The Role of microRNAs and Cell-Free DNAs in Fungal Infections: Systematic Review and Meta-Analysis of the Literature
Abstract
1. Introduction
2. Materials and Methods
2.1. The Literature Search, Selection Process and Data Extraction
2.2. Data Synthesis and Statistical Analysis
2.3. Final Step Analysis Protocol
3. Results
3.1. Search Results
3.2. cfDNA Analyses
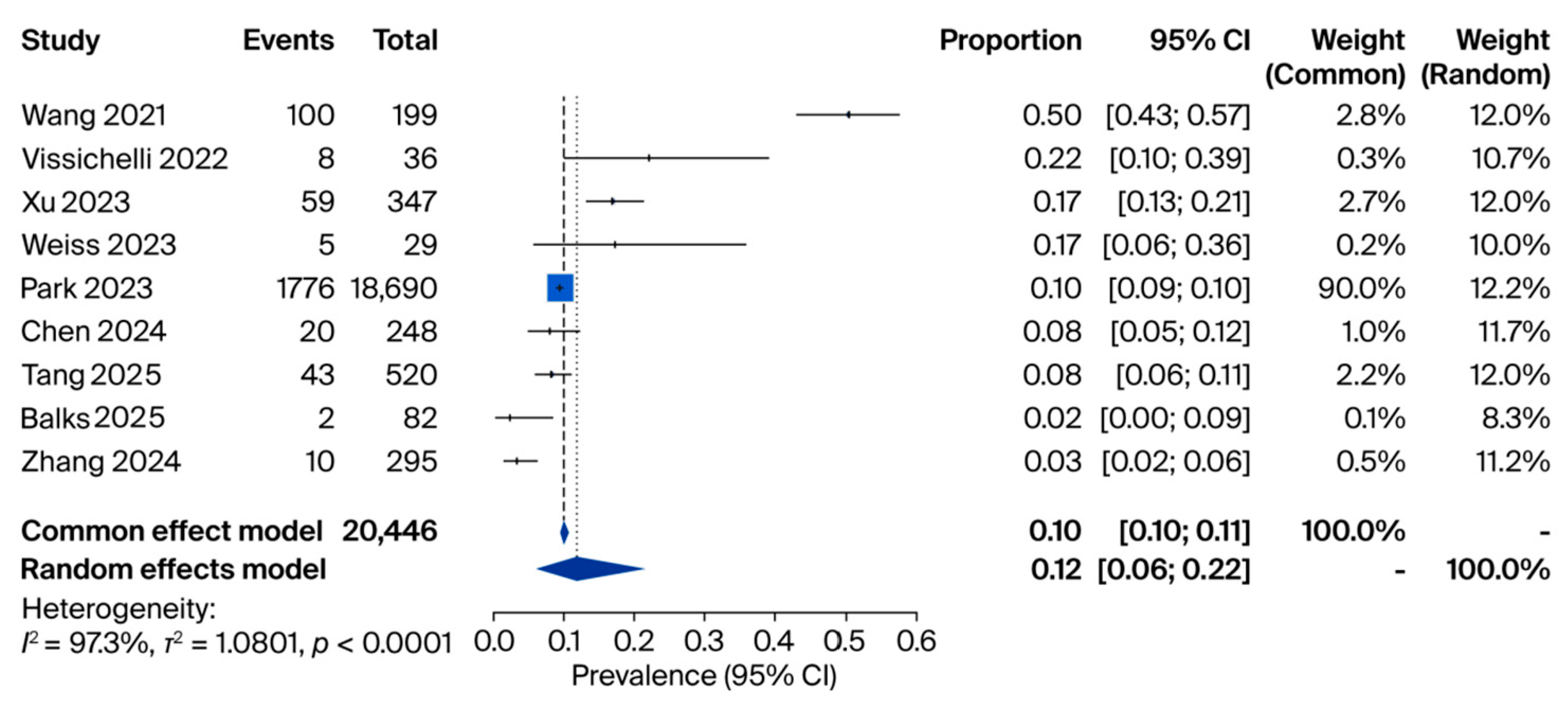
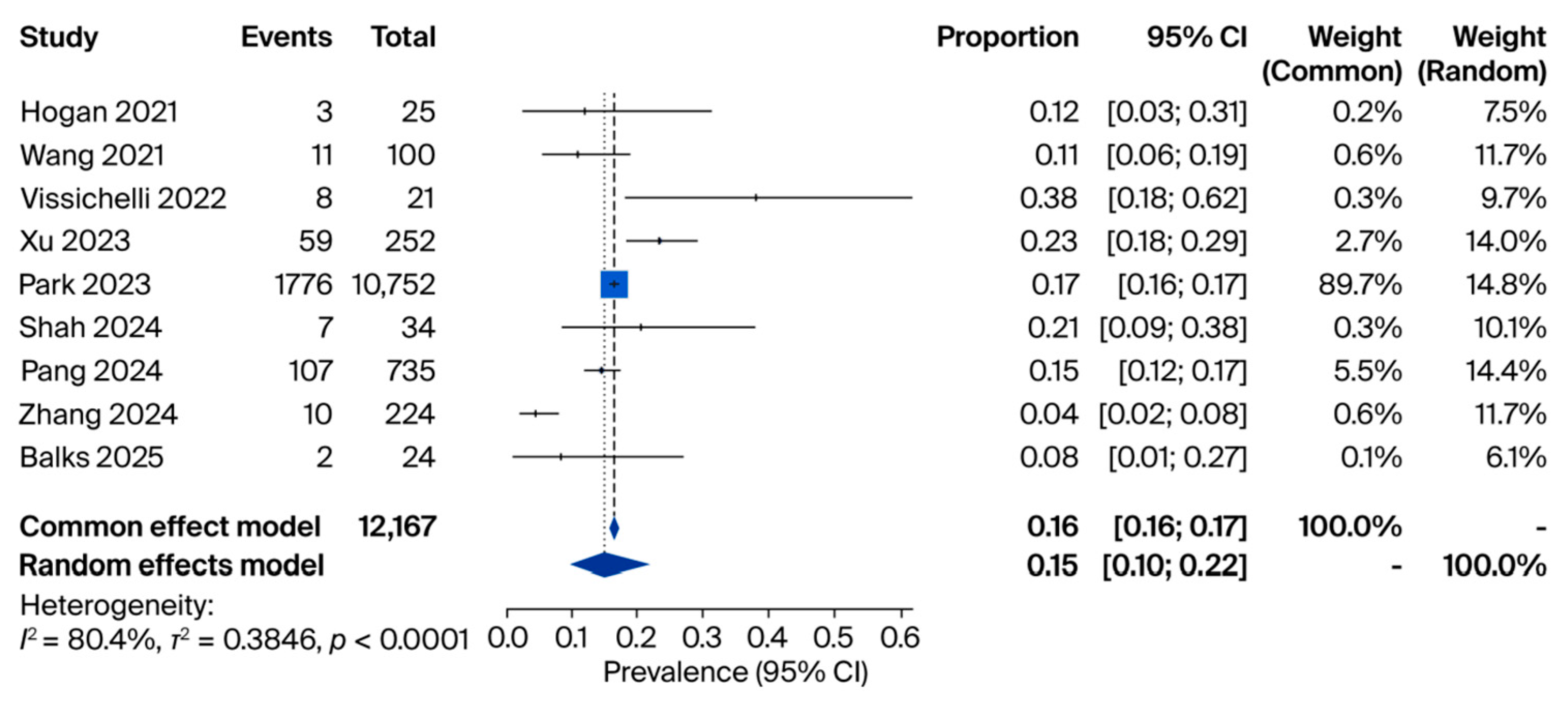

| Study (Year) | Country | Design | Sample Size | miRNA(s) Investigated | Main Finding | Diagnostic Performance |
|---|---|---|---|---|---|---|
| Wang 2010 [29] | China | Case–control | Sepsis: 50, SIRS: 30 | miR-146a, miR-223, miR-126, miR-15b, miR-132, miR-155, and let-7i | miR-146a and miR-223 significantly lower in sepsis; miR-126 lower in sepsis and SIRS vs. controls | miR-223: AUC 0.858 (Sens 80%, Spec 100%); miR-146a: AUC 0.804 (Sens 63.3%, Spec 100%) |
| Da Lacorta Singulani 2017 [30] | Brazil | Case–control | PCM: 4 | 752 miRNAs | Several miRNAs overexpressed (e.g., miR1323p, miR604, miR29b3p); one miRNA underexpressed (miR4233p) | - |
| Attia 2020 [31] | Egypt | Case–control | Sepsis: 50, Control: 20 | miR-146a and miR-150 | Significant correlation between miR-146a and miR-150 expression (r = 0.489, p < 0.001) | - |
| Wei 2020 [32] | China | Case–control | Sepsis: 121, Control: 60 | miR-545 | miR-545 significantly higher in sepsis vs. controls | AUC 0.942 for sepsis diagnosis; AUC 0.740 for 28-day mortality |
| Esawy 2021 [33] | Egypt | Case–control | Asthma: 30, SAFS: 30, ABPA: 30, Control: 30 | miR-21 and miR-132 | miR-21 elevated in all patient groups vs. controls; miR-132 highest in ABPA | ABPA vs. control: Sens 93.3%, Spec 100%; ABPA vs. asthma: Sens 90%, Spec 100%; SAFS: Sens 86.7%, Spec 80% |
| Fidler 2022 [34] | Hungary | Retrospective cohort | IA: 26, Control: 24 | Multiple (incl. hsa-miR-191-5p, hsa-miR-106b-5p, and hsa-miR-15a-5p) | 8 miRNAs downregulated in confirmed IA; 5 miRNAs had perfect discrimination (AUC 1.0) | 5 miRNAs: AUC 1.0; 3 miRNAs: AUC > 0.98 |
3.3. miRNA Analyses
4. Discussion
- (1)
- The role of miRNA and cfDNA in fungal pathogenesis;
- (2)
- Diagnostic options;
- (3)
- Advances in therapeutic strategies;
- (4)
- Future directions and recommendations.
4.1. Role of miRNA and cfDNA in Fungal Pathogenesis
4.2. Diagnostic Options
4.3. Advances in Therapeutic Strategies
4.4. Future Directions and Recommendations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNA | Micro RNA |
| cfDNA | Cell free DNA |
| MVBs | Multivesicular bodies |
| HSP | Heat shock protein |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PROSPERO | Prospective Register of Systematic Reviews |
| CD | Cluster of differentiation |
| IFD | Invasive fungal disease |
| ID | Infectious Disease |
| NOS | Newcastle–Ottawa Scale |
