Central Carbon Metabolism in Candida albicans Biofilms Is Altered by Dimethyl Sulfoxide
Abstract
1. Introduction
2. Materials and Methods
2.1. Candida albicans Strain
2.2. Preparation of Inoculum
2.3. Experimental Groups
- (a)
- Control group: cells were cultured in standard MM medium (replicates: C1a, C2a, C3a, C4a, C5a, C1b, C2b, C3b, C4b, C5b, C1c, C2c, C3c, C4c, and C5c).
- (b)
- DMSO group: cells were cultured in MM medium supplemented with 0.03% (v/v) DMSO (replicates: D1a, D2a, D3a, D4a, D5a, D1b, D2b, D3b, D4b, D5b, D1c, D2c, D3c, D4c, and D5c).
2.4. Formation of Aerobic Biofilms under Continuous Flow
2.5. Extraction of Intracellular Metabolites
2.6. Sampling and Quenching (Fixation of Cellular Metabolism)
2.7. Extraction and Quantification of Intracellular Metabolites
2.8. Quantification of Cell Biomass
2.9. Derivatization with Methyl Chloroformate (MCF)
2.10. Gas Chromatography and Mass Spectrometry
2.11. Statistical Analysis
3. Results
3.1. Principal Component Analysis (PCA)
3.2. Metabolite Analysis
3.3. Pathway Activity Profiling (PAPi) Analysis
4. Discussion
4.1. Lipid Metabolism
4.2. Carbon and Carbohydrate Metabolism
4.3. Amino Acid Metabolism
4.4. Cofactors and Vitamins Metabolism
4.5. Energy Metabolism
4.6. Secondary Metabolism
4.7. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CDC—Centers for Disease Controls and Prevention. Antibiotic Resistance Threats in the United States: Drug-Resistant Candida Species; U.S. Department of Health and Human Services: Washington, DC, USA, 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 10 December 2021).
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Low, A.; Gavriilidis, G.; Larke, N.; B.-Lajoie, M.-R.; Drouin, O.; Stover, J.; Muhe, L.; Easterbrook, P. Incidence of opportunistic infections and the impact of antiretroviral therapy among hiv-infected adults in low- and middle-income countries: A systematic review and meta-analysis. Clin. Infect. Dis. 2016, 62, 1595–1603. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.A.; Hussain, M.A.; Ahmad, Z. Candida albicans: A model organism for studying fungal pathogens. ISRN Microbiol. 2012, 2012, 538694. [Google Scholar] [CrossRef] [PubMed]
- CLSI—Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard, 3rd ed.; CLSI Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; Volume 14. [Google Scholar]
- EUCAST—European Committee on Antibiotic Susceptibility. Method for determination of minimal inhibitory concentration (MIC) by broth dilution of fermentative yeasts, discussion document. Clin. Microbiol. Infect. 2003, 9, 1–8. [Google Scholar]
- Sigma-Aldrich. Fundamental Techniques in Cell Culture, Laboratory Handbook, 4th ed.; Sigma-Aldrich Co., LLC.: St. Louis, MO, USA, 2018; Available online: https://www.sigmaaldrich.com/BR/pt/campaigns/ecacc-handbook-download (accessed on 20 February 2022).
- Brayton, C.F. Dimethyl sulfoxide (DMSO): A review. Cornell Vet. 1986, 76, 61–90. [Google Scholar] [PubMed]
