Botrytis cinerea PMT4 Is Involved in O-Glycosylation, Cell Wall Organization, Membrane Integrity, and Virulence
Abstract
1. Introduction
2. Materials and Methods
2.1. B. cinerea and S. cerevisiae Strains, Growth Conditions, and Genomic DNA Isolation
2.2. Generation of a B. cinerea Δpmt4 Strain
2.3. Real-Time PCR
2.4. Construction of bcpmt4 Complementation Strain
2.5. Construction of bcpmt4 Expression Plasmid for S. cerevisiae Complementation Assays
2.6. Quantification of Cell Wall Carbohydrates
2.7. Sensitivity to Cell Wall Perturbing Agents
2.8. Plant Infection Tests
2.9. Membrane Integrity Assay
2.10. Biofilm Formation and Quantification
2.11. High Hydrostatic Pressure (HHP) Assay
2.12. Preparation of Wall and Soluble Fractions
2.13. Gel Electrophoresis and Glycoprotein Stain
2.14. Statistical Analysis
3. Results
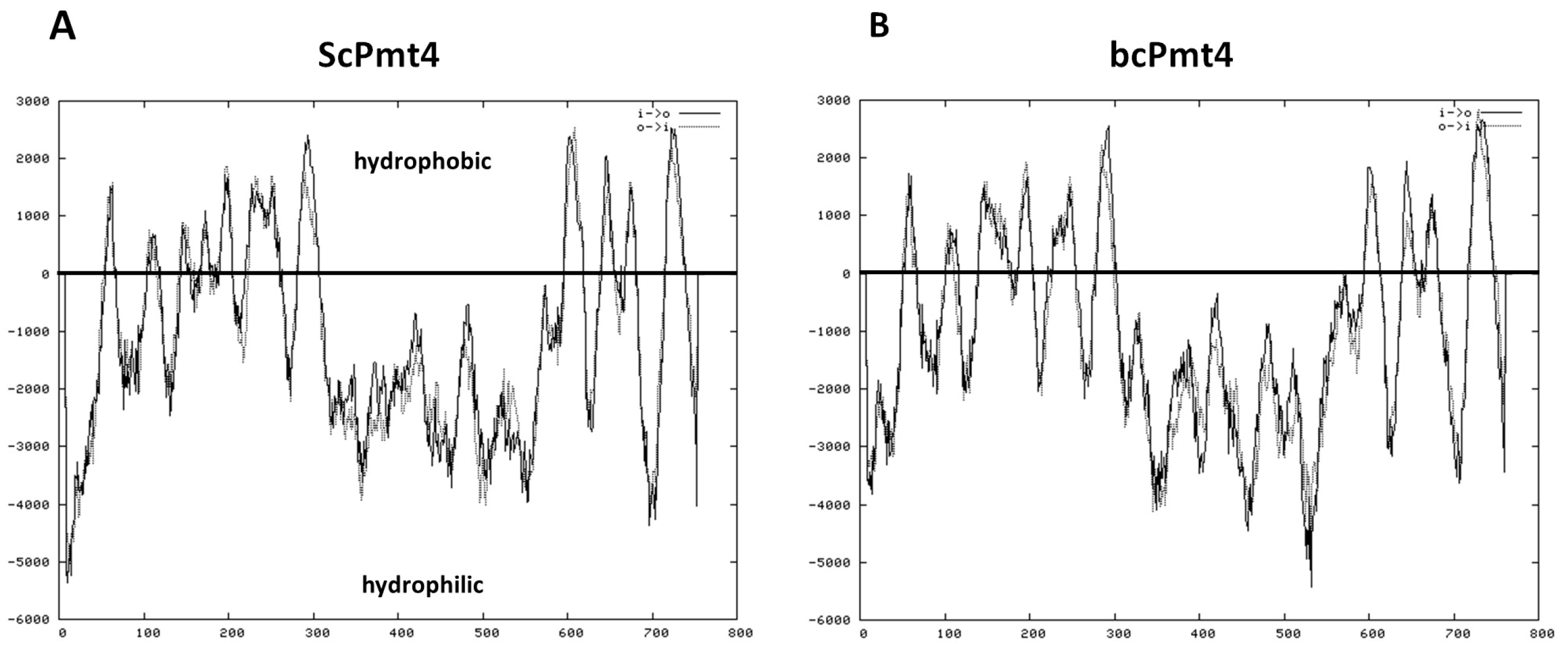
3.1. The B. cinerea PMT4
3.2. The bcpmt4 Complements a S. cerevisiae Δpmt4 Null Mutant
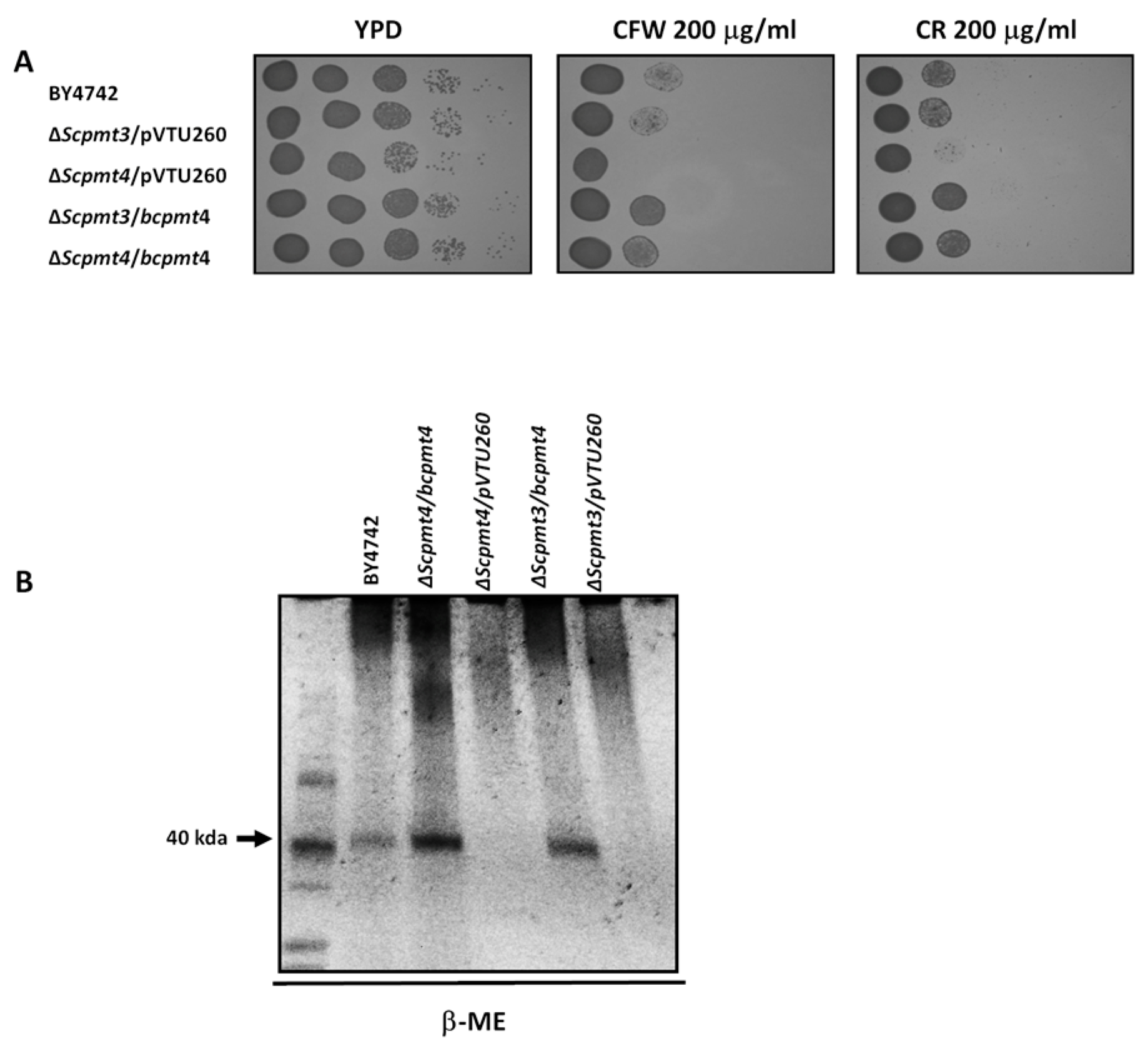
3.3. Deletion of B. cinerea bcpmt4 Led to Alterations in Hyphal Wall Composition and Glycosylation Deficiencies
3.4. Cell Wall Defects of the Δbcpmt4 Mutant
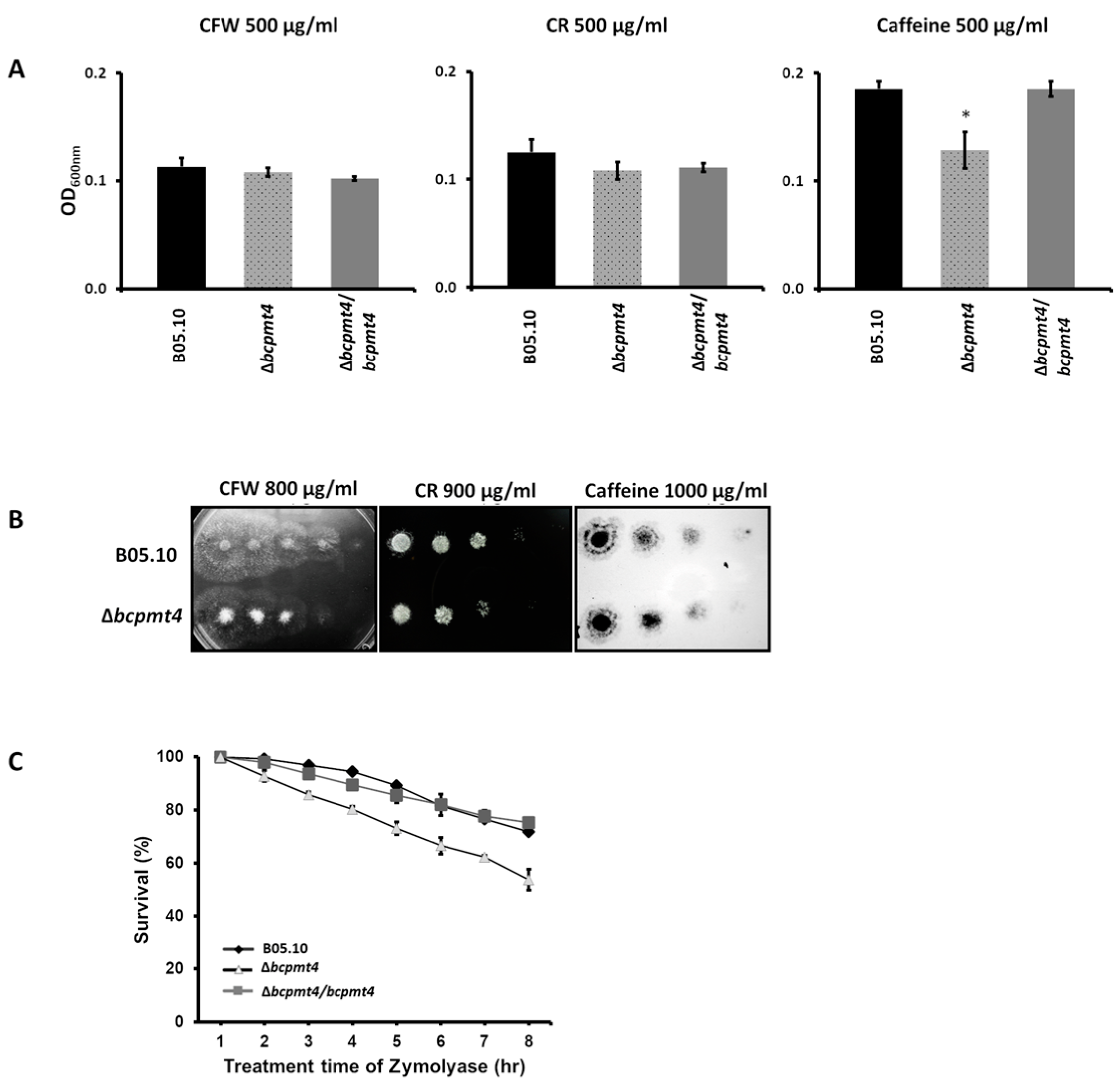
3.5. Membrane Integrity Defects in the Δbcpmt4 Mutant
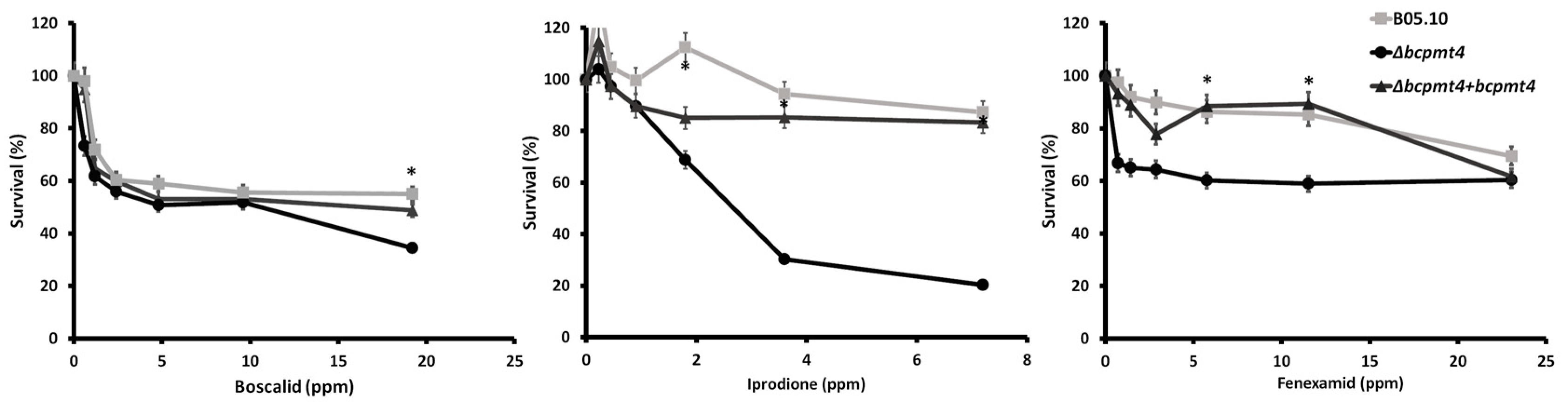
3.6. Deletion of bcpmt4 in B. cinerea Leads to Hypersensitivity to Antifungal Drugs
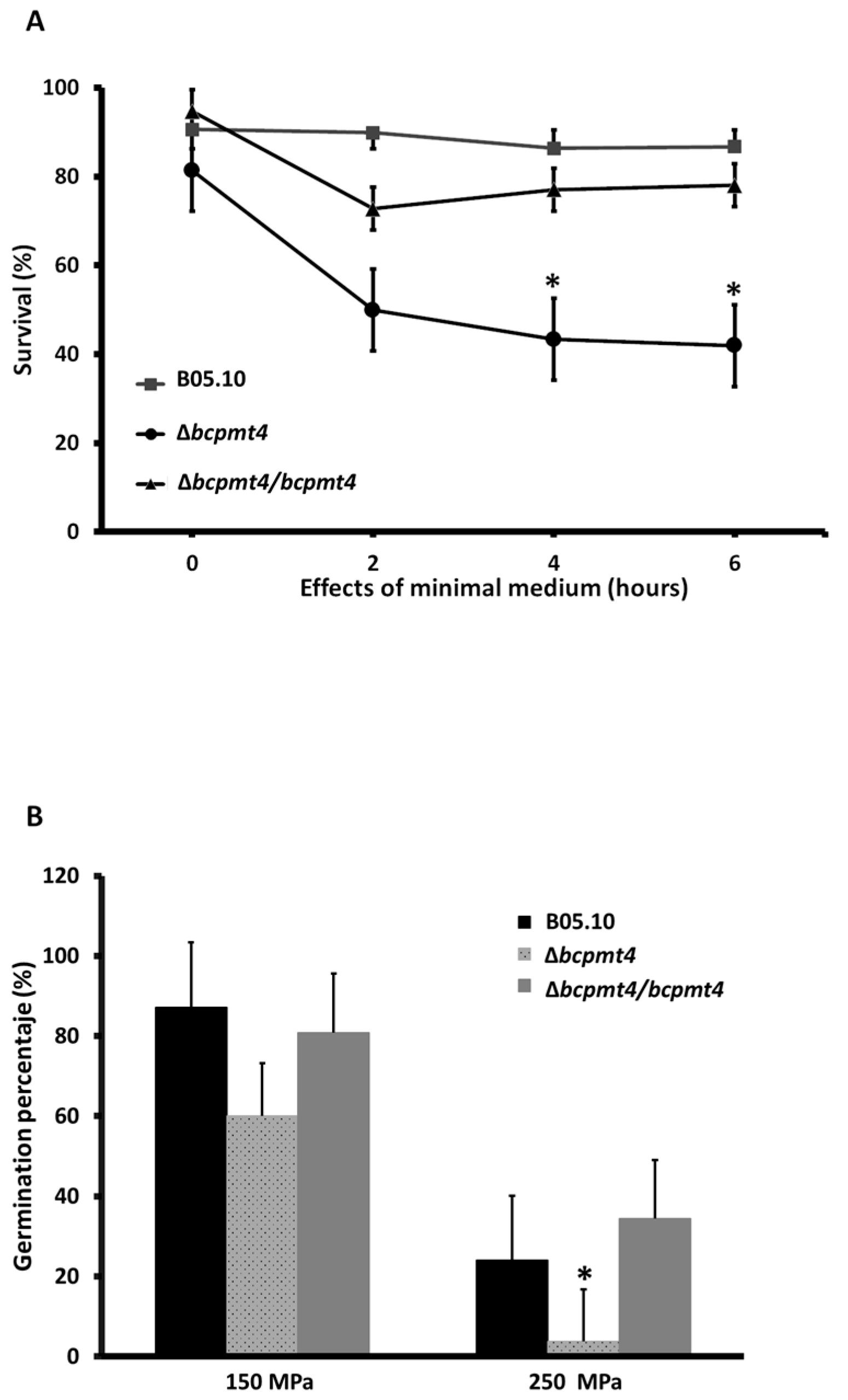
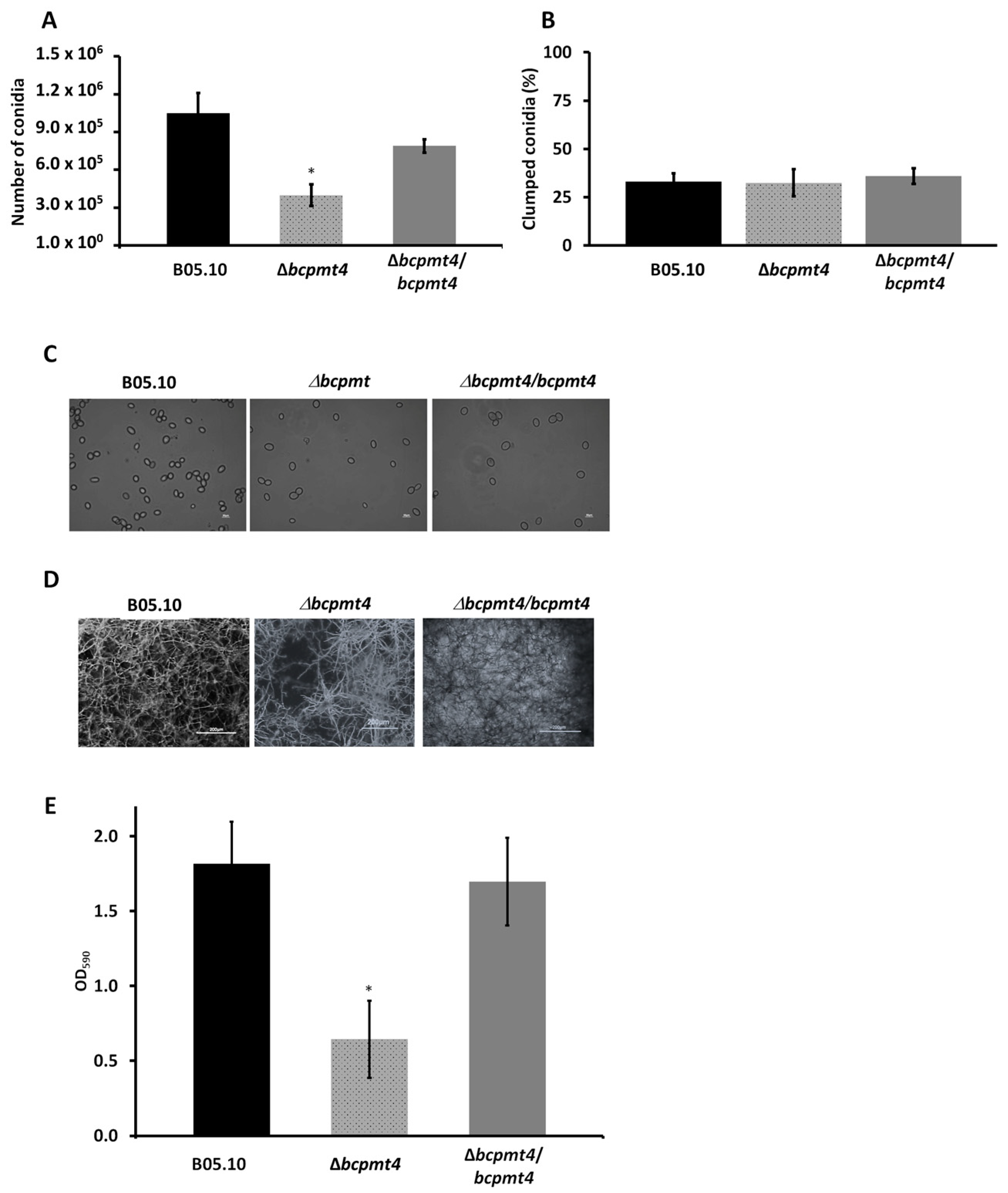
3.7. Lack of bcpmt4 Affects Conidia and Biofilm Formation Capacity
3.8. O-Glycosylation Is Required for Virulence on Fruit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elad, Y.; Williamson, B.; Tudzynski, P.; Delen, N. Botrytis spp. and diseases they cause in agricultural systems—An introduction. In Botrytis: Biology, Pathology and Control; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Fillinger, S.; Elad, Y. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Springer International Publishing: New York, NY, USA, 2016. [Google Scholar]
- Jarvis, W. Botryotinia and Botrytis Species: Taxonomy, Physiology and Pathogenicity—A Guide to the Literature; Agriculture Canada: Ottawa, ON, Canada, 1977. [Google Scholar]
- Notte, A.-M.; Plaza, V.; Marambio-Alvarado, B.; Olivares-Urbina, L.; Poblete-Morales, M.; Silva-Moreno, E.; Castillo, L. Molecular identification and characterization of Botrytis cinerea associated to the endemic flora of semi-desert climate in Chile. Curr. Res. Microb. Sci. 2021, 2, 100049. [Google Scholar] [CrossRef]
- Daryaei, H.; Yousef, A.E.; Balasubramaniam, V.M. Microbiological Aspects of High-Pressure Processing of Food: Inactivation of Microbial Vegetative Cells and Spores. In High Pressure Processing of Food: Principles, Technology and Applications; Balasubramaniam, V.M., Barbosa-Cánovas, G.V., Lelieveld, H.L.M., Eds.; Springer: New York, NY, USA, 2016; pp. 271–294. [Google Scholar]