| JBI | Joanna Briggs Institute (JBI) Critical Appraisal Checklist |
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Vallabhaneni, S.; Mody, R.K.; Walker, T.; Chiller, T. The global burden of fungal diseases. Infect. Dis. Clin. N. Am. 2016, 30, 1–11. [Google Scholar] [CrossRef]
- Benedict, K.; Jackson, B.R.; Chiller, T.; Beer, K.D. Estimation of direct healthcare costs of fungal diseases in the United States. Clin. Infect. Dis. 2019, 68, 1791–1797. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Kimura, M.; Kothari, S.; Gohir, W.; Camargo, J.F.; Husain, S. MicroRNAs in infectious diseases: Potential diagnostic biomarkers and therapeutic targets. Clin. Microbiol. Rev. 2023, 36, e0001523. [Google Scholar] [CrossRef]
- Uslan, D.Z.; Kaur, I.; Garner, O.B.; Yang, S. Incorporating microbial cell-free DNA testing into healthcare-associated invasive fungal infection surveillance: Benefits and challenges. Infect. Control Hosp. Epidemiol. 2025, 46, 445–448. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in pathogen infections: A bridge to deliver molecules and link functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef]
- Rangel-Ramírez, V.V.; González-Sánchez, H.M.; Lucio-García, C. Exosomes: From biology to immunotherapy in infectious diseases. Infect. Dis. 2023, 55, 79–107. [Google Scholar] [CrossRef] [PubMed]
- Croston, T.L.; Lemons, A.R.; Beezhold, D.H.; Green, B.J. MicroRNA Regulation of host immune responses following fungal exposure. Front. Immunol. 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Dix, A.; Czakai, K.; Leonhardt, I.; Schäferhoff, K.; Bonin, M.; Guthke, R.; Einsele, H.; Kurzai, O.; Löffler, J.; Linde, J. Specific and novel microRNAs are regulated as response to fungal infection in human dendritic cells. Front. Microbiol. 2017, 8, 270. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, R.; Zhu, Y.; Hu, L.; Xia, H.; Li, J.; Ye, Y. Metagenomic next-generation sequencing of plasma cell-free DNA improves the early diagnosis of suspected infections. BMC Infect. Dis. 2024, 24, 187. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, W.; Shen, H.; Guo, J.; Wen, D.; Yu, Y.; Wu, W. Plasma microbial Cell-Free DNA sequencing technology for the diagnosis of sepsis in the ICU. Front. Mol. Biosci. 2021, 8, 659390. [Google Scholar] [CrossRef] [PubMed]
- Vissichelli, N.C.; Morales, M.K.; Kolipakkam, B.; Bryson, A.; Sabo, R.T.; Toor, A.A. Cell-free next-generation sequencing impacts diagnosis and antimicrobial therapy in immunocompromised hosts: A retrospective study. Transpl. Infect. Dis. 2023, 25, e13954. [Google Scholar] [CrossRef]
- Kushner, L.E.; Schwenk, H.T.; Qin, F.; Boothroyd, D.; Aftandilian, C. Application of cell-free DNA fungal polymerase chain reaction for invasive fungal disease evaluation in pediatric oncology and stem cell transplant patients. Pediatr. Blood Cancer 2024, 71, e31133. [Google Scholar] [CrossRef]