- Rodríguez-Tudela, J.L.; Cuenca-Estrella, M.; Díaz-Guerra, T.M.; Mellado, E. Standardization of antifungal susceptibility variables for a semiautomated methodology. J. Clin. Microbiol. 2001, 39, 2513–2517. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, M.A. Dimethyl Sulfoxide (DMSO) Inhibits the germination of Candida albicans and the arthrospores of Trichophyton mentagrophytes. Nippon Ishinkin Gakkai Zasshi 2008, 49, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Yee, B.; Tsuyumu, S.; Adams, B.G. Biological effects of dimethyl sulfoxide on yeast. Biochem. Biophys. Res. Commun. 1972, 49, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.W.; Quinn, P.J. Dimethyl sulphoxide: A review of its applications in cell biology. Biosci. Rep. 1994, 14, 259–281. [Google Scholar] [CrossRef]
- Alastruey-Izquierdo, A.; Gómez-López, A.; Arendrup, M.C.; Lass-Florl, C.; Hope, W.W.; Perlin, D.S.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Comparison of dimethyl sulfoxide and water as solvents for echinocandin susceptibility testing by the EUCAST methodology. J. Clin. Microbiol. 2012, 50, 2509–2512. [Google Scholar] [CrossRef]
- Hazen, K.C. Influence of DMSO on antifungal activity during susceptibility testing in vitro. Diagn. Microbiol. Infect. Dis. 2013, 75, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Netíková, L.; Bogusch, P.; Heneberg, P. Czech Ethanol-Free Propolis Extract Displays Inhibitory Activity against a Broad Spectrum of Bacterial and Fungal Pathogens. J. Food Sci. 2013, 78, M1421–M1429. [Google Scholar] [CrossRef] [PubMed]
- Tristram-Nagle, S.; Moore, T.; Petrache, H.I.; Nagle, J.F. DMSO produces a new subgel phase in DPPC: DSC and X-ray diffraction study. Biochim. Biophys. Acta 1998, 1369, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.W.; Quinn, P.J. The effect of dimethyl sulphoxide on the structure and phase behavior of palmitoleoylphosphatidylethanolamine. Biochim. Biophys. Acta 2000, 1509, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Sum, A.K.; de Pablo, J.J. molecular simulation study on the influence of dimethylsulfoxide on the structure of phospholipid bilayers. Biophys. J. 2003, 85, 3636–3645. [Google Scholar] [CrossRef] [PubMed]
- de Ménorval, M.-A.; Mir, L.M.; Fernández, M.L.; Reigada, R. Effects of dimethyl sulfoxide in cholesterol-containing lipid membranes: A comparative study of experiments in silico and with cells. PLoS ONE 2012, 7, e41733. [Google Scholar] [CrossRef] [PubMed]
- Malajczuk, C.J.; Hughes, Z.E.; Mancera, R.L. Molecular dynamics simulations of the interactions of DMSO, mono- and polyhydroxylated cryosolvents with a hydrated phospholipid bilayer. Biochim. Biophys. Acta 2013, 1828, 2041–2055. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Donald, K.G.; Griffiths, D.E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991, 19, 5791. [Google Scholar] [CrossRef]
- Notman, R.; Otter, W.K.D.; Noro, M.G.; Briels, W.; Anwar, J. The permeability enhancing mechanism of DMSO in ceramide bilayers simulated by molecular dynamics. Biophys. J. 2007, 93, 2056–2068. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Song, J.; Pas, J.; Meijer, L.H.; Han, S. DMSO induces dehydration near lipid membrane surfaces. Biophys. J. 2015, 109, 330–339. [Google Scholar] [CrossRef]
- Murata, Y.; Watanabe, T.; Sato, M.; Momose, Y.; Nakahara, T.; Oka, S.-I.; Iwahashi, H. Dimethyl sulfoxide exposure facilitates phospholipid biosynthesis and cellular membrane proliferation in yeast cells. J. Biol. Chem. 2003, 278, 33185–33193. [Google Scholar] [CrossRef] [PubMed]