- De Simone, N.; Pace, B.; Grieco, F.; Chimienti, M.; Tyibilika, V.; Santoro, V.; Capozzi, V.; Colelli, G.; Spano, G.; Russo, P. Botrytis cinerea and Table Grapes: A Review of the Main Physical, Chemical, and Bio-Based Control Treatments in Post-Harvest. Foods 2020, 9, 1138. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Sokorai, K.; Ukuku, D.; Fan, X.; Juneja, V. Effect of high hydrostatic pressure processing on the background microbial loads and quality of cantaloupe puree. Food Res. Int. 2017, 91, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar] [CrossRef] [PubMed]
- de Boer, A.D.; de Groot, P.W.J.; Weindl, G.; Schaller, M.; Riedel, D.; Diez-Orejas, R.; Klis, F.M.; de Koster, C.G.; Dekker, H.L.; Gross, U.; et al. The Candida albicans cell wall protein Rhd3/Pga29 is abundant in the yeast form and contributes to virulence. Yeast 2010, 27, 611–624. [Google Scholar] [CrossRef]
- De Las Penas, A.; Pan, S.J.; Castano, I.; Alder, J.; Cregg, R.; Cormack, B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003, 17, 2245–2258. [Google Scholar] [CrossRef] [PubMed]
- Gelis, S.; de Groot, P.W.J.; Castillo, L.; Moragues, M.D.; Sentandreu, R.; Gomez, M.M.; Valentin, E. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 2012, 49, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Green, C.B.; Oh, S.H.; Zhao, X.M. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—A sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef]
- Laforet, L.; Moreno, I.; Sanchez-Fresneda, R.; Martinez-Esparza, M.; Martinez, J.P.; Arguelles, J.C.; de Groot, P.W.J.; Valentin-Gomez, E. Pga26 mediates filamentation and biofilm formation and is required for virulence in Candida albicans. FEMS Yeast Res. 2011, 11, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Elorza, M.V.; Valentin, E.; Sentandreu, R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006, 6, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Pardo, M.; Monteoliva, L.; Vazquez, P.; Martinez, R.; Molero, G.; Nombela, C.; Gil, C. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 2004, 150, 4157–4170. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Garcia-Maceira, P.; Castillo-Barahona, L.C.; Valentin, E.; Sentandreu, R. Cell wall composition and structure of Yarrowia lipolytica transposon mutants affected in calcofluor sensitivity. Antonie Van Leeuwenhoek 2003, 84, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Kalkkinen, N.; Sareneva, H.; Paakkola, J.; Makarow, M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA 1992, 89, 3671–3675. [Google Scholar] [CrossRef]
- De Groot, P.W.; Hellingwerf, K.J.; Klis, F.M. Genome-wide identification of fungal GPI proteins. Yeast 2003, 20, 781–796. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; Martinez, A.I.; Castillo, L. A Genomic Inventory of Cell Wall Biosynthesis in the Ubiquitous Plant Pathogen Botrytis cinerea. In The Fungal Cell Wall; Mora-Montes, H.M., Ed.; Nova Biomedical: Hauppauge, NY, USA, 2013. [Google Scholar]