- Gracia, M.; Hadley, E.; Ramchandar, N.; Coufal, N.G. Emerging role of plasma microbial cell-free DNA in the diagnosis of pediatric mucormycosis. Pediatr. Infect. Dis. J. 2024, 43, 704–707. [Google Scholar] [CrossRef]
- Xu, C.; Chen, X.; Zhu, G.; Yi, H.; Chen, S.; Yu, Y.; Jiang, E.; Zheng, Y.; Zhang, F.; Wang, J.; et al. Utility of plasma cell-free DNA next-generation sequencing for diagnosis of infectious diseases in patients with hematological disorders. J. Infect. 2023, 86, 14–23. [Google Scholar] [CrossRef]
- Weiss, Z.F.; Pyden, A.D.; Jhaveri, T.A.; Kanjilal, S. The diagnostic and clinical utility of microbial cell-free DNA sequencing in a real-world setting. Diagn. Microbiol. Infect. Dis. 2023, 107, 116004. [Google Scholar] [CrossRef]
- Park, S.Y.; Chang, E.J.; Ledeboer, N.; Messacar, K.; Lindner, M.S.; Venkatasubrahmanyam, S.; Wilber, J.C.; Vaughn, M.L.; Bercovici, S.; Perkins, B.A.; et al. Plasma microbial Cell-Free DNA sequencing from over 15,000 patients identified a broad spectrum of pathogens. J. Clin. Microbiol. 2023, 61, e0185522. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhang, X.; Liu, S.; Yin, Y.; Guo, Y.; Wang, X.; Zhang, Y.; Zhao, C.; Gai, W.; et al. Clinical evaluation of cell-free and cellular metagenomic next-generation sequencing of infected body fluids. J. Adv. Res. 2024, 55, 119–129.20. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Chen, Y.; Tang, X.; Wei, M.; Hu, J.; Zhang, X.; Xiang, D.; Yang, Q.; Han, D. Yield of clinical metagenomics: Insights from real-world practice for tissue infections. EBioMedicine 2025, 111, 105536, Erratum in: EBioMedicine. 2025, 114, 105574.21. [Google Scholar] [CrossRef]
- Balks, J.; Grumaz, S.; Mazzitelli, S.; Neder, U.; Lemloh, L.; Melaku, T.; Glaser, K.; Mueller, A.; Kipfmueller, F. Microbial cell-free DNA-sequencing as an addition to conventional diagnostics in neonatal sepsis. Pediatr. Res. 2025, 97, 614–624. [Google Scholar] [CrossRef]
- Hogan, C.A.; Yang, S.; Garner, O.B.; Green, D.A.; Gomez, C.A.; Dien Bard, J.; Pinsky, B.A.; Banaei, N. Clinical impact of metagenomic next-generation sequencing of plasma cell-free dna for the diagnosis of infectious diseases: A multicenter retrospective cohort study. Clin. Infect. Dis. 2021, 72, 239–245. [Google Scholar] [CrossRef]
- Shah, J.R.; Sohail, M.R.; Lasco, T.; Goss, J.A.; Mohajer, M.A.; Khalil, S. Clinical utility of plasma microbial cell-free DNA sequencing in determining microbiologic etiology of infectious syndromes in solid organ transplant recipients. Ther. Adv. Infect. Dis. 2024, 11, 20499361241308643. [Google Scholar] [CrossRef]
