Review and Current Perspectives on DNA Topoisomerase I and II Enzymes of Fungi as Study Models for the Development of New Antifungal Drugs
Abstract
1. Introduction
2. DNA Topoisomerases: Definition and Classification
3. Characteristics of the Genes That Code for Topo I and II of Fungal Organisms
4. Biology of DNA Topoisomerase I and II Enzymes of Fungal Organisms
5. Structural Characteristics of DNA Topoisomerase I Enzymes of Fungal Organisms
6. Structural Characteristics of DNA Topoisomerase II Enzymes of Fungal Organisms
7. Phylogenetic Analysis of Type I and II DNA Topoisomerases of Fungal Organisms
8. Topo I and II Enzymes of Fungi as Targets for Antifungal Therapy
9. Selected Inhibitors of Topo I and II and Their Mechanism of Action
10. Selected Inhibitors of Topo I and II as Antifungal Agents
| Compound | Binding Energy (kcal/mol) | Residues Interacting with the Ligand | Nucleotide Residues | Polar Interactions | Hydrophobic Interactions |
|---|---|---|---|---|---|
| Topo I of H. sapiens | |||||
| [104,141,142,143] Topotecan | −9.42 | Arg364, Arg488, Lys532, Asp533, Ile535, Asn631, His632, Gln633, Thr718, PTR723 | TGP11, DG12, DC112, DA113 | Arg364 (C-HO) Gln633 (C-HO) His632 (C-C) Thr718 (C-C) DG12 (C-C) DC112 (C-C) | Arg364 (π-cation) Arg364 (π-alkyl) Asp533 (π-anion) |
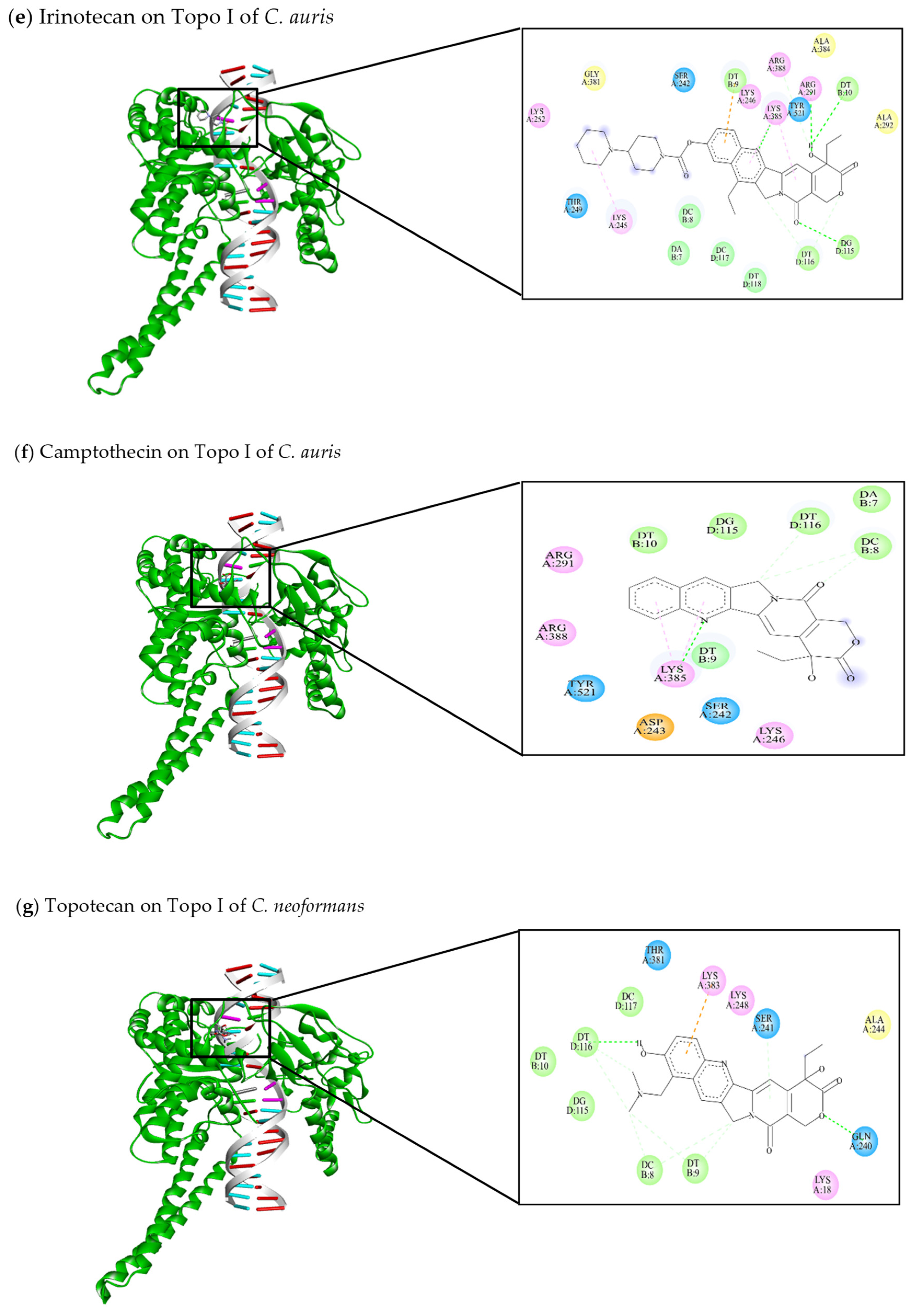
| [144] Irinotecan | −11.47 | Arg364, Leu485, Ala486, Leu487, Arg488, Phe 529, Gly531, Lys532, Asp533, Ile535, Tyr537, Cys630, Leu629, Asn631, His632, Gln633, Thr718, PTR723 | DT10, DG12, TGP11 | Lys532 (C-HO) Lys532 (C-C) Asp533 (C-C) Cys630 (C-C) Thr718 (C-C) TGP11 (C-C) | Lys532 (π-alkyl) Arg364 (π-alkyl) Ile535 (π-alkyl) |
| [46,103,145,146] Camptothecin | −9.3 | Arg364, Arg488, Lys532, Asp533, Ile535, Asn631, His632, Gln633, Thr718, PTR723 | DT10, TGP11, DG12, DA13 | Arg488 (C-HO) Lys532 (C-HO) PTR723 (C-C) | Arg364 (π-cation) Asp533 (π-anion) |
| Topo I of C. auris | |||||
| This review Topotecan | −9.08 | Ser242, Lys245, Lys246, Arg291, Lys329, Lys385, Arg388, Tyr521 | DA7, DC8, DT9, DT10, DA114, DG115, DT116 | Arg291 (C-HO) Lys385 (C-HO) DC8 (C-C) DT9 (C-C) DG115 (C-C) DT116 (C-C) | Lys385 (π-alkyl) Arg388 (π-alkyl) |
| This review Irinotecan | −11.62 | Ser242, Lys245, Lys246, Thr249, Lys252, Arg291, Ala292, Gly381, Ala384, Lys385, Arg388, Tyr521 | DA7, DC8, DT9, DT10, DG115, DT116, DC117, DT118 | Arg291 (C-HO) Lys385 (C-HO) DT10 (C-HO) DG115 (C-HO) DT116 (C-C) | Lys245 (π-alkyl) Arg291 (π-alkyl) DT9 (π-anion) |
| This review Camptothecin | −9.26 | Ser242, Asp243, Lys246, Arg291, Lys385, Arg388, Tyr521 | DA7, DC8, DT9, DT10, DG115, DT116 | Lys385 (C-HO) DC8 (C-C) DT116 (C-C) | Lys385 (π-alkyl) |
| Topo I of C. neoformans | |||||
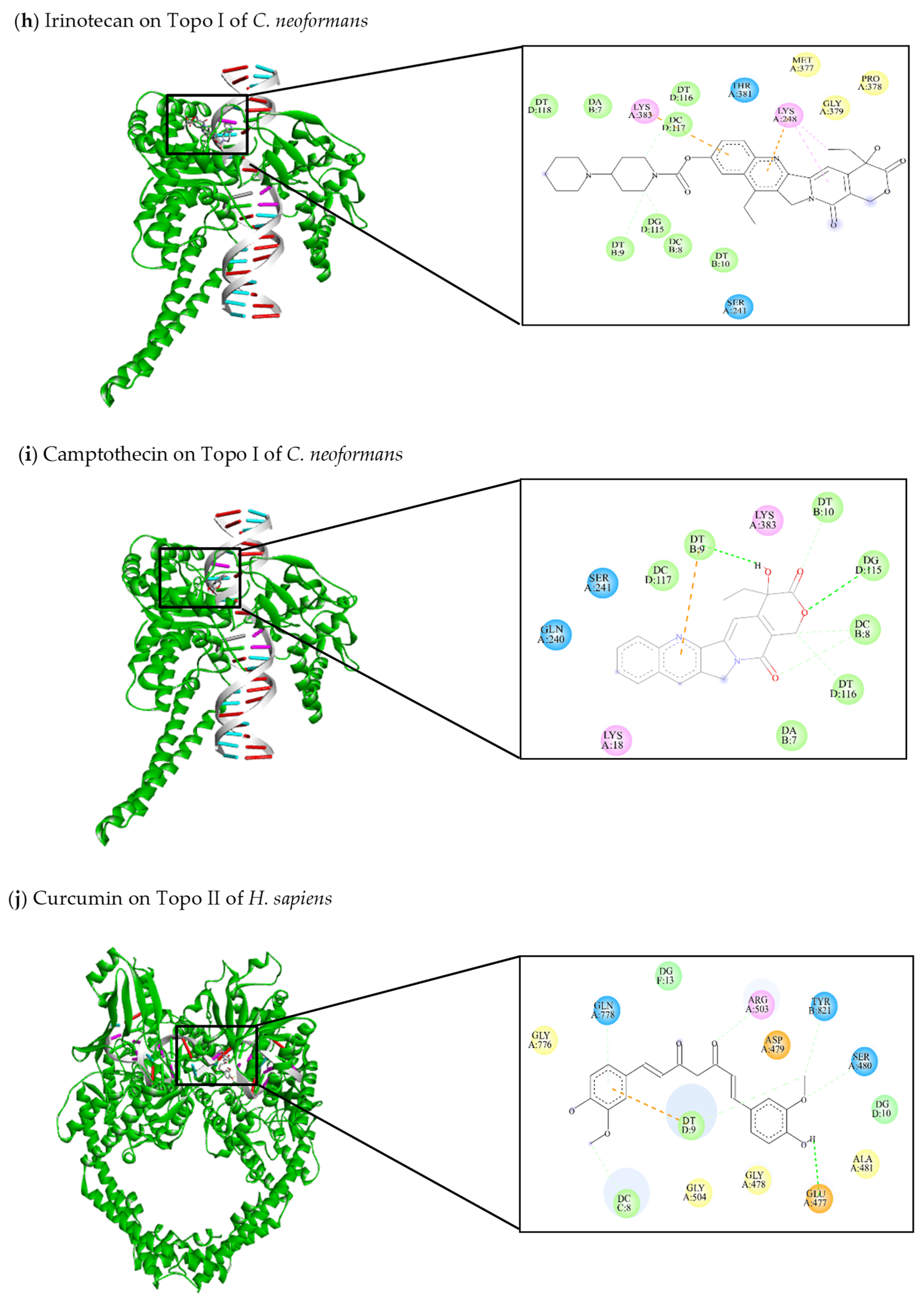
| This review Topotecan | −9.28 | Lys18, Gln240, Ser241, Ala244, Lys248, Thr381, Lys383 | DC8, DT9, DT10, DC117, DG115, DT116 | Gln240 (C-HO) DC8 (C-C) DT9 (C-C) DT116 (C-C) | Lys383 (π-anion) |
| This review Irinotecan | −10.67 | Ser241, Lys248, Met377, Pro378, Gly379, Thr381, Lys383 | DA7, DC8, DT9, DT10, DG115, DT116, DC117, DT118 | DT9 (C-C) DG115 (C-C) DT116 (C-C) | Lys248 (π-alkyl) Lys383 (π-anion) |
| This review Camptothecin | −8.4 | Lys18, Gln240, Ser241, Lys383 | DA7, DC8, DT9, DT10, DG115, DT116, DC117 | DT9 (C-HO) DG115 (C-HO) DC8 (C-C) DT10 (C-C) DT116 (C-C) | DT9 (π-anion) |
| Topo II of H. sapiens | |||||
| [47,144] Curcumin | −8.73 | Glu477, Gly478, Asp479, Ser480, Ala481, Arg503, Gly504, Gly776, Gln778, Tyr821 | DC8, DT9, DG10, DG13 | Glu477 (C-HO) Ser480 (C-C) Gln778 (C-C) Arg503 (C-C) Tyr821 (C-C) DT9 (C-C) | DT9 (π-anion) |
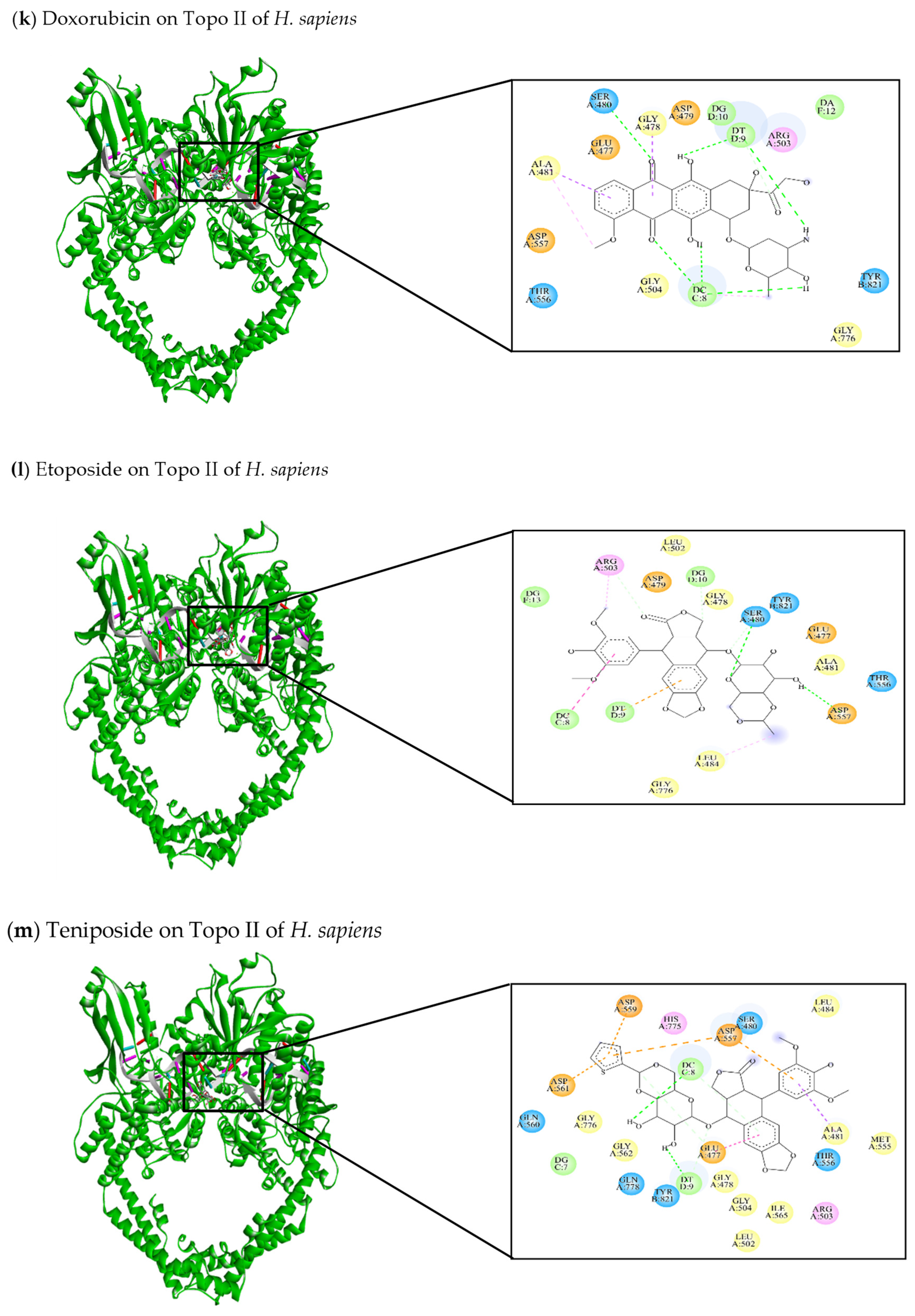
| [145] Doxorubicin | −9.67 | Glu477, Gly478, Asp479, Ser480, Ala481, Arg503, Gly504, Thr556, Asp557, Gly776, Tyr821 | DC8, DT9, DG10, DA12 | Ser480 (C-HO) DC8 (C-HO) DT9 (C-HO) | Gly478 (π-sigma) Ala481 (π-Alkyl) Ala481 (π-sigma) |
| [47,141,145,146] Etoposide | −7.2 | Glu477, Gly478, Asp479, Ser480, Ala481, Leu484, Leu502, Arg503, Thr556, Asp557, Gly776, Tyr821 | DC8, DT9, DG10, DG13 | Ser480 (C-HO) Asp557 (C-HO) Arg503 (C-C) | Leu484 (π-alkyl) Arg503 (π-alkyl) DC8 (π-alkyl) DT9 (π-anion) |
| [103,146] Teniposide | −7.0 | Glu477, Gly478, Ser480, Ala481, Leu484, Leu502, Arg503, Gly504, Met555, Thr556, Asp557, Asp559, Gln560, Asp561, Gly562, Ile565, His775, Gly776, Gln778, Tyr821 | DG7, DC8, DT9 | Glu477 (C-C) DC8 (C-HO) DT9 (C-HO) | Ala481 (π-sigma) Glu477 (π-π stacked) Asp557 (π-anion) Asp559 (π-anion) Asp561 (π-anion) |
| Topo II of C. auris | |||||
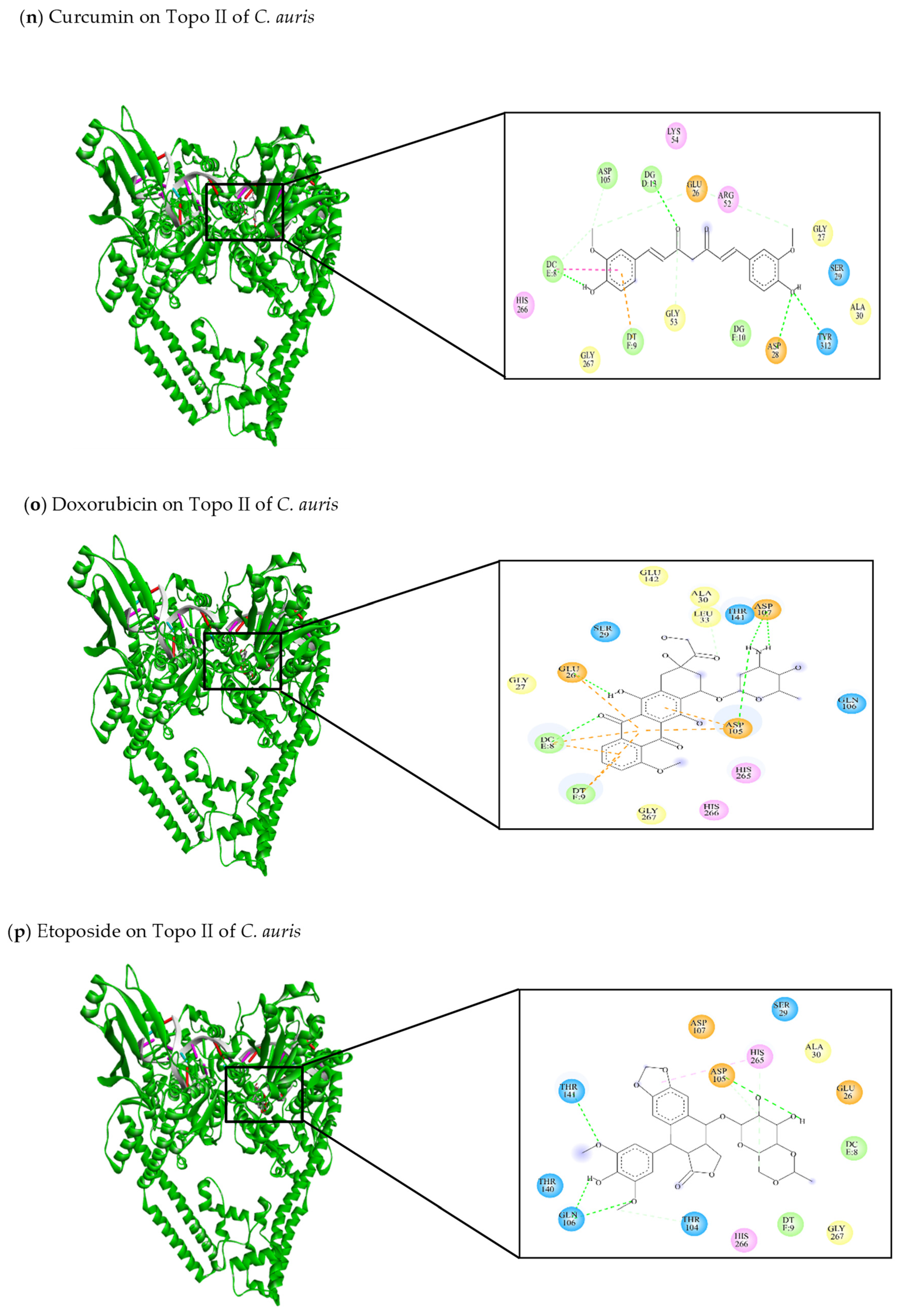
| This review Curcumin | −8.27 | Glu26, Gly27, Asp28, Ser29, Ala30, Arg52, Gly53, Lys54, Asp105, His266, Gly267, Tyr312 | DC8, DT9, DG10, DG13 | Glu26 (C-C) Asp28 (C-HO) Gly53 (C-C) Asp105 (C-C) Tyr312 (C-HO) DC8 (C-HO) DG13 (C-HO) | DC8 (π-π stacked) DT9 (π-anion) |
| This review Doxorubicin | −9.53 | Glu26, Gly27, Ser29, Ala30, Leu33, Asp105, Gln106, Asp107, Thr141, Glu142, His265, His266, Gly267 | DC8, DT9 | Glu26 (C-HO) Asp105 (C-HO) Asp107 (C-HO) Leu33 (C-C) | Glu26 (π-anion) Asp105 (π-anion) Asp107 (π-anion) DC8 (π-anion) DT9 (π-anion) |