- Castillo, L.; Martinez, A.I.; Garcera, A.; Elorza, M.V.; Valentin, E.; Sentandreu, R. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: The repetitive sequence is needed for binding to the cell wall beta-1,3-glucan. Yeast 2003, 20, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Deutzmann, R.; Lehle, L.; Mrsa, V.; Tanner, W. Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 2006, 281, 11523–11529. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Alvarez, A.; Elias-Villalobos, A.; Ibeas, J.I. The O-Mannosyltransferase PMT4 Is Essential for Normal Appressorium Formation and Penetration in Ustilago maydis. Plant Cell 2009, 21, 3397–3412. [Google Scholar] [CrossRef]
- Kriangkripipat, T.; Momany, M. Aspergillus nidulans Protein O-Mannosyltransferases Play Roles in Cell Wall Integrity and Developmental Patterning. Eukaryot. Cell 2009, 8, 1475–1485. [Google Scholar] [CrossRef]
- Zhou, H.; Hu, H.Y.; Zhang, L.J.; Li, R.Y.; Ouyang, H.M.; Ming, J.; Jin, C. O-mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 2007, 6, 2260–2268. [Google Scholar] [CrossRef]
- Mouyna, I.; Kniemeyer, O.; Jank, T.; Loussert, C.; Mellado, E.; Aimanianda, V.; Beauvais, A.; Wartenberg, D.; Sarfati, J.; Bayry, J.; et al. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol. Microbiol. 2010, 76, 1205–1221. [Google Scholar] [CrossRef]
- Willer, T.; Brandl, M.; Sipiczki, M.; Strahl, S. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 2005, 57, 156–170. [Google Scholar] [CrossRef]
- Prill, S.K.H.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schroppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 2005, 55, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Gentzsch, M.; Tanner, W. The PMT gene family: Protein O-glycosylation in Saccharomyces cerevisiae is vital. Embo J. 1996, 15, 5752–5759. [Google Scholar] [CrossRef] [PubMed]
- Castillo, L.; Caivo, E.; Martinez, A.I.; Ruiz-Herrera, J.; Valentin, E.; Lopez, J.A.; Sentandreu, R. A study of the Candida albicans cell wall proteome. Proteomics 2008, 8, 3871–3881. [Google Scholar] [CrossRef] [PubMed]
- Castillo, L.; Martinez, A.I.; Garcera, A.; Garcia-Martinez, J.; Ruiz-Herrera, J.; Valentin, E.; Sentandreu, R. Genomic response programs of Candida albicans following protoplasting and regeneration. Fungal Genet. Biol. 2006, 43, 124–134. [Google Scholar] [CrossRef]
- Olson, G.M.; Fox, D.S.; Wang, P.; Alspaugh, J.A.; Buchanan, K.L. Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Rouabhia, M.; Schaller, M.; Corbucci, C.; Vecchiarelli, A.; Prill, S.K.; Giasson, L.; Ernst, J.F. Virulence of the fungal pathogen Candida albicans requires the five isoforms of protein mannosyltransferases. Infect. Immun. 2005, 73, 4571–4580. [Google Scholar] [CrossRef]
