of Aspergillus uvarum HT4 as a Novel Producer of Extracellular Tannase" Journal of Fungi 11, no. 10: 722. https://doi.org/10.3390/jof11100722
APA StyleArbildi, E., Ovsejevi, K., Roldán, D., Durán, R., Portela, M., Garmendia, G., & Vero, S. (2025). From Isolation to Genomics: Characterization of Aspergillus uvarum HT4 as a Novel Producer of Extracellular Tannase. Journal of Fungi, 11(10), 722. https://doi.org/10.3390/jof11100722