- Pang, F.; Xu, W.; Zhao, H.; Chen, S.; Tian, Y.; Fu, J.; You, Z.; Song, P.; Xian, Q.; Zhao, Q.; et al. Comprehensive evaluation of plasma microbial cell-free DNA sequencing for predicting bloodstream and local infections in clinical practice: A multicenter retrospective study. Front. Cell. Infect. Microbiol. 2024, 13, 1256099. [Google Scholar] [CrossRef]
- Hong, D.K.; Blauwkamp, T.A.; Kertesz, M.; Bercovici, S.; Truong, C.; Banaei, N. Liquid biopsy for infectious diseases: Sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis. 2018, 92, 210–213. [Google Scholar] [CrossRef]
- Armstrong, A.E.; Rossoff, J.; Hollemon, D.; Hong, D.K.; Muller, W.J.; Chaudhury, S. Cell-free DNA next-generation sequencing successfully detects infectious pathogens in pediatric oncology and hematopoietic stem cell transplant patients at risk for invasive fungal disease. Pediatr. Blood Cancer 2019, 66, e27734. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Yu, M.L.; Yu, G.; Bian, J.J.; Deng, X.M.; Wan, X.J.; Zhu, K.M. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef]
- De Lacorte Singulani, J.; De Fátima Da Silva, J.; Gullo, F.P.; Costa, M.C.; Fusco-Almeida, A.M.; Enguita, F.J.; Mendes-Giannini, M.J.S. Preliminary evaluation of circulating microRNAs as potential biomarkers in paracoccidioidomycosis. Biomed. Rep. 2017, 6, 353–357. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Attia, N.M.; El Din, A.; El Sawaf, G.; Abo-Ollo, M.M.; Beak, C.A.A.; Naga, I.S. Diagnostic value of microRNA-150 and microRNA-146a in critically ill patients with suspected sepsis. Egyp J. Med. Microbiol. 2020, 29, 113–119. [Google Scholar][Green Version]
- Wei, B.; Yu, L. Circular RNA PRKCI and microRNA-545 relate to sepsis risk, disease severity and 28-day mortality. Scand. J. Clin. Lab. Investig. 2020, 80, 659–666. [Google Scholar] [CrossRef]
- Esawy, M.M.; Baioumy, S.A.; Ismail, N.A.; Shabana, M.A. Role of circulating microRNA-132 in allergic bronchopulmonary aspergillosis: A case-control study. Immunobiology 2021, 226, 152074. [Google Scholar] [CrossRef]
- Fidler, G.; Szilágyi-Rácz, A.A.; Dávid, P.; Tolnai, E.; Rejtő, L.; Szász, R.; Póliska, S.; Biró, S.; Paholcsek, M. Circulating microRNA sequencing revealed miRNome patterns in hematology and oncology patients aiding the prognosis of invasive aspergillosis. Sci. Rep. 2022, 12, 7144. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Q.; Gai, W.; Guo, Y.; Zheng, Y. A comprehensive evaluation of plasma metagenomics sequencing for the diagnosis of suspected infection in pediatric patients with hematologic diseases. Front. Cell. Infect. Microbiol. 2025, 15, 1584214. [Google Scholar] [CrossRef]