- Willger, S.D.; Ernst, J.F.; Alspaugh, J.A.; Lengeler, K.B. Characterization of the PMT gene family in Cryptococcus neoformans. PLoS ONE 2009, 4, e6321. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Ding, W.; Wang, B.; Zhou, H.; Ouyang, H.; Ming, J.; Jin, C. Reduced expression of the O-mannosyltransferase 2 (AfPmt2) leads to deficient cell wall and abnormal polarity in Aspergillus fumigatus. Glycobiology 2010, 20, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Gorka-Niec, W.; Pniewski, M.; Kania, A.; Perlinska-Lenart, U.; Palamarczyk, G.; Kruszewska, J.S. Disruption of Trichoderma reesei gene encoding protein O-mannosyltransferase I results in a decrease of the enzyme activity and alteration of cell wall composition. Acta Biochim. Pol. 2008, 55, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Hamaguchi, T.; Sameshima, Y.; Goto, M.; Furukawa, K. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 2004, 150, 1973–1982. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Sameshima, Y.; Koga, T.; Kim, H.; Goto, M.; Furukawa, K. Protein O-mannosyltransferase A of Aspergillus awamori is involved in O-mannosylation of glucoamylase I. Microbiology 2005, 151 Pt 11, 3657–3667. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; Brito, N.; Frias, M.; Gonzalez, C. Botrytis cinerea Protein O-Mannosyltransferases Play Critical Roles in Morphogenesis, Growth, and Virulence. PLoS ONE 2013, 8, e65924. [Google Scholar] [CrossRef] [PubMed]
- Buttner, P.; Koch, F.; Voigt, K.; Quidde, T.; Risch, S.; Blaich, R.; Bruckner, B.; Tudzynski, P. Variations in Ploidy among Isolates of Botrytis cinerea—Implications for Genetic and Molecular Analyses. Curr. Genet. 1994, 25, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Quidde, T.; Osbourn, A.E.; Tudzynski, P. Detoxification of alpha-tomatine by Botrytis cinerea. Physiol. Mol. Plant Pathol. 1998, 52, 151–165. [Google Scholar] [CrossRef]
- Cotoras, M.; Folch, C.; Mendoza, L. Characterization of the antifungal activity on Botrytis cinerea of the natural diterpenoids kaurenoic acid and 3beta-hydroxy-kaurenoic acid. J. Agric. Food Chem. 2004, 52, 2821–2826. [Google Scholar] [CrossRef] [PubMed]
- Plaza, V.; Lagues, Y.; Carvajal, M.; Perez-Garcia, L.A.; Mora-Montes, H.M.; Canessa, P.; Larrondo, L.F.; Castillo, L. bcpmr1 encodes a P-type Ca2+/Mn2+-ATPase mediating cell-wall integrity and virulence in the phytopathogen Botrytis cinerea. Fungal Genet. Biol. 2015, 76, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Kars, I.; McCalman, M.; Wagemakers, L.; Van Kan, J.A.L. Functional analysis of Botrytis cinerea pectin methylesterase genes by PCR-based targeted mutagenesis: Bcpme1 and Bcpme2 are dispensable for virulence of strain B05.10. Mol. Plant Pathol. 2005, 6, 641–652. [Google Scholar] [CrossRef]