| This review Etoposide | −7.48 | Glu26, Ser29, Ala30, Thr104, Asp105, Gln106, Asp107, Thr140, Thr141, His265, His266, Gly267 | DC8, DT9 | Thr104 (C-C) Asp105 (C-HO) Gln106 (C-HO) Thr141 (C-HO) | His265 (π-alkyl) |
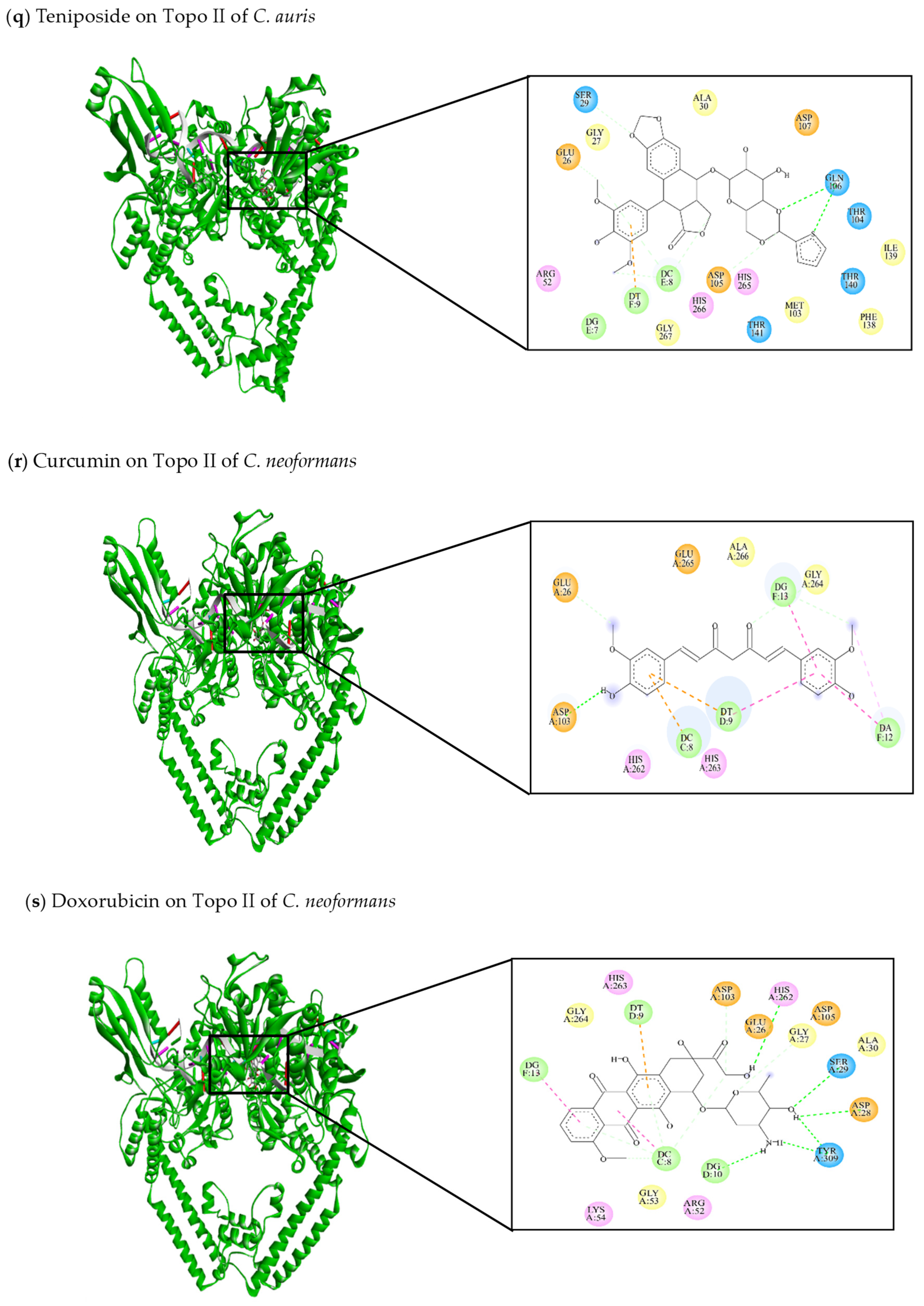
| This review Teniposide | −8.54 | Glu26, Gly27, Ser29, Ala30, Arg52, Met103, Thr104, Asp105, Gln106, Asp107, Phe138, Ile139, Thr140, Thr141, His265, His266, Gly267 | DG7, DC8, DT9 | Glu26 (C-C) Ser29 (C-C) Gln106 (C-HO) DC8 (C-C) | DT9 (π-anion) |
| Topo II of C. neoformans | |||||
| This review Curcumin | −8.4 | Glu26, Asp103, His262, His263, Gly264, Glu265, Ala266 | DC8, DT9, DA12, DG13 | Glu26 (C-C) Asp103 (C-HO) DG13 (C-C) | DC8 (π-anion) DT9 (π-anion) DA12 (π-alkyl) DA12 (π-π stacked) DT9 (π-π stacked) |
| This review Doxorubicin | −11.04 | Glu26, Gly27, Asp28, Ser29, Ala 30, Arg52, Gly53, Lys54, Asp103, Asp105, His262, His263, Gly264, Tyr309 | DC8, DT9, DG10, DG13 | Gly27 (C-C) Asp28 (C-HO) Ser29 (C-HO) Asp103 (C-C) His262 (C-HO) Tyr309 (C-HO) DC8 (C-C) | DC8 (π-alkyl) DT9 (π-anion) DG13 (π-alkyl) |
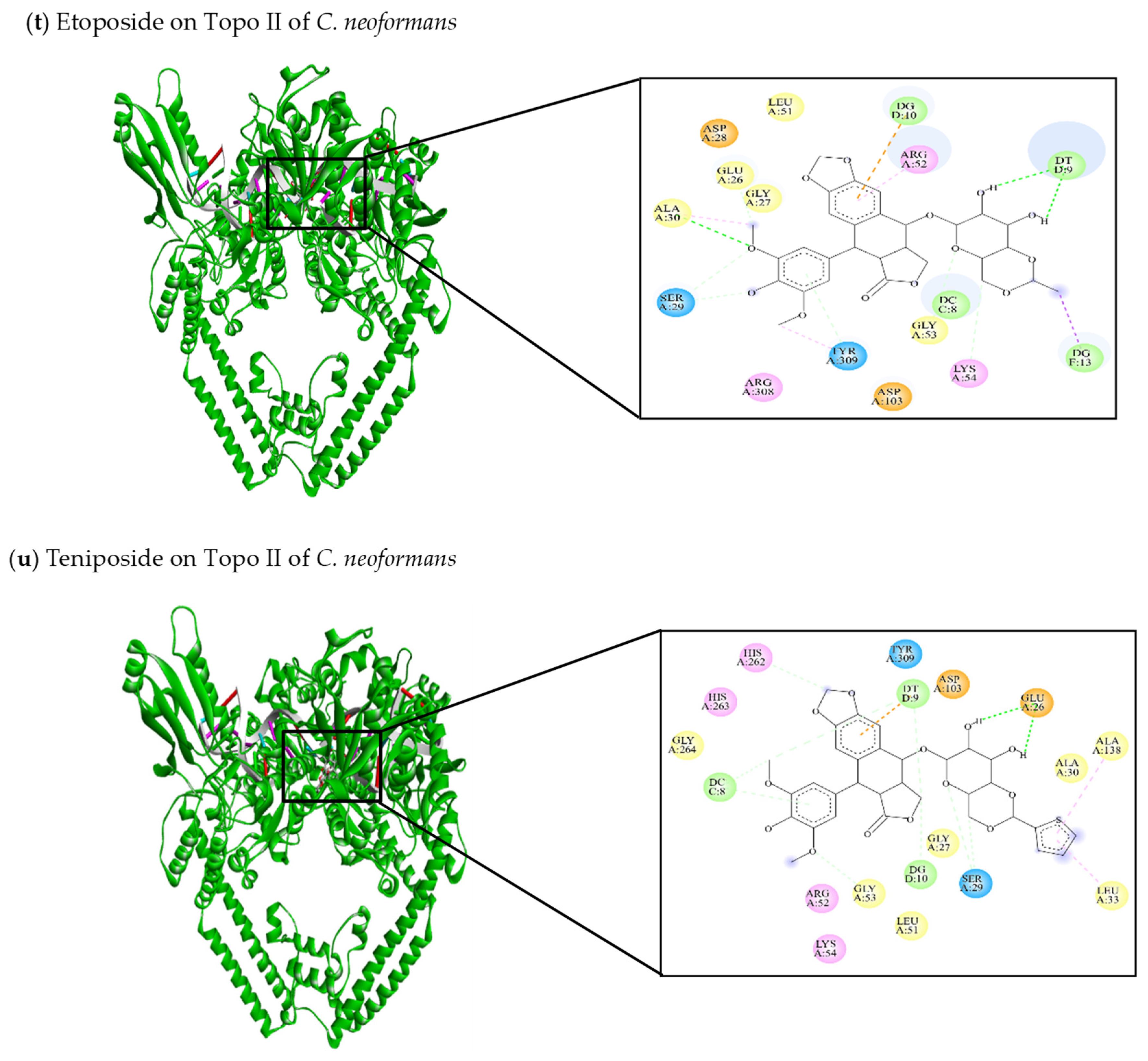
| This review Etoposide | −9.4 | Glu26, Gly27, Asp28, Ser29, Ala30, Leu51, Arg52, Gly53, Lys54, Asp103, Arg308, Tyr309 | DC8, DT9, DG10, DG13 | Ser29 (C-C) Ala30 (C-HO) Tyr309 (C-C) DC8 (C-C) DT9 (C-HO) | Ala30 (π-alkyl) Arg52 (π-alkyl) Tyr309 (π-alkyl) DG13 (π-sigma) |
| This review Teniposide | −10.58 | Glu26, Gly27, Ser29, Ala30, Leu33, Leu51, Arg52, Gly53, Lys54, Asp103, Ala138, His262, His263, Gly264, Tyr309 | DC8, DT9, DG10 | Glu26 (C-HO) Ser29 (C-C) Gly53 (C-C) His262 (C-C) DC8 (C-C) DG10 (C-C) | Leu33 (π-alkyl) Ala138 (π-alkyl) DT9 (π-anion) |
11. Therapeutic Potential of Topo I and II Inhibitors in Fungal Organisms
12. Future Perspectives on Topo I and II Enzymes as Alternative Targets for Antifungal Drugs
13. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Angarone, M. Fungal infections in cancer patients. Cancer Treat. Res. 2014, 161, 129–155. [Google Scholar] [PubMed]
- Garbee, D.D.; Pierce, S.S.; Manning, J. Opportunistic fungal infections in critical care units. Crit. Care Nurs. Clin. N. Am. 2017, 29, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef]
- Suleyman, G.; Alangaden, G.J. Nosocomial fungal infections: Epidemiology, infection control, and prevention. Infect. Dis. Clin. N. Am. 2021, 35, 1027–1053. [Google Scholar] [CrossRef]
- Spallone, A.; Schwartz, I.S. Emerging fungal infections. Infect. Dis. Clin. N. Am. 2021, 35, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.G.; Diekema, D.J. What Is New in Fungal Infections? Mod. Pathol. 2023, 36, 100187. [Google Scholar] [CrossRef] [PubMed]
- Mavor, A.L.; Thewes, S.; Hube, B. Systemic fungal infections caused by Candida species: Epidemiology, infection process and virulence attributes. Curr. Drug Targets 2005, 6, 863–874. [Google Scholar] [CrossRef]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Molecular mechanisms in Candida albicans and beyond. Chem. Rev. 2021, 121, 3390–3411. [Google Scholar] [CrossRef]
- Ismadi, Y.K.M.; Mohamad, S.; Harun, A. Species-specific PCR primers for simultaneous detection of Aspergillus fumigatus, Aspergillus terreus, Candida albicans and Candida glabrata in invasive fungal infections. Malays. J. Pathol. 2023, 45, 397–403. [Google Scholar]
- Ruhnke, M. Epidemiology of Candida albicans infections and role of non-Candida-albicans yeasts. Curr. Drug Targets 2006, 7, 495–504. [Google Scholar] [CrossRef]
- Silva, L.N.; de Mello, T.P.; de Souza Ramos, L.; Branquinha, M.H.; dos Santos, A.L. New and promising chemotherapeutics for emerging infections involving drug-resistant non-albicans Candida species. Curr. Top. Med. Chem. 2019, 19, 2527–2553. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gaviria, M.; Ramírez-Sotelo, U.; Mora-Montes, H.M. Non-albicans Candida Species: Immune Response, Evasion Mechanisms, and New Plant-Derived Alternative Therapies. J. Fungi 2022, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Finn, T. Multidrug-resistant Candida haemulonii and C. Auris, tel aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gaviria, M.; Martínez-Álvarez, J.A.; Chávez-Santiago, J.O.; Mora-Montes, H.M. Candida haemulonii Complex and Candida auris: Biology, Virulence Factors, Immune Response, and Multidrug Resistance. Infect. Drug Resist. 2023, 16, 1455–1470. [Google Scholar] [CrossRef]
- Francisco, E.C.; de Jong, A.W.; Colombo, A.L. Candida haemulonii species complex: A mini-review. Mycopathologia 2023, 188, 909–917. [Google Scholar] [CrossRef]
- Ruiz de Alegría-Puig, C.; Rodríguez-Lozano, J.; Agüero-Balbin, J. Identification and antifungal susceptibility testing of Candida haemulonii complex isolated from clinical samples. Folia Microbiol. 2024, 69, 165–171. [Google Scholar] [CrossRef]
- Góralska, K.; Blaszkowska, J.; Dzikowiec, M. Neuroinfections caused by fungi. Infection 2018, 46, 443–459. [Google Scholar] [CrossRef]