- Gaytán, B.D.; Loguinov, A.V.; De La Rosa, V.Y.; Lerot, J.-M.; Vulpe, C.D. Functional genomics indicates yeast requires Golgi/ER transport, chromatin remodeling, and DNA repair for low dose DMSO tolerance. Front. Genet. 2013, 4, 154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Needham, D.L.; Coffin, M.; Rooker, A.; Hurban, P.; Tanzer, M.M.; Shuster, J.R. Microarray analyses of the metabolic responses of Saccharomyces cerevisiae to organic solvent dimethyl sulfoxide. J. Ind. Microbiol. Biotechnol. 2003, 30, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, N.; Ma, X.; Jiang, L. The transcriptional control machinery as well as the cell wall integrity and its regulation are involved in the detoxification of the organic solvent dimethyl sulfoxide in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 13, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Gombert, A.K. Análise do metaboloma: Uma ferramenta biotecnológica emergente na era pós-genômica. Biotecnol. Cienc. Desenvolv. 2006, 36, 58–69. [Google Scholar]
- Selow, M.L.C.; Rymovicz, A.U.M.; Ribas, C.R.; Saad, R.S.; Rosa, R.T.; Rosa, E.A.R. Growing Candida albicans biofilms on paper support and dynamic conditions. Mycopathologia 2015, 180, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatisation followed by gas chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Aggio, R.B.M.; Ruggiero, K.; Villas-Bôas, S.G. Pathway activity profiling (PAPi): From the metabolite profile to the metabolic pathway activity. Bioinformatics 2010, 26, 2969–2976. [Google Scholar] [CrossRef]
- Hansen, J. Inactivation of MXR1 abolishes formation of dimethyl sulfide from dimethyl ulfoxide in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1999, 65, 3915–3919. [Google Scholar] [CrossRef]
- Le, D.T.; Lee, B.C.; Marino, S.M.; Zhang, Y.; Fomenko, D.E.; Kaya, A.; Hacioglu, E.; Kwak, G.-H.; Koc, A.; Kim, H.-Y.; et al. Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 4354–4364. [Google Scholar] [CrossRef] [PubMed]
- Kwak, G.-H.; Choi, S.-H.; Kim, H.-Y. Dimethyl sulfoxide elevates hydrogen peroxide-mediated cell death in Saccharomyces cerevisiae by inhibiting the antioxidant function of methionine sulfoxide reductase A. BMB Rep. 2010, 43, 622–628. [Google Scholar] [CrossRef] [PubMed]
- CGD—Candida Genome Database. 2019. Available online: http://www.candidagenome.org (accessed on 11 July 2022).
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of fluconazole resistance in Candida albicans biofilms: Phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [PubMed]
- Lattif, A.A.; Mukherjee, P.K.; Chandra, J.; Roth, M.R.; Welti, R.; Rouabhia, M.; Ghannoum, M.A. Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology 2011, 157, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Garbarino, J.; Padamsee, M.; Wilcox, L.; Oelkers, P.M.; D’Ambrosio, D.; Ruggles, K.V.; Ramsey, N.; Jabado, O.; Turkish, A.; Sturley, S.L. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 2009, 284, 30994–31005. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Büttner, S. Lipids and cell death in yeast. FEMS Yeast Res. 2014, 14, 179–197. [Google Scholar] [CrossRef] [PubMed]
- León-García, M.C.; Ríos-Castro, E.; López-Romero, E.; Cuéllar-Cruz, M. Evaluation of cell wall damage by dimethyl sulfoxide in Candida species. Res. Microbiol. 2017, 168, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.; Poolman, B. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 1994, 269, 8022–8028. [Google Scholar] [CrossRef]