- Aguayo, C.; Riquelme, J.; Valenzuela, P.D.T.; Hahn, M.; Silva Moreno, E. Bchex virulence gene of Botrytis cinerea: Characterization and functional analysis. J. Gen. Plant Pathol. 2011, 77, 230–238. [Google Scholar] [CrossRef]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR: Methods and Applications; Meuer, S., Wittwer, C., Nakagawara, K.-I., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.S.; Thiewes, H.; van Kan, J.A.L. The D-galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet. Biol. 2011, 48, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Diaz-Jimenez, D.F.; Lopez-Romero, E.; Zinker, S.; Ponce-Norla, P.; Kullberg, B.J.; Brown, A.J.P.; Odds, F.C.; et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef]
- van der Vaart, J.M.; Caro, L.H.; Chapman, J.W.; Klis, F.M.; Verrips, C.T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1995, 177, 3104–3110. [Google Scholar] [CrossRef] [PubMed]
- Yadav, J.; Muend, S.; Zhang, Y.; Rao, R. A phenomics approach in yeast links proton and calcium pump function in the Golgi. Mol. Biol. Cell 2007, 18, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Berndt, P.; Hahn, M. Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 2006, 59, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Torres-Ossandon, M.J.; Vega-Galvez, A.; Salas, C.E.; Rubio, J.; Silva-Moreno, E.; Castillo, L. Antifungal activity of proteolytic fraction (P1G10) from (Vasconcellea cundinamarcensis) latex inhibit cell growth and cell wall integrity in Botrytis cinerea. Int. J. Food Microbiol. 2019, 289, 7–16. [Google Scholar] [CrossRef]
- Torres-Ossandon, M.J.; Castillo, L.; Uribe, E.; Bilbao-Sainz, C.; Ah-Hen, K.S.; Vega-Galvez, A. Combined Effect of High Hydrostatic Pressure and Proteolytic Fraction P1G10 from Vasconcellea cundinamarcensis Latex against Botrytis cinerea in Grape Juice. Foods 2023, 12, 3400. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.E.; Tabilo-Munizaga, G.; Perez-Won, M.; Maluenda, D.; Roco, T. Effect of high hydrostatic pressure (HHP) treatments on microbiological shelf-life of chilled Chilean jack mackerel (Trachurus murphyi). Innov. Food Sci. Emerg. Technol. 2015, 29, 107–112. [Google Scholar] [CrossRef]
- Davis, M.R., Jr.; Goldberg, J.B. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J. Vis. Exp. 2012, 63, 3916. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Scheinost, A. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 1999, 274, 9068–9075. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.H.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Brown, A.J.P.; Odds, F.C.; et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Bermejo, C.; Garcia, R.; Rodriguez-Pena, J.M. Genomics in the detection of damage in microbial systems: Cell wall stress in yeast. Clin. Microbiol. Infect. 2009, 15 (Suppl. 1), 44–46. [Google Scholar] [CrossRef] [PubMed]