- Yar Saglam, A.S.; Kalkanci, A.; Usta Salimi, D.D.; Escan, F.; Fidan, I.; Guzel Tunccan, O. MicroRNA expression profile of alveolar epithelial cells infected with Aspergillus fumigatus. J. Appl. Genet. 2024, 65, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Lu, H.; Jiang, B.; Zheng, Y.; Chen, P.; Liu, X.; Huang, J. Virulence factors released by extracellular vesicles from Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 2025, 15, 1572520. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.T.; Cheng, Z.; Hu, Z.D.; Zhou, L.; Zhong, R.Q. Upregulated miR-155 inhibits inflammatory response induced by C. albicans in human monocytes derived dendritic cells via targeting p65 and BCL-10. Ann. Transl. Med. 2019, 7, 758. [Google Scholar] [CrossRef]
- Li, X.; Gong, Y.; Lin, X.; Lin, Q.; Luo, J.; Yu, T.; Xu, J.; Chen, L.; Xu, L.; Hu, Y. Down-regulation of microRNA-155 suppressed Candida albicans induced acute lung injury by activating SOCS1 and inhibiting inflammation response. J. Microbiol. 2022, 60, 402–410. [Google Scholar] [CrossRef]
- Zhang, M.; Xia, Z.; Yang, X.; Ao, J.; Yang, R. Specific microRNA/mRNA expression profiles and novel immune regulation mechanisms are induced in THP-1 macrophages by in vitro exposure to Trichosporon asahii. Mycoses 2021, 64, 831–840. [Google Scholar] [CrossRef]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; May, R.C. Extracellular vesicles of human pathogenic fungi. Curr. Opin. Microbiol. 2019, 52, 90–99. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Sun, T.; Ding, C. Pathogen-host interaction repertoire at proteome and posttranslational modification levels during fungal infections. Front. Cell. Infect. Microbiol. 2021, 11, 774340. [Google Scholar] [CrossRef]
- Jiao, P.; Wang, X.P.; Luoreng, Z.M.; Yang, J.; Jia, L.; Ma, Y.; Wei, D.W. miR-223: An effective regulator of immune cell differentiation and inflammation. Int. J. Biol. Sci. 2021, 17, 2308–2322. [Google Scholar] [CrossRef]
- Al-Nakhle, H.H.; Khateb, A.M. Comprehensive in silico characterization of the coding and non-coding SNPs in human Dectin-1 gene with the potential of high-risk pathogenicity associated with fungal infections. Diagnostics 2023, 13, 1785. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 10. [Google Scholar] [CrossRef]
- Tan, D.; Li, G.; Fu, W.; Lei, C. Exosomes: The next frontier in vaccine development and delivery. Front. Immunol. 2024, 15, 1435426. [Google Scholar] [CrossRef]
- Douglas, C.M. Fungal beta(1,3)-D-glucan synthesis. Med. Mycol. 2001, 39 (Suppl. S1), 55–66. [Google Scholar] [CrossRef] [PubMed]
- Lieu, A.; Zimmet, A.N.; Pozdol, J.; Kushner, L.E.; Ho, D.; Banaei, N. Concordance of noninvasive plasma cell-free DNA with invasive diagnostics for diagnosis of invasive fungal disease. Clin. Infect. Dis. 2025, 80, 1095–1102. [Google Scholar] [CrossRef]
- Cai, X.; Sun, C.; Zhong, H.; Cai, Y.; Cao, M.; Wang, L.; Sun, W.; Tao, Y.; Ma, G.; Huang, B.; et al. The value of metagenomic next-generation sequencing with different nucleic acid extracting methods of cell-free DNA or whole-cell DNA in the diagnosis of non-neutropenic pulmonary aspergillosis. Front. Cell. Infect. Microbiol. 2024, 14, 1398190. [Google Scholar] [CrossRef] [PubMed]