- Miura, S.; Zou, W.; Ueda, M.; Tanaka, A. Screening of genes involved in isooctane tolerance in Saccharomyces cerevisiae by using mRNA differential display. Appl. Environ. Microbiol. 2000, 66, 4883–4889. [Google Scholar] [CrossRef]
- Dashko, S.; Zhou, N.; Compagno, C.; Piškur, J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014, 14, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Sandai, D.; Yin, Z.; Selway, L.; Stead, D.; Walker, J.; Leach, M.D.; Bohovych, I.; Ene, I.V.; Kastora, S.; Budge, S.; et al. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 2012, 3, e00495-12. [Google Scholar] [CrossRef] [PubMed]
- Askew, C.; Sellam, A.; Epp, E.; Hogues, H.; Mullick, A.; Nantel, A.; Whiteway, M. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009, 5, e1000612. [Google Scholar] [CrossRef]
- Setiadi, E.R.; Doedt, T.; Cottier, F.; Noffz, C.; Ernst, J.F. Transcriptional Response of Candida albicans to hypoxia: Linkage of oxygen sensing and Efg1p-regulatory Networks. J. Mol. Biol. 2006, 361, 399–411. [Google Scholar] [CrossRef]
- Synnott, J.M.; Guida, A.; Mulhern-Haughey, S.; Higgins, D.G.; Butler, G. Regulation of the Hypoxic Response in Candida albicans. Eukaryot. Cell 2010, 9, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Marttila, E.; Bowyer, P.; Sanglard, D.; Uittamo, J.; Kaihovaara, P.; Salaspuro, M.; Richardson, M.; Rautemaa, R. Fermentative 2-carbon metabolism produces carcinogenic levels of acetaldehyde in Candida albicans. Mol. Oral Microbiol. 2013, 28, 281–291. [Google Scholar] [CrossRef]
- Rozpȩdowska, E.; Galafassi, S.; Johansson, L.; Hagman, A.; Piškur, J.; Compagno, C. Candida albicans—A pre-whole genome duplication yeast—Is predominantly aerobic and a poor ethanol producer. FEMS Yeast Res. 2010, 11, 285–291. [Google Scholar] [CrossRef]
- Pronk, J.T.; Steensmays, H.Y.; Van, J.P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 1996, 12, 1607–1633. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef]
- Brown, A.J.; Gow, N.A. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999, 7, 333–338. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Navarathna, D.H.M.L.P.; Roberts, D.D.; Cooper, J.T.; Atkin, A.L.; Petro, T.M.; Nickerson, K.W. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 2009, 77, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-L.; Cannon, R.D.; Villas-Bôas, S.G. Metabolome analysis during the morphological transition of Candida albicans. Metabolomics 2012, 8, 1204–1217. [Google Scholar] [CrossRef]
- Jiménez-López, C.; Collette, J.R.; Brothers, K.M.; Shepardson, K.M.; Cramer, R.A.; Wheeler, R.T.; Lorenz, M.C. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot. Cell 2013, 12, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.B.; López, M.C.; García, S.; Schmidt, A.; Spaltmann, F.; Ruiz-Herrera, J.; Dominguez, A. Control of filament formation in Candida albicans by polyamine levels. Infect. Immun. 1999, 67, 4870–4878. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y.; Fukumatsu, M.; Ogasawara, A.; Watanabe, T.; Mikami, T.; Matsumoto, T. Hyphae formation of Candida albicans is regulated by polyamines. Biol. Pharm. Bull. 2004, 27, 890–892. [Google Scholar] [CrossRef] [PubMed]