- von Lilienfeld-Toal, M.; Wagener, J.; Einsele, H.; Cornely, O.A.; Kurzai, O. Invasive fungal infection: New treatments to meet new challenges. Dtsch. Arztebl. Int. 2019, 116, 271. [Google Scholar]
- Godoy, M.C.; Dalla Pria, H.R.F.; Truong, M.T.; Shroff, G.S.; Marom, E.M. Invasive fungal pneumonia in immunocompromised patients. Radiol. Clin. N. Am. 2022, 60, 497–506. [Google Scholar] [CrossRef]
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.; Colombo, A.L.; Hoenigl, M.; Cornely, O.A. Defining and managing COVID-19-associated pulmonar aspergilosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef] [PubMed]
- Casalini, G.; Giacomelli, A.; Ridolfo, A.; Gervasoni, C.; Antinori, S. Invasive fungal infections complicating COVID-19: A narrative review. J. Fungi 2021, 7, 921. [Google Scholar] [CrossRef] [PubMed]
- Hoenigl, M.; Seidel, D.; Sprute, R.; Cunha, C.; Oliverio, M.; Goldman, G.H.; Ibrahim, A.S.; Carvalho, A. COVID-19-associated fungal infections. Nat. Microbiol. 2022, 7, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Vinayagamoorthy, K.; Pentapati, K.C.; Prakash, H. Prevalence, risk factors, treatment and outcome of multidrug resistance Candida auris infections in coronavirus disease (COVID-19) patients: A systematic review. Mycoses 2022, 65, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Kariyawasam, R.M.; Dingle, T.C.; Kula, B.E.; Vandermeer, B.; Sligl, W.I.; Schwartz, I.S. Defining COVID-19–associated pulmonary aspergillosis: Systematic review and meta-analysis. Clin. Microbiol. Infect. 2022, 28, 920–927. [Google Scholar] [CrossRef]
- Khojasteh, S.; Jafarzdeh, J.; Hosseini, S.A.; Hagan, I.; Turki, H.; Gharehbolagh, S.A.; Abastabar, M.; Mahmoudi, S. Candida auris and COVID-19: A health threatening combination. Curr. Med. Mycol. 2022, 8, 44. [Google Scholar] [CrossRef]
- Tsai, C.S.; Lee, S.S.J.; Chen, W.C.; Tseng, C.H.; Lee, N.Y.; Chen, P.L.; Li, M.C.; Syue, L.S.; Lo, C.L.; Ko, W.C.; et al. COVID-19-associated candidiasis and the emerging concern of Candida auris infections. J. Microbiol. Immunol. Infect. 2023, 56, 672–679. [Google Scholar] [CrossRef]
- Özbek, L.; Topçu, U.; Manay, M.; Esen, B.H.; Bektas, S.N.; Aydın, S.; Özdemir, B.; Khostelidi, S.N.; Klimko, N.; Cornely, O.; et al. COVID-19-associated mucormycosis: A systematic review and meta-analysis of 958 cases. Clin. Microbiol. Infect. 2023, 29, 722–731. [Google Scholar] [CrossRef]
- Usuda, D.; Kato, M.; Sugawara, Y.; Shimizu, R.; Inami, T.; Tsuge, S.; Sakurai, R.; Kawai, K.; Matsubara, S.; Tanaka, R.; et al. Secondary pulmonary infection by Fusarium solani and Aspergillus niger during systemic steroid treatment for COVID-19: A case report. World J. Clin. Cases 2023, 11, 6280. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Sharma, C.; Chowdhary, A. Molecular bases of antifungal resistance in filamentous fungi. Int. J. Antimicrob. Agents 2017, 50, 607–616. [Google Scholar] [CrossRef]
- Wiederhold, N.P. Antifungal resistance: Current trends and future strategies to combat. Infect. Drug Resist. 2017, 10, 249–259. [Google Scholar] [CrossRef]
- Pereira, R.; dos Santos Fontenelle, R.O.; De Brito, E.H.S.; De Morais, S.M. Biofilm of Candida albicans: Formation, regulation and resistance. J. Appl. Microbiol. 2021, 131, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Campoy, S.; Adrio, J.L. Antifungals. Biochem. Pharmacol. 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Kontoyiannis, D.P. Resistance to antifungal drugs. Infect. Dis. Clin. N. Am. 2021, 35, 279–311. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.; Johnson, C.; Berman, J.; Coste, A.T.; Cuomo, C.A.; Perlin, D.S.; Bicanic, T.; Harrison, T.S.; Wiederhold, N.; Bromley, M.; et al. The importance of antimicrobial resistance in medical mycology. Nat. Commun. 2022, 13, 5352. [Google Scholar] [CrossRef]
- Lupetti, A.; Danesi, R.; Campa, M.; Del Tacca, M.; Kelly, S. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 2022, 8, 76–81. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Wiederhold, N.P.; Patterson, T.F. Emergence of azole resistance in Aspergillus. Semin. Respir. Crit. Care Med. 2015, 36, 673–680. [Google Scholar] [CrossRef]
- Dannaoui, E. Antifungal resistance in mucorales. Int. J. Antimicrob. Agents 2017, 50, 617–621. [Google Scholar] [CrossRef]
- García-Rubio, R.; Cuenca-Estrella, M.; Mellado, E. Triazole resistance in Aspergillus species: An emerging problem. Drugs 2017, 77, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Ellsworth, M.; Ostrosky-Zeichner, L. Isavuconazole: Mechanism of action, clinical efficacy, and resistance. J. Fungi 2020, 6, 324. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cantero, A.; Lopez-Fernandez, L.; Guarro, J.; Capilla, J. Azole resistance mechanisms in Aspergillus: Update and recent advances. Int. J. Antimicrob. Agents 2020, 55, 105807. [Google Scholar] [CrossRef] [PubMed]