- Das Gupta, M.; Fliesser, M.; Springer, J.; Breitschopf, T.; Schlossnagel, H.; Schmitt, A.L.; Kurzai, O.; Hünniger, K.; Einsele, H.; Löffler, J. Aspergillus fumigatus induces microRNA-132 in human monocytes and dendritic cells. Int. J. Med. Microbiol. 2014, 304, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Gohir, W.; Klement, W.; Singer, L.G.; Palmer, S.M.; Mazzulli, T.; Keshavjee, S.; Husain, S. Identifying host microRNAs in bronchoalveolar lavage samples from lung transplant recipients infected with Aspergillus. J. Heart Lung Transpl. 2020, 39, 1228–1237. [Google Scholar] [CrossRef]
- Bregón-Villahoz, M.; Menéndez-Manjón, P.; Carrano, G.; Díez-Villalba, A.; Arrieta-Aguirre, I.; Fernandez-de-Larrinoa, I.; Moragues, M.D. Candida albicans cDNA library screening reveals novel potential diagnostic targets for invasive candidiasis. Diagn. Microbiol. Infect. Dis. 2024, 109, 116311. [Google Scholar] [CrossRef]
- Ghasemian, S.O. Application of exosomes-derived mesenchymal stem cells in treatment of fungal diseases: From basic to clinical sciences. Front. Fungal Biol. 2021, 2, 736093. [Google Scholar] [CrossRef] [PubMed]
- Pakshir, K.; Badali, H.; Nami, S.; Mirzaei, H.; Ebrahimzadeh, V.; Morovati, H. Interactions between immune response to fungal infection and microRNAs: The pioneer tuners. Mycoses 2020, 63, 4–20. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Santosh, S.S.; Ahamed, M.A.; Sathiyaseelan, A.; Sultan, G.; Irfan, N.; Ali, D.M.; Wang, M.H. Bioinformatics strategies for studying the molecular mechanisms of fungal extracellular vesicles with a focus on infection and immune responses. Brief. Bioinform. 2022, 23, bbac250. [Google Scholar] [CrossRef]
- Chen, G.; Bai, Y.; Li, Z.; Wang, F.; Fan, X.; Zhou, X. Bacterial extracellular vesicle-coated multi-antigenic nanovaccines protect against drug-resistant Staphylococcus aureus infection by modulating antigen processing and presentation pathways. Theranostics 2020, 10, 7131–7149. [Google Scholar] [CrossRef]
- Zhao, N.; Song, Y.; Xie, X.; Zhu, Z.; Duan, C.; Nong, C.; Wang, H.; Bao, R. Synthetic biology-inspired cell engineering in diagnosis, treatment, and drug development. Signal Transduct. Target. Ther. 2023, 8, 112. [Google Scholar] [CrossRef]
- Vargas, G.; Honorato, L.; Guimarães, A.J.; Rodrigues, M.L.; Reis, F.C.G.; Vale, A.M.; Ray, A.; Nosanchuk, J.D.; Nimrichter, L. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell Microbiol. 2020, 22, e13238. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.A.; Nanjappa, S.G. Advances in dendritic-cell-based vaccines against respiratory fungal infections. Vaccines 2024, 12, 981. [Google Scholar] [CrossRef]