- Brawley, J.V.; Ferro, A.J. Polyamine biosynthesis during germination of yeast ascospores. J. Bacteriol. 1979, 140, 649–654. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, Z.; Chen, X.; Yao, X.; Zhao, L.; Wang, H.; Yan, L.; Wu, H.; Chai, Y.; Jiang, Y. Effect of amphotericin B on the metabolic profiles of Candida albicans. J. Proteome Res. 2013, 12, 2921–2932. [Google Scholar] [CrossRef]
- Kamthan, M.; Mukhopadhyay, G.; Chakraborty, N.; Chakraborty, S.; Datta, A. Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet. Biol. 2012, 49, 369–378. [Google Scholar] [CrossRef]
- Chen, H.; Fujita, M.; Feng, Q.; Clardy, J.; Fink, G.R. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 2004, 101, 5048–5052. [Google Scholar] [CrossRef]
- Alem, M.A.S.; Oteef, M.D.Y.; Flowers, T.H.; Douglas, L.J. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot. Cell 2006, 5, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gomariz, M.; Perumal, P.; Mekala, S.; Nombela, C.; Chaffin, W.L.; Gil, C. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics 2009, 9, 2230–2252. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Punekar, N.S. The metabolism of 4-aminobutyrate (GABA) in fungi. Mycol. Res. 1997, 101, 403–409. [Google Scholar] [CrossRef]
- Ramos, F.; El Guezzar, M.; Grenson, M.; Wiame, J. Mutations affecting the enzymes involved in the utilization of 4-aminobutyric acid as nitrogen source by the yeast Saccharomyces cerevisiae. JBIC J. Biol. Inorg. Chem. 1985, 149, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.T.; Fang, T.K.; Rovinsky, S.A.; Turano, F.J.; Moye-Rowley, W.S. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 244–250. [Google Scholar] [CrossRef] [PubMed]
- McNemar, M.D.; Gorman, J.A.; Buckley, H.R. Isolation of a gene encoding a putative polyamine transporter from Candida albicans, GPT1. Yeast 2001, 18, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Dervartanian, D.; Veeger, C. Studies on succinate dehydrogenase: I. Spectral properties of the purified enzyme and formation of enzyme-competitive inhibitor complexes. Biochim. Biophys. Acta Spec. Sect. Enzym. Subj. 1964, 92, 233–247. [Google Scholar] [CrossRef]
- Aoki, S.; Ito-Kuwa, S. The appearance and characterization of cyanide-resistant respiration in the fungus Candida albicans. Microbiol. Immunol. 1984, 28, 393–406. [Google Scholar] [CrossRef]
- Huh, W.-K.; Kang, S.-O. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 2001, 356, 595–604. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Murphy, M.P.; Troxler, R.F.; Oppenheim, F.G. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim. Biophys. Acta Bioenerg. 2002, 1556, 73–80. [Google Scholar] [CrossRef]
- Ruy, F.; Vercesi, A.E.; Kowaltowski, A.J. Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. J. Bioenerg. Biomembr. 2006, 38, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Chabrier-Roselló, Y.; Giesselman, B.R.; De Jesús-Andino, F.J.; Foster, T.H.; Mitra, S.; Haidaris, C.G. Inhibition of electron transport chain assembly and function promotes photodynamic killing of Candida. J. Photochem. Photobiol. B Biol. 2010, 99, 117–125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Duan, Q.; Wang, T.; Ahmed, M.; Zhang, N.; Li, Y.; Li, L.; Yao, X. Mitochondrial respiratory chain inhibitors involved in ROS production induced by acute high concentrations of iodide and the effects of SOD as a protective factor. Oxidative Med. Cell. Longev. 2015, 2015, 217670. [Google Scholar] [CrossRef] [PubMed]