- Pasula, S.; Chandrasekar, P.H. Azole resistance in Aspergillus species: Promising therapeutic options. Expert. Opin. Pharmacother. 2021, 22, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Lewis, R.E.; Kontoyiannis, D.P. Investigational antifungal agents for invasive mycoses: A clinical perspective. Clin. Infect. Dis. 2022, 75, 534–544. [Google Scholar] [CrossRef]
- Shen, L.L.; Baranowski, J.; Fostel, J.; Montgomery, D.A.; Lartey, P.A. DNA topoisomerases from pathogenic fungi: Targets for the discovery of antifungal drugs. Antimicrob. Agents Chemother. 1992, 36, 2778–2784. [Google Scholar] [CrossRef]
- Tagle-Olmedo, T.; Andrade-Pavón, D.; Martínez-Gamboa, A.; Gómez-García, O.; García-Sierra, F.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Inhibitors of DNA topoisomerases I and II applied to Candida dubliniensis reduce growth, viability, the generation of petite mutants and toxicity, while acting synergistically with fluconazole. FEMS Yeast Res. 2021, 21, foab023. [Google Scholar] [CrossRef]
- Kondaka, K.; Gabriel, I. Targeting DNA Topoisomerase II in Antifungal Chemotherapy. Molecules 2022, 27, 7768. [Google Scholar] [CrossRef]
- Gellert, M. DNA topoisomerases. Annu. Rev. Biochem. 1981, 50, 879–910. [Google Scholar] [CrossRef]
- Wang, J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996, 65, 635–692. [Google Scholar] [CrossRef]
- Champoux, J.J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA topoisomerases. EcoSal Plus 2015, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.S.; Nikolaou, C.; Roca, J. Structure and chromosomal organization of yeast genes regulated by Topoisomerase II. Int. J. Mol. Sci. 2018, 19, 134. [Google Scholar] [CrossRef]
- Forterre, P.; Gribaldo, S.; Gadelle, D.; Serre, M.C. Origin and evolution of DNA topoisomerases. Biochimie 2007, 89, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Roca, J. The mechanisms of DNA topoisomerases. Trends Biochem. Sci. 1995, 20, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Wigley, D.B. Structure and mechanism of DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 185–208. [Google Scholar] [CrossRef]
- Bugreev, D.V.; Nevinsky, G.A. Structure and mechanism of action of type IA DNA topoisomerases. Biochemistry 2009, 74, 1467–1481. [Google Scholar] [CrossRef]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Chikamori, K.; G Grozav, A.; Kozuki, T.; Grabowski, D.; Ganapathi, R.; K Ganapathi, M. DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy. Curr. Cancer Drug Targets 2010, 10, 758–771. [Google Scholar] [CrossRef]
- Madabhushi, R. The roles of DNA topoisomerase IIβ in transcription. Int. J. Mol. Sci. 2018, 19, 1917. [Google Scholar] [CrossRef]
- Hirsch, J.; Klostermeier, D. What makes a type IIA topoisomerase a gyrase or a Topo IV? Nucleic Acids Res. 2021, 49, 6027–6042. [Google Scholar] [CrossRef]
- McKie, S.J.; Desai, P.R.; Seol, Y.; Allen, A.M.; Maxwell, A.; Neuman, K.C. Topoisomerase VI is a chirally-selective, preferential DNA decatenase. eLife 2022, 11, e67021. [Google Scholar] [CrossRef] [PubMed]
- Kunze, N.; Yang, G.C.; Dölberg, M.; Sundarp, R.; Knippers, R.; Richter, A. Structure of the human type I DNA topoisomerase gene. J. Biol. Chem. 1991, 266, 9610–9616. [Google Scholar] [CrossRef]
- Austin, C.A.; Marsh, K.L. Eukaryotic DNA topoisomerase IIβ. Bioessays 1998, 20, 215–226. [Google Scholar] [CrossRef]
- Deneke, J.; Burgin, A.B.; Wilson, S.L.; Chaconas, G. Catalytic residues of the telomere resolvase ResT: A pattern similar to, but distinct from, tyrosine recombinases and type IB topoisomerases. J. Biol. Chem. 2004, 279, 53699–53706. [Google Scholar] [CrossRef] [PubMed]
- Brochier-Armanet, C.; Gribaldo, S.; Forterre, P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol. Direct. 2008, 3, 54. [Google Scholar] [CrossRef]
- Jiang, W.; Gerhold, D.; Kmiec, E.B.; Hauser, M.; Becker, J.M.; Koltin, Y. The topoisomerase I gene from Candida albicans. Microbiology 1997, 143, 377–386. [Google Scholar] [CrossRef][Green Version]
- Goto, T.; Wang, J.C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc. Natl. Acad. Sci. USA 1985, 82, 7178–7182. [Google Scholar] [CrossRef]
- Thrash, C.; Bankier, A.T.; Barrell, B.G.; Sternglanz, R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc. Natl. Acad. Sci. USA 1985, 82, 4374–4378. [Google Scholar] [CrossRef]
- Uemura, T.; Morino, K.; Uzawa, S.; Shiozaki, K.; Yanagida, M. Cloning and sequencing of Schizosaccharomyces pombe DNA topoisomerase I gene, and effect of gene disruption. Nucleic Acids Res. 1987, 15, 9727–9739. [Google Scholar] [CrossRef][Green Version]
- Uemura, T.; Yanagida, M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: Single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984, 3, 1737–1744. [Google Scholar] [CrossRef]
- Christman, M.F.; Dietrich, F.S.; Fink, G.R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 1988, 55, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, P.; Bjornsti, M.A. Mechanisms of DNA Topoisomerase I-Induced Cell Killing in the Yeast: Saccharomyces cerevisiae. Ann. N. Y. Acad. Sci. 2000, 922, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Losasso, C.; Cretaio, E.; Palle, K.; Pattarello, L.; Bjornsti, M.A.; Benedetti, P. Alterations in linker flexibility suppress DNA topoisomerase I mutant-induced cell lethality. J. Biol. Chem. 2007, 282, 9855–9864. [Google Scholar] [CrossRef]