- Huda, M.N.; Nurunnabi, M. Potential application of exosomes in vaccine development and delivery. Pharm. Res. 2022, 39, 2635–2671. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Mah, J.; Budvytiene, I.; Ho, D.Y.; Schwenk, H.T.; Banaei, N. Dynamics and prognostic value of plasma cell-free DNA PCR in patients with invasive aspergillosis and mucormycosis. J. Clin. Microbiol. 2024, 62, e0039424. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Disease/Condition | Regulation | Reported/Predicted Function |
|---|---|---|---|
| miR-146a | Sepsis, Invasive aspergillosis | Downregulated | Regulates TLR signaling, controls inflammation and pathogen recognition |
| miR-223 | Sepsis, Aspergillosis | Downregulated | Neutrophil activation, cytokine regulation, pathogen clearance |
| miR-132 | ABPA, Invasive aspergillosis | Upregulated | Th2 immune response, inflammation modulation |
| miR-155 | Candida albicans, Aspergillus spp. | Upregulated | Macrophage polarization, enhances fungal clearance |
| miR-21 | ABPA, Asthma, Aspergillosis (in vitro) | Upregulated | Suppresses IL-6/TNF-α, promotes immune evasion |
| miR-545 | Sepsis | Upregulated | Associated with severity and mortality prediction |
| miR-191-5p, miR-106b-5p, miR-15a-5p | Invasive aspergillosis (hematology/oncology patients) | Downregulated | High discriminatory power for diagnosis in immunocompromised patients |
| miR-21-5p, miR-145-5p, miR-583, miR-3978, miR-4488, miR-4454 | Aspergillus fumigatus infection (A549 in vitro) | Downregulated | Differential expression associated with host response in epithelial cells |
| miR-186-5p, miR-490-5p, miR-26a-5p, miR-26b-5p, miR-424-5p, miR-548d-3p, miR-196a-5p, miR-150-5p, miR-17-5p, miR-99b-5p | Aspergillus fumigatus infection (A549 in vitro) | Upregulated | Differential expression associated with host response in epithelial cells |
| Study (Author, Year) | Design | Tool Used | Risk of Bias/Quality Score | Comments |
|---|---|---|---|---|
| Armstrong, 2019 [28] | Prospective cohort (IFD) | QUADAS-2 | Moderate risk | Small sample size, Standardized test protocol |
| Balks, 2025 [23] | Prospective cohort (Sepsis) | QUADAS-2 | Low risk | Clear patient selection; appropriate index test |
| Chen, 2024 [21] | Prospective cohort (Suspected infectious diseases) | QUADAS-2 | Moderate risk | Sample location differences, temporal variations in treatment protocols |
| Gracia, 2024 [17] | Case series (Mucormycosis) | QUADAS-2 | Moderate risk | Small sample size, Clear patient selection |
| Hogan, 2021 [24] | Retrospective cohort (ID) | QUADAS-2 | Low risk | Standardized test protocol, Multicenter |
| Hong, 2018 [27] | Retrospective (IFD) | QUADAS-2 | Moderate risk | Small sample size, Standardized test protocol |
| Kushner, 2024 [16] | Retrospective (IFD) | QUADAS-2 | Low risk | Clear patient selection; appropriate index test |
| Pang, 2024 [26] | Retrospective cohort (ID) | QUADAS-2 | Low risk | Clear patient selection; Multicenter |