- Otzen, C.; Bardl, B.; Jacobsen, I.D.; Nett, M.; Brock, M. Candida albicans utilizes a modified β-oxidation pathway for the degradation of toxic Propionyl-CoA. J. Biol. Chem. 2014, 289, 8151–8169. [Google Scholar] [CrossRef]
- Dantas, A.D.S.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef]
- Guedouari, H.; Gergondey, R.; Bourdais, A.; Vanparis, O.; Bulteau, A.; Camadro, J.; Auchère, F. Changes in glutathione-dependent redox status and mitochondrial energetic strategies are part of the adaptive response during the filamentation process in Candida albicans. Biochim. Biophys. Acta 2014, 1842, 1855–1869. [Google Scholar] [CrossRef]
- Sundaram, A.; Grant, C.M. Oxidant-specific regulation of protein synthesis in Candida albicans. Fungal Genet. Biol. 2014, 67, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Abegg, M.A.; Gil Alabarse, P.V.; Schüller, K.; Benfato, M.S. Glutathione levels in and total antioxidant capacity of Candida sp. cells exposed to oxidative stress caused by hydrogen peroxide. Rev. Soc. Bras. Med. Trop. 2012, 45, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Szkopińska, A. Ubiquinone. Biosynthesis of quinone ring and its isoprenoid side chain. Intracellular localization. Acta Biochim. Pol. 2000, 47, 469–480. [Google Scholar] [CrossRef]
- Diezmann, S.; Cox, C.J.; Schönian, G.; Vilgalys, R.J.; Mitchell, T.G. Phylogeny and evolution of medical species of Candida and related taxa: A multigenic analysis. J. Clin. Microbiol. 2004, 42, 5624–5635. [Google Scholar] [CrossRef]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 2004, 1660, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Naseem, S.; Sharma, S.; Konopka, J.B. Flavodoxin-like proteins protect Candida albicans from oxidative stress and promote virulence. PLoS Pathog. 2015, 11, e1005147. [Google Scholar] [CrossRef] [PubMed]
- Homer, N.Z.; Reglinski, J.; Sowden, R.; Spickett, C.M.; Wilson, R.; Walker, J.J. Dimethylsulfoxide oxidizes glutathione in vitro and in human erythrocytes: Kinetic analysis by 1H NMR. Cryobiology 2005, 50, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Kashino, G.; Prise, K.M.; Suzuki, K.; Matsuda, N.; Kodama, S.; Suzuki, M.; Nagata, K.; Kinashi, Y.; Masunaga, S.-I.; Ono, K.; et al. Effective suppression of bystander effects by DMSO treatment of irradiated CHO cells. J. Radiat. Res. 2007, 48, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Sadowska-Bartosz, I.; Pączka, A.; Mołoń, M.; Bartosz, G. Dimethyl sulfoxide induces oxidative stress in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Firestone, B.Y.; Koser, S.A. Growth promoting effect of some biotin analogues for Candida albicans. J. Bacteriol. 1960, 79, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Hussin, N.A.; Pathirana, R.U.; Hasim, S.; Tati, S.; Scheib-Owens, J.A.; Nickerson, K.W. Biotin auxotrophy and biotin enhanced germ tube formation in Candida albicans. Microorganisms 2016, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.T.; Loewus, F.A. Concerning the presence and formation of ascorbic acid in yeasts. Plant Sci. 1985, 38, 65–69. [Google Scholar] [CrossRef]
- Nick, J.A.; Leung, C.T.; Loewus, F.A. Isolation and identification of erythroascorbic acid in Saccharomyces cerevisiae and Lypomyces starkeyi. Plant Sci. 1986, 46, 181–187. [Google Scholar] [CrossRef]