- Heldrich, J.; Sun, X.; Vale-Silva, L.A.; Markowitz, T.E.; Hochwagen, A. Topoisomerases modulate the timing of meiotic DNA breakage and chromosome morphogenesis in Saccharomyces cerevisiae. Genetics 2020, 215, 59–73. [Google Scholar] [CrossRef]
- Gerhold, D.; Thiyagarajan, M.; Kmiec, E.B. The topoisomerase I gene from Ustilago maydis: Sequence, disruption and mutant phenotype. Nucleic Acids Res. 1994, 22, 3773–3778. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choder, M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991, 5, 2315–2326. [Google Scholar] [CrossRef]
- Del Poeta, M.; Chen, S.F.; Von Hoff, D.; Dykstra, C.C.; Wani, M.C.; Manikumar, G.; Perfect, J.R. Comparison of in vitro activities of camptothecin and nitidine derivatives against fungal and cancer cells. Antimicrob. Agents Chemother. 1999, 43, 2862–2868. [Google Scholar] [CrossRef]
- Lynn, R.M.; Bjornsti, M.A.; Caron, P.R.; Wang, J.C. Peptide sequencing and site-directed mutagenesis identify tyrosine-727 as the active site tyrosine of Saccharomyces cerevisiae DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 1989, 86, 3559–3563. [Google Scholar] [CrossRef]
- Sharma, A.; Mondragón, A. DNA topoisomerases. Curr. Opin. Struct. Biol. 1995, 5, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Reguera, R.M.; Pérez-Pertejo, Y.; Redondo, C.M.; Díaz-González, R.; Balaña-Fouce, R. La ADN topoisomerasa tipo I de protozoos patógenos como Diana terapéutica de fármacos antitumorales. Medicina 2007, 67, 747–757. [Google Scholar]
- Aravind, L.; Leipe, D.D.; Koonin, E.V. Toprim—A conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998, 26, 4205–4213. [Google Scholar] [CrossRef] [PubMed]
- Allemand, F.; Mathy, N.; Brechemier-Baey, D.; Condon, C. The 5S rRNA maturase, ribonuclease M5, is a Toprim domain family member. Nucleic Acids Res. 2005, 33, 4368–4376. [Google Scholar] [CrossRef]
- Fu, G.; Wu, J.; Liu, W.; Zhu, D.; Hu, Y.; Deng, J.; Wang, D.C. Crystal structure of DNA gyrase B′ domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009, 37, 5908–5916. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.M. Type II DNA topoisomerases. Curr. Opin. Struct. Biol. 1998, 8, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Erdős, M.; Lányi, Á.; Balázs, G.; Casanova, J.L.; Boisson, B.; Maródi, L. Inherited TOP2B mutation: Possible confirmation of mutational hotspots in the TOPRIM domain. J. Clin. Immunol. 2021, 41, 817–819. [Google Scholar] [CrossRef]
- Gadelle, D.; Filie, J.; Buhler, C.; Forterre, P. Phylogenomics of type II DNA topoisomerases. Bioessays 2003, 25, 232–242. [Google Scholar] [CrossRef]
- Coelho, J.; Martins, C.; Ferreira, F.; Leitao, A. African swine fever virus ORF P1192R codes for a functional type II DNA topoisomerase. Virology 2015, 474, 82–93. [Google Scholar] [CrossRef]
- Kato, M.; Ozeki, M.; Kikuchi, A.; Kanbe, T. Phylogenetic relationship and mode of evolution of yeast DNA topoisomerase II gene in the pathogenic Candida species. Gene 2001, 272, 275–281. [Google Scholar] [CrossRef]
- Shamsizadeh, F.; Pchelin, I.M.; Makimura, K.; Alshahni, M.M.; Satoh, K.; Katiraee, F.; Rezaei-Matehhkolaei, A. DNA topoisomerase 2 gene polymorphism in dermatophytes. Mycoses 2020, 63, 694–703. [Google Scholar] [CrossRef]
- Fostel, J.M.; Montgomery, D.A.; Shen, L.L. Characterization of DNA topoisomerase I from Candida albicans as a target for drug discovery. Antimicrob. Agents Chemother. 1992, 36, 2131–2138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fostel, J.; Montgomery, D.; Lartey, P. Comparison of responses of DNA topoisomerase I from Candida albicans and human cells to four new agents which stimulate topoisomerase-dependent DNA nicking. FEMS Microbiol. Lett. 1996, 138, 105–111. [Google Scholar] [CrossRef][Green Version]
- Van Dross, R.T.; Rao, K.V.; Eisenberg, E.; Sanders, M.M. Cloning and characterization of the Aspergillus nidulans DNA topoisomerase I gene. Gene 1997, 203, 169–174. [Google Scholar] [CrossRef]
- Kim, K.H.; Akashi, T.; Mizuguchi, I.; Kikuchi, A. Cloning and characterization of the gene encoding Aspergillus nidulans DNA topoisomerase II. Gene 1999, 236, 293–301. [Google Scholar] [CrossRef]
- Nitiss, J.L. Yeast as a genetic model system for studying topoisomerase inhibitors. Adv. Pharmacol. 1994, 29, 201–226. [Google Scholar]
- Zheng, H.; Yu, Y.S. TOP2 gene is involved in the pathogenicity of Candida albicans. Mol. Cell. Biochem. 2012, 364, 45–52. [Google Scholar] [CrossRef]
- Reid, R.J.; Benedetti, P.; Bjornsti, M.A. Yeast as a model organism for studying the actions of DNA topoisomerase-targeted drugs. Biochim. Biophys. Acta 1998, 1400, 289–300. [Google Scholar] [CrossRef]