- Proszynski, T.J.; Simons, K.; Bagnat, M. O-glycosylation as a sorting determinant for cell surface delivery in yeast. Mol. Biol. Cell 2004, 15, 1533–1543. [Google Scholar] [CrossRef]
- Ram, A.F.; Wolters, A.; Ten Hoopen, R.; Klis, F.M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 1994, 10, 1019–1030. [Google Scholar] [CrossRef]
- Bravim, F.; de Freitas, J.M.; Fernandes, A.A.; Fernandes, P.M. High hydrostatic pressure and the cell membrane: Stress response of Saccharomyces cerevisiae. Ann. N. Y. Acad. Sci. 2010, 1189, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Calvo, M.; Prieto, M.; Lopez, M.; Alvarez-Ordonez, A. Effects of high hydrostatic pressure on Escherichia coli ultrastructure, membrane integrity and molecular composition as assessed by FTIR spectroscopy and microscopic imaging techniques. Molecules 2014, 19, 21310–21323. [Google Scholar] [CrossRef] [PubMed]
- Peltroche-Llacsahuanga, H.; Goyard, S.; d’Enfert, C.; Prill, S.K.; Ernst, J.F. Protein O-mannosyltransferase isoforms regulate biofilm formation in Candida albicans. Antimicrob. Agents Chemother. 2006, 50, 3488–3491. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bates, S.; Hall, R.A.; Cheetham, J.; Netea, M.G.; MacCallum, D.M.; Brown, A.J.; Odds, F.C.; Gow, N.A. Role of the Candida albicans MNN1 gene family in cell wall structure and virulence. BMC Res. Notes 2013, 6, 294. [Google Scholar] [CrossRef] [PubMed]
- Pinchai, N.; Juvvadi, P.R.; Fortwendel, J.R.; Perfect, B.Z.; Rogg, L.E.; Asfaw, Y.G.; Steinbach, W.J. The Aspergillus fumigatus P-Type Golgi Apparatus Ca2+/Mn2+ ATPase PmrA Is Involved in Cation Homeostasis and Cell Wall Integrity but Is Not Essential for Pathogenesis. Eukaryot. Cell 2010, 9, 472–476. [Google Scholar] [CrossRef]
- Bates, S.; MacCallum, D.M.; Bertram, G.; Munro, C.A.; Hughes, H.B.; Buurman, E.T.; Brown, A.J.P.; Odds, F.C.; Gow, N.A.R. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 2005, 280, 23408–23415. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rodriguez, N.; Manzano-Lopez, J.; Munoz-Bravo, M.; Fernandez-Garcia, E.; Muniz, M.; Wellinger, R.E. Manganese redistribution by calcium-stimulated vesicle trafficking bypasses the need for P-type ATPase function. J. Biol. Chem. 2015, 290, 9335–9347. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.C.; Katoh-Fukui, R.; Moto, K.; Ribas, J.C.; Ishiguro, J. Schizosaccharomyces pombe Pmr1p is essential for cell wall integrity and is required for polarized cell growth and cytokinesis. Eukaryot. Cell 2004, 3, 1124–1135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fujikawa, T.; Kuga, Y.; Yano, S.; Yoshimi, A.; Tachiki, T.; Abe, K.; Nishimura, M. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 2009, 73, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Arias, M.J.; Defosse, T.A.; Dementhon, K.; Csonka, K.; Mellado-Mojica, E.; Dias Valerio, A.; Gonzalez-Hernandez, R.J.; Courdavault, V.; Clastre, M.; Hernandez, N.V.; et al. Disruption of Protein Mannosylation Affects Candida guilliermondii Cell Wall, Immune Sensing, and Virulence. Front. Microbiol. 2016, 7, 1951. [Google Scholar] [CrossRef]