- Huh, W.; Kim, S.; Yang, K.; Seok, Y.; Hah, Y.C.; Kang, S. Characterisation of D-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur. J. Biochem. 1994, 225, 1073–1079. [Google Scholar] [CrossRef]
- Kim, S.-T.; Huh, W.-K.; Kim, J.-Y.; Hwang, S.-W.; Kang, S.-O. D-Arabinose dehydrogenase and biosynthesis of erythroascorbic acid in Candida albicans. Biochim. Biophys. Acta 1996, 1297, 1–8. [Google Scholar] [CrossRef]
- Hébert, A.; Forquin-Gomez, M.-P.; Roux, A.; Aubert, J.; Junot, C.; Heilier, J.-F.; Landaud, S.; Bonnarme, P.; Beckerich, J.-M. New insights into sulfur metabolism in yeasts as revealed by studies of Yarrowia lipolytica. Appl. Environ. Microbiol. 2013, 79, 1200–1211. [Google Scholar] [CrossRef]
- Hébert, A.; Casaregola, S.; Beckerich, J.-M. Biodiversity in sulfur metabolism in hemiascomycetous yeasts. FEMS Yeast Res. 2011, 11, 366–378. [Google Scholar] [CrossRef]
- Yurimoto, H.; Oku, M.; Sakai, Y. Yeast methylotrophy: Metabolism, gene regulation and peroxisome homeostasis. Int. J. Microbiol. 2011, 2011, 101298. [Google Scholar] [CrossRef]
- Negruta, O.; Csutak, O.; Stoica, I.; Rusu, E.; Vassu, T. Methylotrophic yeasts: Diversity and methanol metabolism. Rom. Biotechnol. Lett. 2010, 15, 5369–5375. [Google Scholar]
- Klein, S.M.; Cohen, G.; Cederbaum, A.I. Production of formaldehyde during metabolism of dimethyl sulfoxide by hydroxyl radical-generating systems. Biochemistry 1981, 20, 6006–6012. [Google Scholar] [CrossRef]
- Dufour, N.; Rao, R.P. Secondary metabolites and other small molecules as intercellular pathogenic signals. FEMS Microbiol. Lett. 2011, 314, 10–17. [Google Scholar] [CrossRef]
- Strack, D.; Vogt, T.; Schliemann, W. Recent advances in betalain research. Phytochemistry 2003, 62, 247–269. [Google Scholar] [CrossRef]
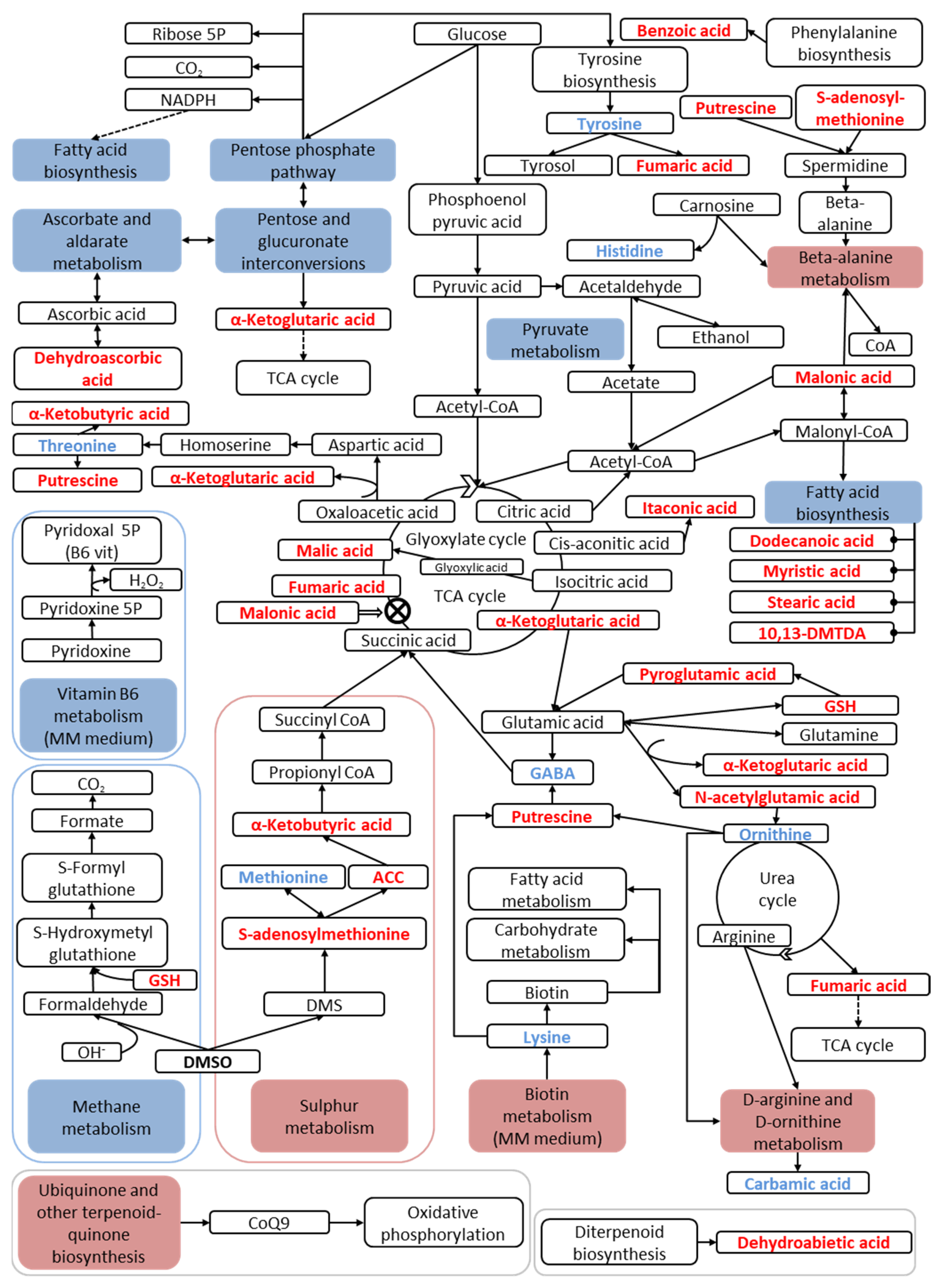
| Classification of Metabolites | N° | Intracellular Metabolites |
|---|---|---|
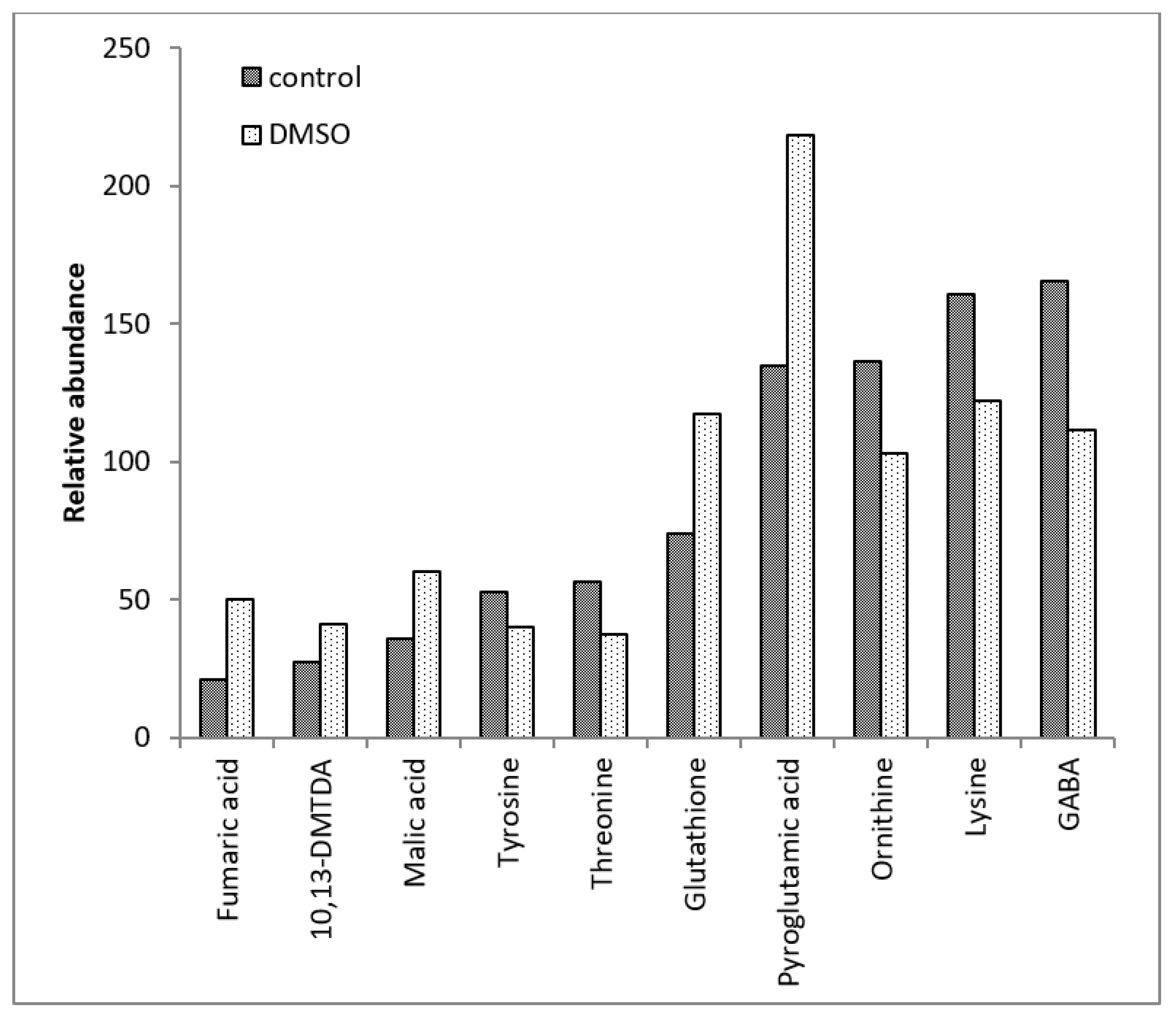
| Amino acids and their isoforms | 22 | Alanine, asparagine, aspartic acid, beta-alanine, cysteine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, norvaline, ornithine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine |
| Amino acid derivatives | 6 | Creatinine, cystathionine, N-acetylglutamic acid, putrescine, pyroglutamic acid, S-adenosylmethionine |
| TCA cycle intermediates | 8 | Alpha-ketoglutaric acid, alpha-ketobutyric acid, cis-aconitic acid, citric acid, fumaric acid, isocitric acid, malic acid, succinic acid |
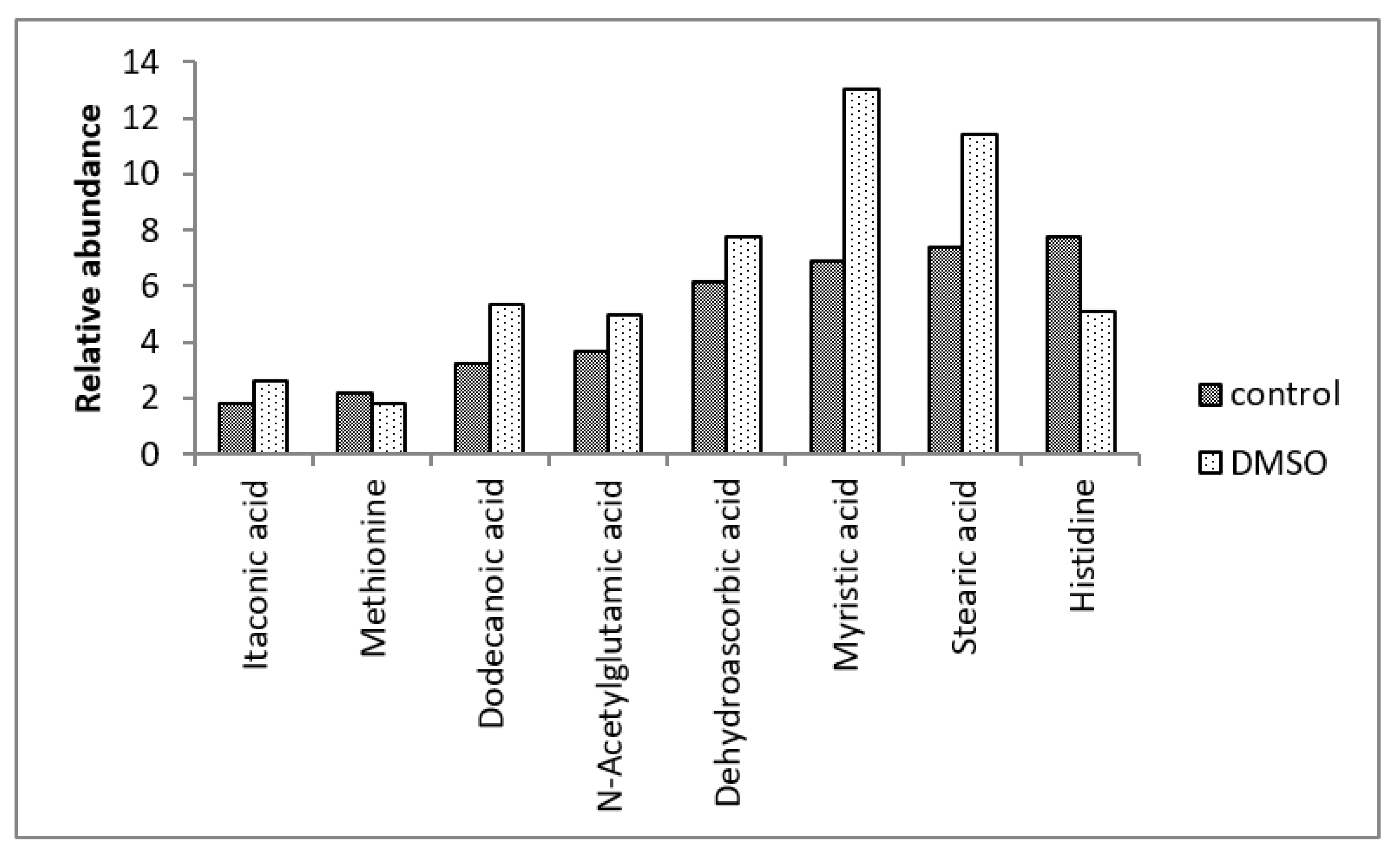
| Fatty acids | 17 | 10,13-dimethyl tetradecanoic acid (10,13-DMTDA), 9-heptadecenoic acid, adipic acid, caprinoic acid, caprylic acid, decanoic acid, docosanoic acid, dodecanoic acid, eicosanoic acid, gamma-linoleic acid, hexanoic acid, myristic acid, palmitelaidic acid, pentadecanoic acid, stearic acid, trans-vaccenic |
| Glycolytic intermediates | 2 | 2-phosphoenolpyruvic acid, pyruvic acid |
| Cofactors and vitamins | 6 | Dehydroascorbic acid, gamma-aminobutyric acid (GABA), glutathione, NADP/NADPH, nicotinamide, nicotinic acid |
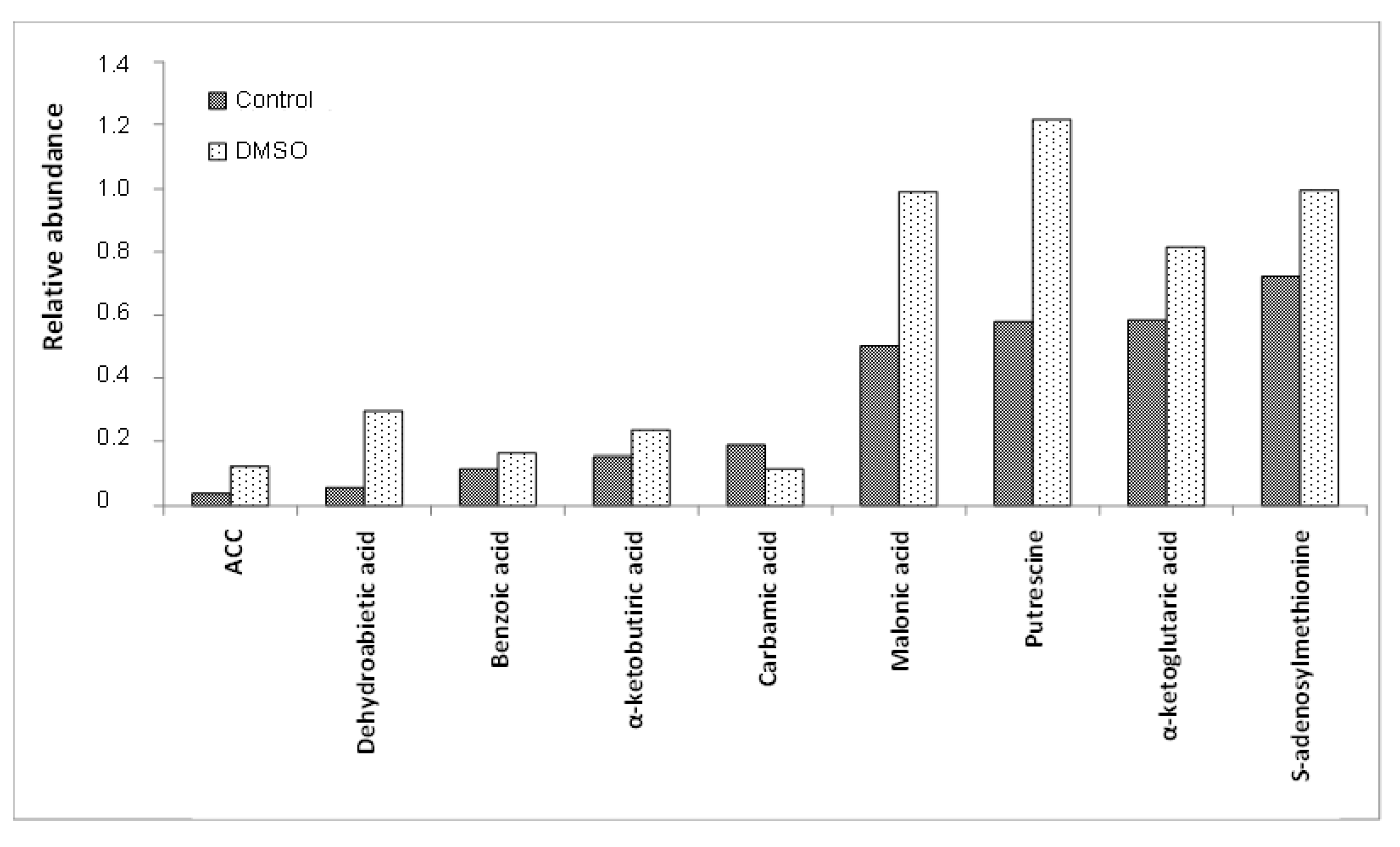