- Nitiss, J.L.; Nitiss, K.C. Yeast systems for demonstrating the targets of anti-topoisomerase II agents. In DNA Topoisomerase Protocols; Springer: Berlin, Germany, 2001; pp. 315–327. [Google Scholar]
- Woo, M.H.; Vance, J.R.; Bjornsti, M.A. Studying DNATopoisomerase I-Targeted Drugs in the Yeast: Saccharomyces cerevisiae. Methods Mol. Biol. 2001, 95, 303–313. [Google Scholar] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Protein Sci. 2016, 86, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Pavón, D.; Gómez-García, O. Etoposide and Camptothecin Reduce Growth, Viability, the Generation of Petite Mutants, and Recognize the Active Site of DNA Topoisomerase I and II Enzymes in Candida glabrata. Indian J. Microbiol. 2021, 61, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Pourquier, P.; Fan, Y.I.; Strumberg, D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1998, 1400, 83–106. [Google Scholar] [CrossRef] [PubMed]
- Bjornsti, M.A.; Benedetti, P.; Viglianti, G.A.; Wang, J.C. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: Restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989, 49, 6318–6323. [Google Scholar] [PubMed]
- Madden, K.R.; Champoux, J.J. Overexpression of human topoisomerase I in baby hamster kidney cells: Hypersensitivity of clonal isolates to camptothecin. Cancer Res. 1992, 52, 525–532. [Google Scholar] [PubMed]
- Pommier, Y. Diversity of DNA topoisomerases I and inhibitors. Biochimie 1998, 80, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Fujimori, A.; Pommier, Y. Eukaryotic DNA topoisomerases i. Biochim. Biophys. Acta 1995, 1262, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Desai, S.D.; LI, T.K.; Mao, Y.; Sun, M.E.I.; SIM, S.P. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 2000, 922, 1–10. [Google Scholar] [CrossRef]
- Hsiang, Y.H.; Liu, L.F. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988, 48, 1722–1726. [Google Scholar]
- Del Bino, G.; Lassota, P.; Darzynkiewicz, Z. The S-phase cytotoxicity of camptothecin. Exp. Cell Res. 1991, 193, 27–35. [Google Scholar] [CrossRef]
- Kollmannsberger, C.; Mross, K.; Jakob, A.; Kanz, L.; Bokemeyer, C. Topotecan–a novel topoisomerase I inhibitor: Pharmacology and clinical experience. Oncology 1999, 56, 1–12. [Google Scholar] [CrossRef]
- Liu, L.F. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 1989, 58, 351–375. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Huang, X.S.; Wu, J.F.; Yang, L.; Zheng, Y.T.; Shen, Y.M.; Li, X. Discovery of novel topoisomerase II inhibitors by medicinal chemistry approaches. J. Med. Chem. 2018, 61, 8947–8980. [Google Scholar] [CrossRef] [PubMed]
- Cortés, F.; Pastor, N. Induction of endoreduplication by topoisomerase II catalytic inhibitors. Mutagenesis 2003, 18, 105–112. [Google Scholar] [CrossRef]
- Andoh, T.; Ishida, R. Catalytic inhibitors of DNA topoisomerase II. Biochim. Biophys. Acta 1998, 1400, 155–171. [Google Scholar] [CrossRef]
- Long, B.H. Mechanisms of action of teniposide (VM-26) and comparison with etoposide (VP-16). Semin. Oncol. 1992, 19, 3–19. [Google Scholar]
- Shen, L.L.; Fostel, J.M. DNA topoisomerase inhibitors as antifungal agents. Adv. Pharmacol. 1994, 29, 227–244. [Google Scholar]
- Nitiss, J.L.; Liu, Y.X.; Hsiung, Y. A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: A genetic system for determining the targets of topoisomerase II inhibitors. Cancer Res. 1993, 53, 89–93. [Google Scholar]
- Ishida, R.; Hamatake, M.; Wasserman, R.A.; Nitiss, J.L.; Wang, J.C.; Andoh, T. DNA topoisomerase II is the molecular target of bisdioxopiperazine derivatives ICRF-159 and ICRF-193 in Saccharomyces cerevisiae. Cancer Res. 1995, 55, 2299–2303. [Google Scholar]
- Anizon, F.; Belin, L.; Moreau, P.; Sancelme, M.; Voldoire, A.; Prudhomme, M.; Meyer, T. Syntheses and biological activities (topoisomerase inhibition and antitumor and antimicrobial properties) of rebeccamycin analogues bearing modified sugar moieties and substituted on the imide nitrogen with a methyl group. J. Med. Chem. 1997, 40, 3456–3465. [Google Scholar] [CrossRef]
- Hufford, C.D.; Liu, S.; Clark, A.M.; Oguntimein, B.O. Anticandidal activity of eupolauridine and onychine, alkaloids from Cleistopholis patens. J. Nat. Prod. 1987, 50, 961–964. [Google Scholar] [CrossRef]
- Khan, S.I.; Nimrod, A.C.; Mehrpooya, M.; Nitiss, J.L.; Walker, L.A.; Clark, A.M. Antifungal activity of eupolauridine and its action on DNA topoisomerases. Antimicrob. Agents Chemother. 2002, 46, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Taghavi-Moghadam, S.; Kwong, C.D.; Secrist III, J.A.; Khan, S.I.; Clark, A.M. The synthesis and biological evaluation of alkyl and benzyl naphthyridinium analogs of eupolauridine as potential antimicrobial and cytotoxic agents. Bioorg. Med. Chem. 2016, 24, 6119–6130. [Google Scholar] [CrossRef]
- Jannatipour, M.; Liu, Y.X.; Nitiss, J.L. The top2-5 mutant of yeast topoisomerase II encodes an enzyme resistant to etoposide and amsacrine. J. Biol. Chem. 1993, 268, 18586–18592. [Google Scholar] [CrossRef]