- Popolo, L.; Gilardelli, D.; Bonfante, P.; Vai, M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Ram, A.F.; Groos, E.M.; Kollar, R.; Montijn, R.C.; Van Den Ende, H.; Llobell, A.; Cabib, E.; Klis, F.M. Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. J. Bacteriol. 1997, 179, 6279–6284. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Ponce-Noyola, P.; Villagomez-Castro, J.C.; Gow, N.A.; Flores-Carreon, A.; Lopez-Romero, E. Protein glycosylation in Candida. Future Microbiol. 2009, 4, 1167–1183. [Google Scholar] [CrossRef]
- Agaphonov, M.O.; Packeiser, A.N.; Chechenova, M.B.; Choi, E.S.; Ter-Avanesyan, M.D. Mutation of the homologue of GDP-mannose pyrophosphorylase alters cell wall structure, protein glycosylation and secretion in Hansenula polymorpha. Yeast 2001, 18, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.; Sommer, S.S.; Hensel, A.; Bussey, H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J. Cell Biol. 1990, 110, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Bussey, H. The yeast KRE9 gene encodes an O glycoprotein involved in cell surface beta-glucan assembly. Mol. Cell. Biol. 1993, 13, 6346–6356. [Google Scholar] [CrossRef]
- de Nobel, J.G.; Klis, F.M.; Priem, J.; Munnik, T.; van den Ende, H. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast 1990, 6, 491–499. [Google Scholar] [CrossRef]
- Lipke, P.N. What We Do Not Know about Fungal Cell Adhesion Molecules. J. Fungi 2018, 4, 59. [Google Scholar] [CrossRef]
- Munro, C.A.; Bates, S.; Buurman, E.T.; Hughes, H.B.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef]
- Doss, R.P.; Potter, S.W.; Chastagner, G.A.; Christian, J.K. Adhesion of Nongerminated Botrytis cinerea Conidia to Several Substrata. Appl. Environ. Microbiol. 1993, 59, 1786–1791. [Google Scholar] [CrossRef] [PubMed]
- Doss, R.P.; Potter, S.W.; Soeldner, A.H.; Christian, J.K.; Fukunaga, L.E. Adhesion of germlings of Botrytis cinerea. Appl. Environ. Microbiol. 1995, 61, 260–265. [Google Scholar] [CrossRef]
- de Groot, P.W.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in human fungal pathogens: Glue with plenty of stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 2004, 1666, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Marin-Menguiano, M.; Moreno-Sanchez, I.; Barrales, R.R.; Fernandez-Alvarez, A.; Ibeas, J.I. N-glycosylation of the protein disulfide isomerase Pdi1 ensures full Ustilago maydis virulence. PLoS Pathog. 2019, 15, e1007687. [Google Scholar] [CrossRef] [PubMed]