| Park, 2023 [20] | Descriptive (ID) | STROBE | Moderate/High Reporting Quality | Standardized test, Multicenter, Big sample size |
| Shah, 2024 [25] | Retrospective (ID) | NOS | 6/9 stars | Clear patient selection, Standardized test |
| Tang, 2025 [22] | Cross-sectional (ID) | QUADAS-2 | Low risk | Standardized test, Clear sample selection |
| Vissichelli, 2023 [15] | Retrospective observational study (ID) | NOS | 6/9 stars | Limited sample size, Sample location differences |
| Wang, 2021 [14] | Prospective cohort (Sepsis) | QUADAS-2 | Low risk | Strong design, Clear sample selection |
| Weiss, 2023 [19] | Retrospective cohort (ID) | JBI | Moderate risk | Small sample size, appropriate index test |
| Xu, 2023 [18] | Retrospective cohort (ID) | QUADAS-2 | Low risk | Clear patient selection; appropriate index test |
| Zhang, 2024 [12] | Retrospective cohort (ID) | QUADAS-2 | Moderate risk | |
| Wang, 2010 [29] | Case–control (Sepsis) | QUADAS-2 | Low risk | Clear patient selection; appropriate index test |
| Attia, 2020 [31] | Case–control (Sepsis) | QUADAS-2 | Moderate risk | Small sample size; unclear blinding |
| Wei, 2020 [32] | Case–control (Sepsis) | NOS | 7/9 stars | Good selection and comparability; limited outcome follow-up |
| Esawy, 2021 [33] | Case–control (ABPA) | NOS | 6/9 stars | Adequate selection; limited sample size |
| Fidler, 2022 [34] | Retrospective cohort (IA) | NOS | 8/9 stars | Strong design; potential retrospective bias |
| Da Lacorta Singulani, 2017 [30] | Case–control (PCM) | NOS | 5/9 stars | Very small sample; exploratory findings |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalkanci, A.; Bozdag, F.; Fidan, I.; Guzel Tunccan, O.; Cetintepe, S.P.; Ilhan, M.N. The Role of microRNAs and Cell-Free DNAs in Fungal Infections: Systematic Review and Meta-Analysis of the Literature. J. Fungi 2025, 11, 718. https://doi.org/10.3390/jof11100718
Kalkanci A, Bozdag F, Fidan I, Guzel Tunccan O, Cetintepe SP, Ilhan MN. The Role of microRNAs and Cell-Free DNAs in Fungal Infections: Systematic Review and Meta-Analysis of the Literature. Journal of Fungi. 2025; 11(10):718. https://doi.org/10.3390/jof11100718
Chicago/Turabian StyleKalkanci, Ayse, Fatma Bozdag, Isil Fidan, Ozlem Guzel Tunccan, Sultan Pinar Cetintepe, and Mustafa Necmi Ilhan. 2025. "The Role of microRNAs and Cell-Free DNAs in Fungal Infections: Systematic Review and Meta-Analysis of the Literature" Journal of Fungi 11, no. 10: 718. https://doi.org/10.3390/jof11100718
APA StyleKalkanci, A., Bozdag, F., Fidan, I., Guzel Tunccan, O., Cetintepe, S. P., & Ilhan, M. N. (2025). The Role of microRNAs and Cell-Free DNAs in Fungal Infections: Systematic Review and Meta-Analysis of the Literature. Journal of Fungi, 11(10), 718. https://doi.org/10.3390/jof11100718