- Knab, A.M.; Fertala, J.; Bjornsti, M.A. Mechanisms of camptothecin resistance in yeast DNA topoisomerase I mutants. J. Biol. Chem. 1993, 268, 22322–22330. [Google Scholar] [CrossRef] [PubMed]
- Knab, A.M.; Fertala, J.; Bjornsti, M.A. A Camptothecin-resistant DNA Topoisomerase I Mutant Exhibits Altered Sensitivities to Other DNA Topoisomerase Poisons. J. Biol. Chem. 1995, 270, 6141–6148. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.J.; Kauh, E.A.; Bjornsti, M.A. Camptothecin sensitivity is mediated by the pleiotropic drug resistance network in yeast. J. Biol. Chem. 1997, 272, 12091–12099. [Google Scholar] [CrossRef]
- Keller, B.A.; Patel, S.; Fisher, L.M. Molecular cloning and expression of the Candida albicans TOP2 gene allows study of fungal DNA topoisomerase II inhibitors in yeast. Biochem. J. 1997, 324, 329–339. [Google Scholar] [CrossRef]
- Fostel, J.; Montgomery, D. Identification of the aminocatechol A-3253 as an in vitro poison of DNA topoisomerase I from Candida albicans. Antimicrob. Agents Chemother. 1995, 39, 586–592. [Google Scholar] [CrossRef][Green Version]
- Kwok, S.C.; Schelenz, S.; Wang, X.; Steverding, D. In vitro effect of DNA topoisomerase inhibitors on Candida albicans. Med. Mycol. 2010, 48, 155–160. [Google Scholar] [CrossRef]
- Steverding, D.; Evans, P.; Msika, L.; Riley, B.; Wallington, J.; Schelenz, S. In vitro antifungal activity of DNA topoisomerase inhibitors. Med. Micol. 2012, 50, 333–336. [Google Scholar] [CrossRef]
- Fan, Y.; Weinstein, J.N.; Kohn, K.W.; Shi, L.M.; Pommier, Y. Molecular modeling studies of the DNA− topoisomerase I ternary cleavable complex with camptothecin. J. Med. Chem. 1998, 41, 2216–2226. [Google Scholar] [CrossRef]
- Staker, B.L.; Feese, M.D.; Cushman, M.; Pommier, Y.; Zembower, D.; Stewart, L.; Burgin, A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I− DNA covalent complex. J. Med. Chem. 2005, 48, 2336–2345. [Google Scholar] [CrossRef]
- Basili, S.; Moro, S. Novel camptothecin derivatives as topoisomerase I inhibitors. Expert Opin. Ther. Pat. 2009, 19, 555–574. [Google Scholar] [CrossRef]
- Wu, C.C.; Li, Y.C.; Wang, Y.R.; Li, T.K.; Chan, N.L. On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs. Nucleic Acids Res. 2013, 41, 10630–10640. [Google Scholar] [CrossRef]
- Kumar, A.; Bora, U. Molecular docking studies of curcumin natural derivatives with DNA topoisomerase I and II-DNA complexes. Interdiscip. Sci. 2014, 6, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wang, G.; Li, J.; Meng, G.; Liu, Z.; Dong, M.; Zhang, Q. Synthesis of novel 10-hydroxycamptothecin derivatives utilizing topotecan hydrochloride as ortho-quinonemethide precursor. Bioorg. Med. Chem. 2015, 23, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Del Castillo, E.; Gómez-García, O.; Andrade-Pavón, D.; Villa-Tanaca, L.; Ramírez-Apan, T.; Nieto-Camacho, A.; Gómez, E. Dibutyltin (IV) complexes derived from L-DOPA: Synthesis, molecular docking, cytotoxic and antifungal activity. Chem. Pharm. Bull. 2018, 66, 1104–1113. [Google Scholar] [CrossRef]
- Andrade, D.; Gómez, O.; Álvarez-Toledano, C. Exploring the binding mode of triflamide derivatives at the active site of Topo I and Topo II enzymes: In silico analysis and precise molecular docking. J. Chem. Sci. 2020, 132, 50. [Google Scholar]
- Singh, S.; Das, T.; Awasthi, M.; Pandey, V.P.; Pandey, B.; Dwivedi, U.N. DNA topoisomerase-directed anticancerous alkaloids: ADMET-based screening, molecular docking, and dynamics simulation. Biotechnol. Appl. Biochem. 2016, 63, 125–137. [Google Scholar] [CrossRef]
- Song, Z.L.; Wang, M.J.; Li, L.; Wu, D.; Wang, Y.H.; Yan, L.T.; Lee, K.H. Design, synthesis, cytotoxic activity and molecular docking studies of new 20 (S)-sulfonylamidine camptothecin derivatives. Eur. J. Med. Chem. 2016, 115, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Drwal, M.N.; Agama, K.; Wakelin, L.P.; Pommier, Y.; Griffith, R. Exploring DNA topoisomerase I ligand space in search of novel anticancer agents. PLoS ONE 2011, 6, e25150. [Google Scholar] [CrossRef] [PubMed]
- Laco, G.S.; Collins, J.R.; Luke, B.T.; Kroth, H.; Sayer, J.M.; Jerina, D.M.; Pommier, Y. Human topoisomerase I inhibition: Docking camptothecin and derivatives into a structure-based active site model. Biochemistry 2002, 41, 1428–1435. [Google Scholar] [CrossRef] [PubMed]
- El-Zoghbi, M.S.; El-Sebaey, S.A.; Al-Ghulikah, H.A.; Sobh, E.A. Design, synthesis, docking, and anticancer evaluations of new thiazolo [3, 2-a] pyrimidines as topoisomerase II inhibitors. J. Enzym. Inhib. Med. Chem. 2023, 38, 2175209. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Li, T.K.; Farh, L.; Lin, L.Y.; Lin, T.S.; Yu, Y.J.; Chan, N.L. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 2011, 333, 459–462. [Google Scholar] [CrossRef]
- Singh, S.; Pandey, V.P.; Yadav, K.; Yadav, A.; Dwivedi, U.N. Natural products as anti-cancerous therapeutic molecules targeted towards topoisomerases. Curr. Protein Pept. Sci. 2020, 21, 1103–1142. [Google Scholar] [CrossRef]
- Gabriel, I. ‘Acridines’ as new horizons in antifungal treatment. Molecules 2020, 25, 1480. [Google Scholar] [CrossRef]