- Scarlata, S. The effect of hydrostatic pressure on membrane-bound proteins. Braz. J. Med. Biol. Res. 2005, 38, 1203–1208. [Google Scholar] [CrossRef]
- Sun, J.Z.; Pang, C.Y.; Cheng, X.; Yang, B.Y.; Jin, B.B.; Jin, L.; Qi, Y.X.; Sun, Y.; Chen, X.; Liu, W.D.; et al. Investigation of the antifungal activity of the dicarboximide fungicide iprodione against Bipolaris maydis. Pestic. Biochem. Phys. 2023, 190, 105319. [Google Scholar] [CrossRef]
- Fillinger, S.; Leroux, P.; Auclair, C.; Barreau, C.; Al Hajj, C.; Debieu, D. Genetic analysis of fenhexamid-resistant field isolates of the phytopathogenic fungus Botrytis cinerea. Antimicrob. Agents Chemother. 2008, 52, 3933–3940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.L.; Liu, C.; Tang, B.; Ren, Z.; Wang, G.L.; Liu, W. Quantitative proteomics analysis reveals important roles of N-glycosylation on ER quality control system for development and pathogenesis in Magnaporthe oryzae. PLoS Pathog. 2020, 16, e1008355. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, L.A.; Csonka, K.; Flores-Carreon, A.; Estrada-Mata, E.; Mellado-Mojica, E.; Nemeth, T.; Lopez-Ramirez, L.A.; Toth, R.; Lopez, M.G.; Vizler, C.; et al. Role of Protein Glycosylation in Candida parapsilosis Cell Wall Integrity and Host Interaction. Front. Microbiol. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]


| Strain | Glucose | Mannose | Glucosamine |
|---|---|---|---|
| B05.10 | 0.469 ± 0.03 (46.9 ± 3%) * | 0.340 ± 0.01 (34.0 ± 1%) * | 0.007 ± 0.00 (7.0%) * |
| Δbcpmt4 | 0.771 ± 0.04 (77.1 ± 4%) * | 0.039 ± 0.06 (39.0 ± 6%) * | 0.035 ± 0.02 (3.5%) * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plaza, V.; Pasten, A.; López-Ramírez, L.A.; Mora-Montes, H.M.; Rubio-Astudillo, J.; Silva-Moreno, E.; Castillo, L. Botrytis cinerea PMT4 Is Involved in O-Glycosylation, Cell Wall Organization, Membrane Integrity, and Virulence. J. Fungi 2025, 11, 71. https://doi.org/10.3390/jof11010071
Plaza V, Pasten A, López-Ramírez LA, Mora-Montes HM, Rubio-Astudillo J, Silva-Moreno E, Castillo L. Botrytis cinerea PMT4 Is Involved in O-Glycosylation, Cell Wall Organization, Membrane Integrity, and Virulence. Journal of Fungi. 2025; 11(1):71. https://doi.org/10.3390/jof11010071
Chicago/Turabian StylePlaza, Verónica, Alice Pasten, Luz A. López-Ramírez, Héctor M. Mora-Montes, Julia Rubio-Astudillo, Evelyn Silva-Moreno, and Luis Castillo. 2025. "Botrytis cinerea PMT4 Is Involved in O-Glycosylation, Cell Wall Organization, Membrane Integrity, and Virulence" Journal of Fungi 11, no. 1: 71. https://doi.org/10.3390/jof11010071
APA StylePlaza, V., Pasten, A., López-Ramírez, L. A., Mora-Montes, H. M., Rubio-Astudillo, J., Silva-Moreno, E., & Castillo, L. (2025). Botrytis cinerea PMT4 Is Involved in O-Glycosylation, Cell Wall Organization, Membrane Integrity, and Virulence. Journal of Fungi, 11(1), 71. https://doi.org/10.3390/jof11010071