| Others | 27 | 1-aminocyclopropane-1-carboxylic acid (ACC), 2-aminoadipic acid, 2-hydroxybutyric acid, 2-isopropylmalic acid, 3-methyl-2-oxopentanoic acid, 4-aminobenzoic acid, 4-hydroxyphenylacetic acid, 4-hydroxyphenylethanol (tyrosol), 5-oxotetrahydrofuran-2- benzoic acid, carbamic acid, carboxylic acid, citramalic acid, dehydroabietic acid, dibutylphthalate (DBP), DL-hydroxyglutaramate, dodecane, EDTA, glutaric acid, glyceric acid, glyoxylic acid, itaconic acid, lactic acid, levulinic acid, malonic acid, oxalic acid, para-toluic acid, quinic acid |
| Total no. of identified metabolites | 88 |
| Classification of Metabolites | Intracellular Metabolites Altered by DMSO | |
|---|---|---|
| Upregulated | Downregulated | |
| Amino acids and their isoforms | - | Histidine, lysine, methionine, ornithine, threonine, tyrosine |
| Amino acid derivatives | N-acetylglutamic acid, putrescine, pyroglutamic acid, S-adenosylmethionine | - |
| TCA cycle intermediates | Alpha-ketobutyric acid, alpha-ketoglutaric acid, fumaric acid, malic acid | - |
| Fatty acids | 10,13-dimethyl tetradecanoic acid (10,13-DMTDA), dodecanoic acid, myristic acid, stearic acid | - |
| Cofactors and vitamins | Dehydroascorbic acid, glutathione | Gamma-aminobutyric acid (GABA) |
| Others | 1-aminocyclopropane-1-carboxylic acid (ACC), benzoic acid, dehydroabietic acid, itaconic acid, malonic acid | Carbamic acid |
| Total no. of altered metabolites | 19 | 8 |
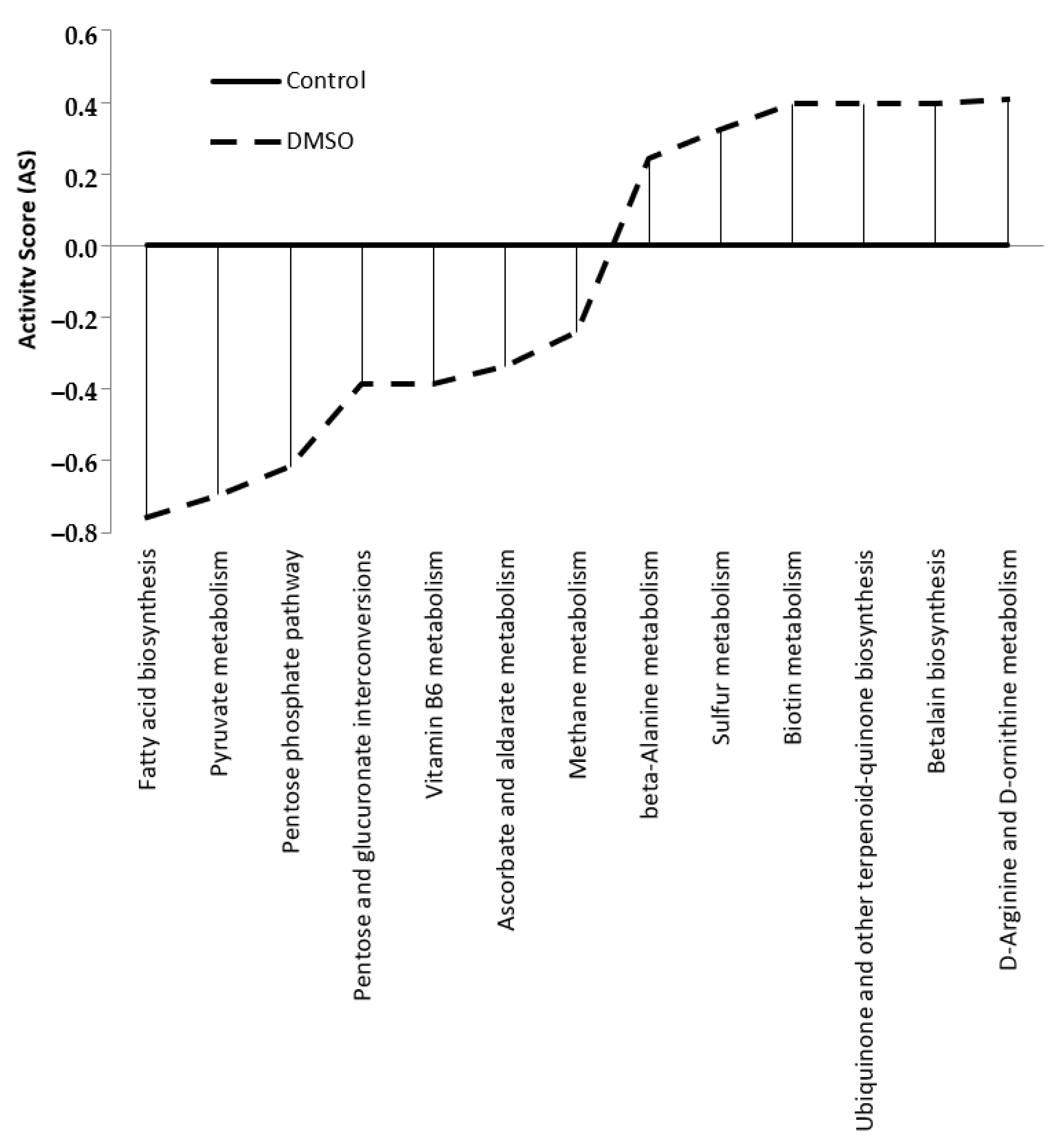
| Metabolic Pathway Groups | Metabolic Pathways Altered by DMSO | |
|---|---|---|
| Upregulated | Downregulated | |
| Amino acid metabolism | Beta-Alanine metabolism D-arginine and D-ornithine metabolism | - |
| Lipid metabolism | - | Fatty acid biosynthesis |
| Carbohydrate metabolism | - | Pentose phosphate pathway Pentose and glucuronate interconversions Ascorbate and aldarate metabolism |
| Carbon metabolism | - | Pyruvate metabolism |
| Energy metabolism | Sulphur metabolism | Methane metabolism |
| Cofactor and vitamin metabolism | Biotin metabolism Ubiquinone and other terpene-quinones biosynthesis | Vitamin B6 Metabolism |
| Secondary metabolites metabolism | Betalain biosynthesis | - |
| Total | 6 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arruda, M.F.C.; da Silva Ramos, R.C.P.; de Oliveira, N.S.; Rosa, R.T.; Stuelp-Campelo, P.M.; Bianchini, L.F.; Villas-Bôas, S.G.; Rosa, E.A.R. Central Carbon Metabolism in Candida albicans Biofilms Is Altered by Dimethyl Sulfoxide. J. Fungi 2024, 10, 337. https://doi.org/10.3390/jof10050337
Arruda MFC, da Silva Ramos RCP, de Oliveira NS, Rosa RT, Stuelp-Campelo PM, Bianchini LF, Villas-Bôas SG, Rosa EAR. Central Carbon Metabolism in Candida albicans Biofilms Is Altered by Dimethyl Sulfoxide. Journal of Fungi. 2024; 10(5):337. https://doi.org/10.3390/jof10050337
Chicago/Turabian StyleArruda, Maria Fernanda Cordeiro, Romeu Cassiano Pucci da Silva Ramos, Nicoly Subtil de Oliveira, Rosimeire Takaki Rosa, Patrícia Maria Stuelp-Campelo, Luiz Fernando Bianchini, Silas Granato Villas-Bôas, and Edvaldo Antonio Ribeiro Rosa. 2024. "Central Carbon Metabolism in Candida albicans Biofilms Is Altered by Dimethyl Sulfoxide" Journal of Fungi 10, no. 5: 337. https://doi.org/10.3390/jof10050337
APA StyleArruda, M. F. C., da Silva Ramos, R. C. P., de Oliveira, N. S., Rosa, R. T., Stuelp-Campelo, P. M., Bianchini, L. F., Villas-Bôas, S. G., & Rosa, E. A. R. (2024). Central Carbon Metabolism in Candida albicans Biofilms Is Altered by Dimethyl Sulfoxide. Journal of Fungi, 10(5), 337. https://doi.org/10.3390/jof10050337