- de Oliveira, D.B.C.; Silva, L.B.; da Silva, B.V.; Borges, T.C.; Marques, B.C.; dos Santos, M.B.; Andrade, A.A. A new acridone with antifungal properties against Candida spp. and dermatophytes, and antibiofilm activity against C. albicans. J. Appl. Microbiol. 2019, 127, 1362–1372. [Google Scholar] [CrossRef]

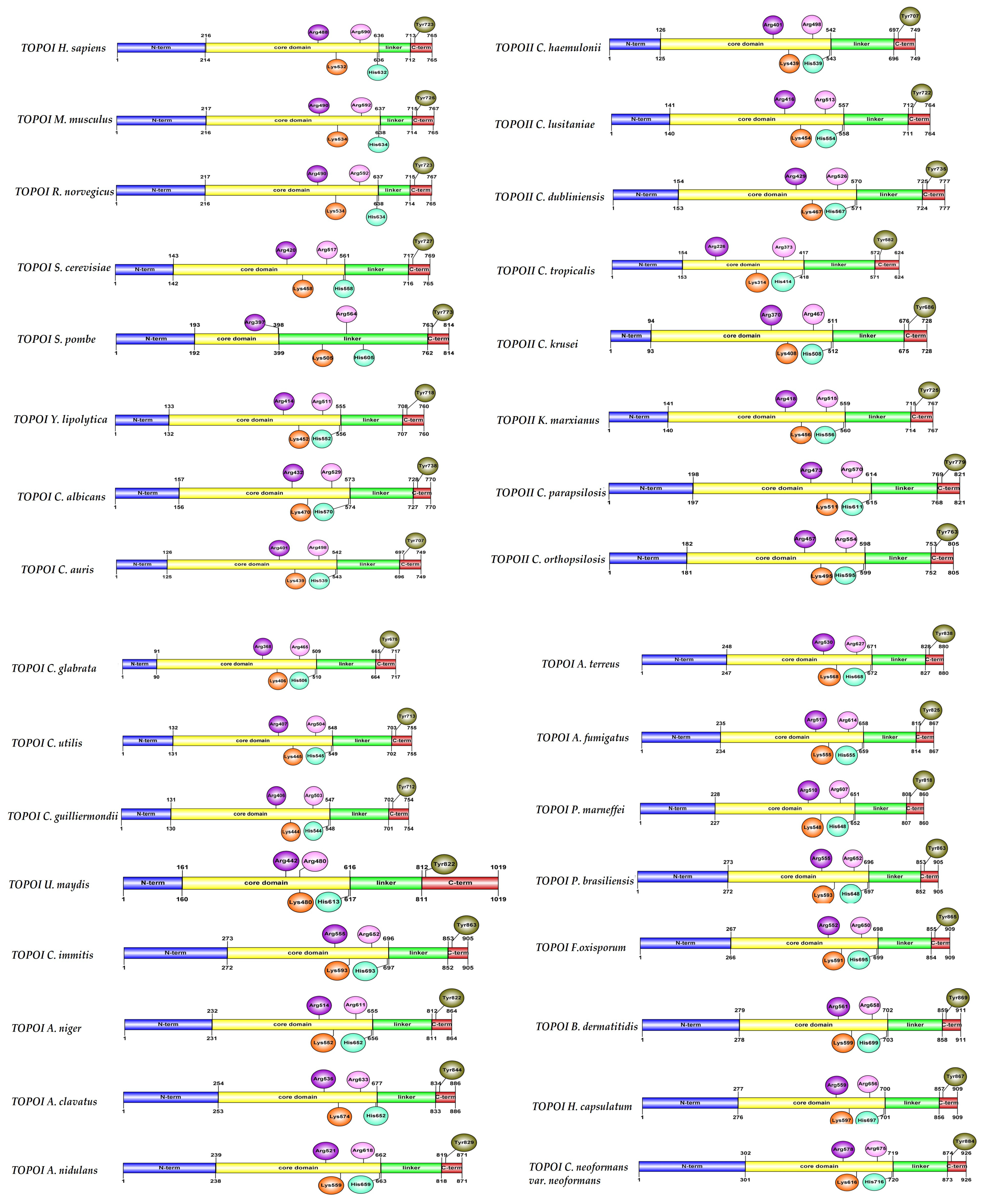

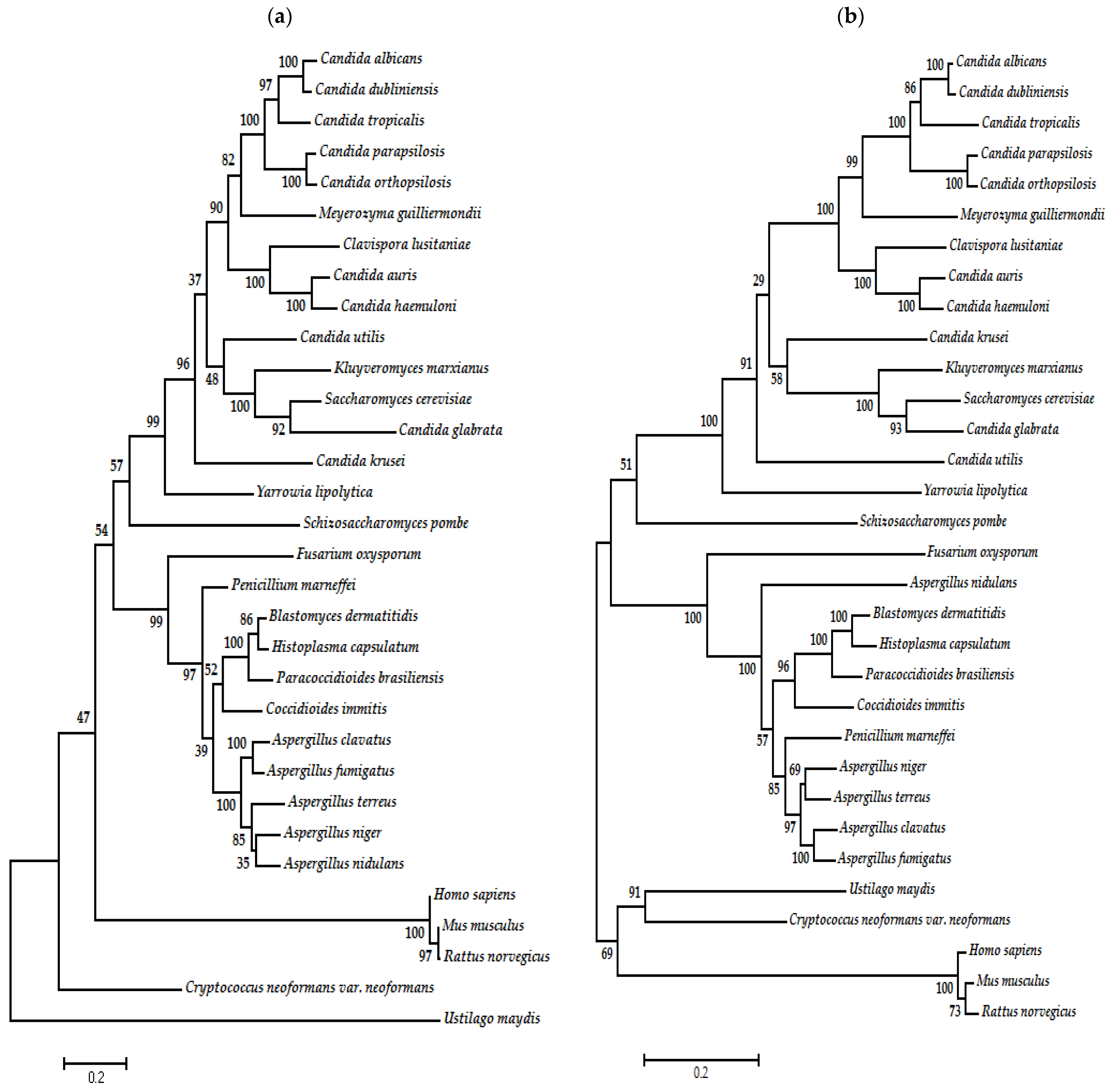
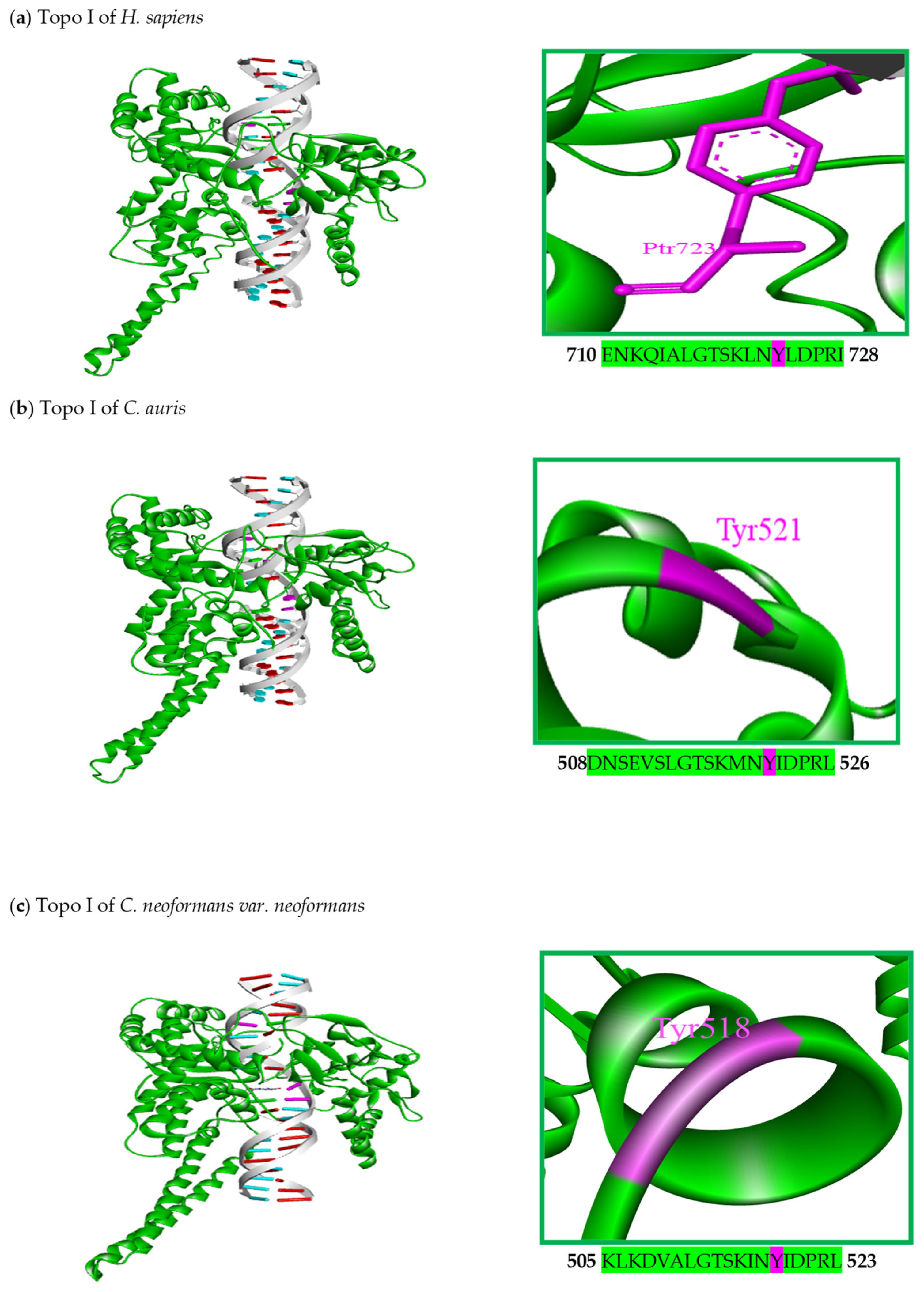
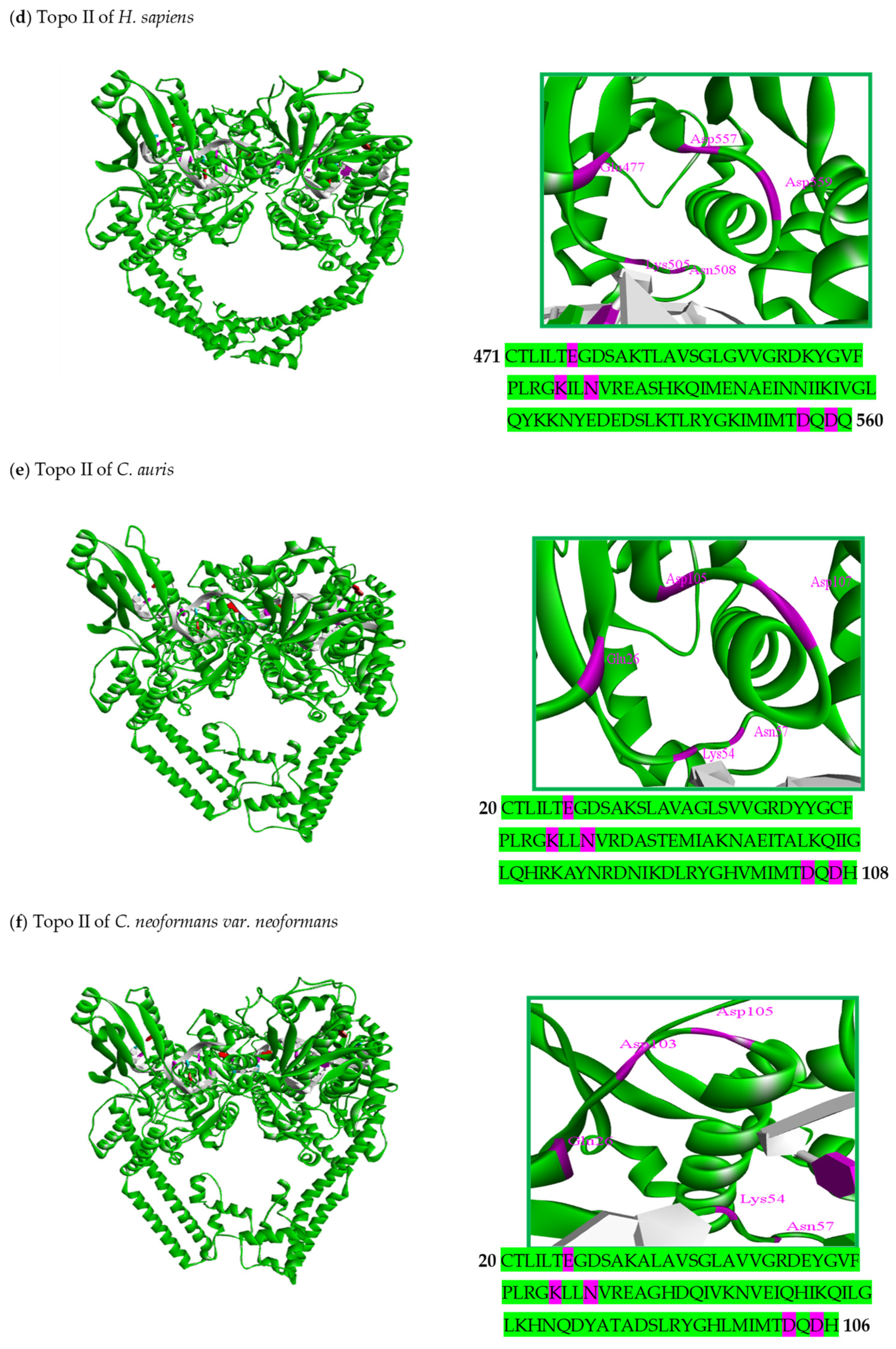

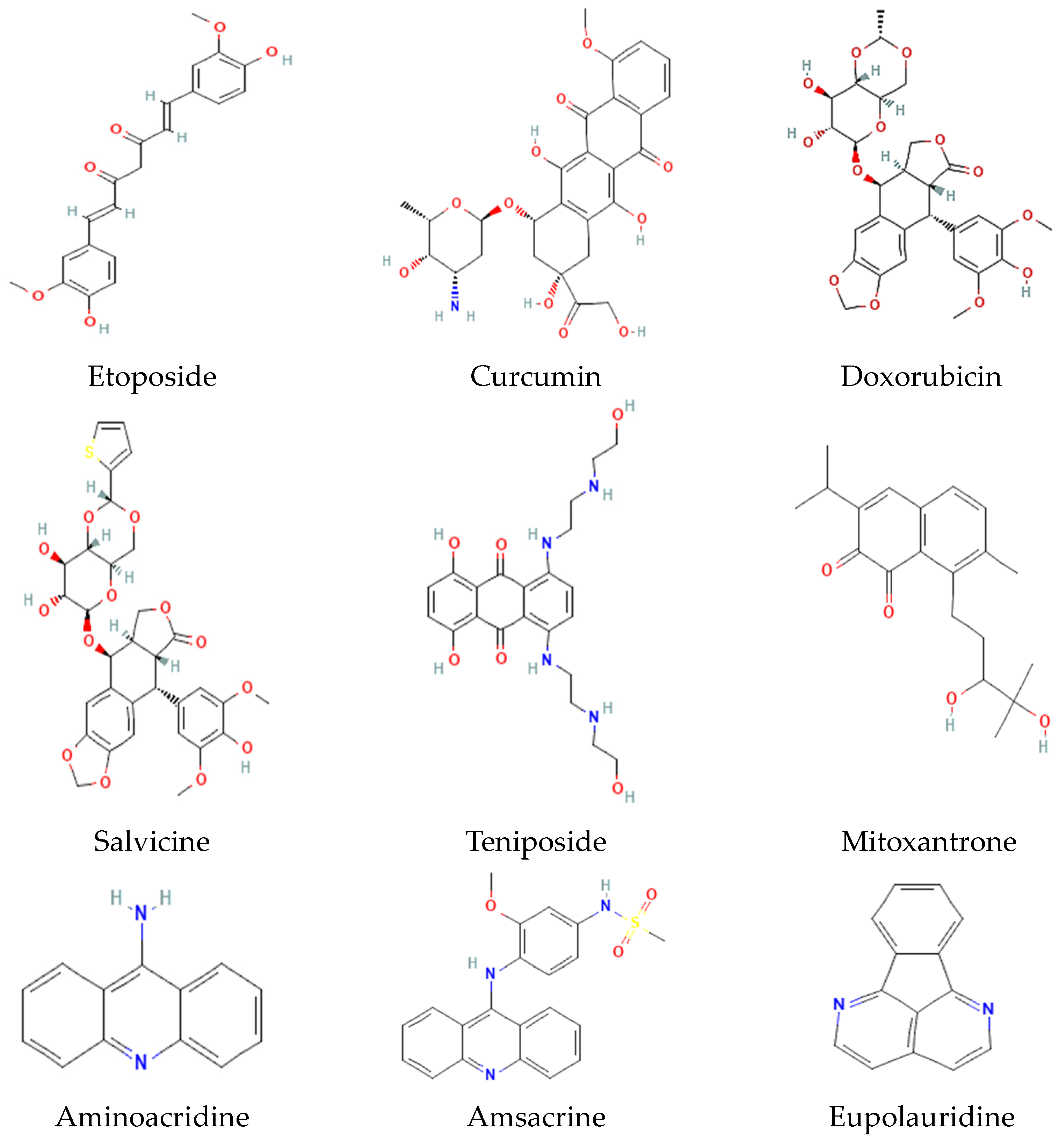


| Organism | Accession Number | Family | Gene Length (bp) | Protein Size (aa) | Accession Number | Family | Gene Length (bp) | Protein Size (aa) |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | NP_003277 | Topo I | 3734 | 765 | NP_001058 | Topo II | 5695 | 1531 |
| Mus musculus | NP_033434 | Topo I | 3859 | 767 | XP_006533217 | Topo II | 5233 | 1527 |
| Rattus norvegicus | NP_072137 | Topo I | 2304 | 767 | P41516 | Topo II | 4581 | 1526 |
| Saccharomyces cerevisiae | NP_014637 | Topo I | 2310 | 769 | AAM005481 | Topo II | 4287 | 1428 |
| Schizosaccharomyces pombe | NP_596209 | Topo I | 2761 | 814 | NP_595805 | Topo II | 6626 | 1485 |
| Yarrowia lipolytica | KAB8283935 | Topo I | 2283 | 760 | RDW22562 | Topo II | 6513 | 1537 |
| Candida albicans | XP_714305 | Topo I | 2343 | 780 | KHC76731 | Topo II | 4386 | 1461 |
| Candida auris | PSK79030 | Topo I | 2250 | 749 | PSK77735 | Topo II | 4377 | 1458 |
| Candida haemulonii | XP_025340562 | Topo I | 2250 | 749 | XP_025343359 | Topo II | 4377 | 1458 |
| Clavispora lusitaniae | OVF09121 | Topo I | 2295 | 764 | OVF06965 | Topo II | 4230 | 1409 |
| Candida dubliniensis | XP_002420888 | Topo I | 2334 | 777 | XP_002420251 | Topo II | 4386 | 1461 |
| Candida tropicalis | XP_002548660 | Topo I | 2334 | 777 | XP_002549527 | Topo II | 4197 | 1398 |
| Candida krusei | AWT08596 | Topo I | 2187 | 728 | XP_029322551 | Topo II | 4272 | 1423 |
| Kluyveromyces marxianus | BAP69451 | Topo I | 2304 | 767 | XP_022678261 | Topo II | 4350 | 1449 |
| Candida parapsilosis | KAF6043449 | Topo I | 2466 | 821 | CCE43879 | Topo II | 4332 | 1443 |
| Candida orthopsilosis | XP_003870756 | Topo I | 2418 | 805 | XP_003867280 | Topo II | 4422 | 1473 |
| Candida glabrata | XP_445795 | Topo I | 2154 | 717 | KTB13451 | Topo II | 4221 | 1406 |
| Candida utilis | XP_020072437 | Topo I | 2772 | 755 | XP_020068929 | Topo II | 4324 | 1416 |
| Meyerozyma guilliermondii | EDK40177 | Topo I | 2265 | 754 | XP_001483297 | Topo II | 4203 | 1400 |
| Ustilago maydis | XP_011388171 | Topo I | 3060 | 1019 | XP_011389948 | Topo II | 4557 | 1518 |
| Coccidioides immitis | XP_001245381 | Topo I | 3335 | 905 | TPX24629 | Topo II | 5157 | 1718 |
| Aspergillus niger | GAQ33639 | Topo I | 2595 | 864 | XP_001392968 | Topo II | 5268 | 1707 |
| Aspergillus clavatus | XP_001269805 | Topo I | 2661 | 886 | XP_001268423 | Topo II | 5208 | 1735 |
| Aspergillus nidulans | AAO19447 | Topo I | 2990 | 871 | XP_663010 | Topo II | 5130 | 1709 |
| Aspergillus terreus | GES60315 | Topo I | 2643 | 880 | XP_001212600 | Topo II | 5199 | 1732 |
| Aspergillus fumigatus | KAF4263330 | Topo I | 2604 | 867 | XP_751245 | Topo II | 4983 | 1660 |
| Penicillium marneffei | KFX43083 | Topo I | 2583 | 860 | XP_002145834 | Topo II | 5216 | 1696 |
| Paracoccidioides brasiliensis | ODH53586 | Topo I | 2718 | 905 | XP_010761698 | Topo II | 5850 | 1949 |
| Fusarium oxysporum | XP_031047401 | Topo I | 4701 | 909 | EWZ41880 | Topo II | 5214 | 1737 |
| Blastomyces dermatitidis | EEQ86368 | Topo I | 2736 | 911 | EGE78098 | Topo II | 5415 | 1804 |
| Histoplasma capsulatum | EEH11244 | Topo I | 2730 | 909 | XP_001536323 | Topo II | 5427 | 1808 |
| Cryptococcus neoformans var. neoformans | XP_572925 | Topo I | 2953 | 926 | XP_566700 | Topo II | 4182 | 1272 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade-Pavón, D.; Gómez-García, O.; Villa-Tanaca, L. Review and Current Perspectives on DNA Topoisomerase I and II Enzymes of Fungi as Study Models for the Development of New Antifungal Drugs. J. Fungi 2024, 10, 629. https://doi.org/10.3390/jof10090629
Andrade-Pavón D, Gómez-García O, Villa-Tanaca L. Review and Current Perspectives on DNA Topoisomerase I and II Enzymes of Fungi as Study Models for the Development of New Antifungal Drugs. Journal of Fungi. 2024; 10(9):629. https://doi.org/10.3390/jof10090629
Chicago/Turabian StyleAndrade-Pavón, Dulce, Omar Gómez-García, and Lourdes Villa-Tanaca. 2024. "Review and Current Perspectives on DNA Topoisomerase I and II Enzymes of Fungi as Study Models for the Development of New Antifungal Drugs" Journal of Fungi 10, no. 9: 629. https://doi.org/10.3390/jof10090629
APA StyleAndrade-Pavón, D., Gómez-García, O., & Villa-Tanaca, L. (2024). Review and Current Perspectives on DNA Topoisomerase I and II Enzymes of Fungi as Study Models for the Development of New Antifungal Drugs. Journal of Fungi, 10(9), 629. https://doi.org/10.3390/jof10090629






